Significance
We report the first functional reconstitution of neuromuscular (NMJ) glutamate receptors from the fruit fly Drosophila. The identification of these receptors enabled tremendous insight into the mechanisms of synapse assembly and development. However, analysis of animals with mutant receptors is complicated by compound phenotypes; studies on isolated receptors are necessary to identify the structural elements and auxiliary proteins important for receptor assembly, surface delivery, and function. We show that Neto is an essential component required for the function of Drosophila NMJ receptors expressed in Xenopus oocytes, and use this system to examine subunit dependence and function. We find that Drosophila NMJ receptors have ligand-binding properties and structural features strikingly different from vertebrate glutamate receptors.
Keywords: Drosophila, NMJ, Neto, glutamate receptors, crystallography
Abstract
The Drosophila larval neuromuscular junction (NMJ), at which glutamate acts as the excitatory neurotransmitter, is a widely used model for genetic analysis of synapse function and development. Despite decades of study, the inability to reconstitute NMJ glutamate receptor function using heterologous expression systems has complicated the analysis of receptor function, such that it is difficult to resolve the molecular basis for compound phenotypes observed in mutant flies. We find that Drosophila Neto functions as an essential component required for the function of NMJ glutamate receptors, permitting analysis of glutamate receptor responses in Xenopus oocytes. In combination with a crystallographic analysis of the GluRIIB ligand binding domain, we use this system to characterize the subunit dependence of assembly, channel block, and ligand selectivity for Drosophila NMJ glutamate receptors.
The amino acid l-glutamate is the major neurotransmitter at vertebrate excitatory central synapses and at the neuromuscular junction (NMJ) of insects and crustaceans (1–3). Many vertebrate AMPA and kainate receptors, as well as GluRI from the worm Caenorhabditis elegans and AvGluR1 from the rotifer Adineta vaga, can form functional homomeric ion channels (1, 4, 5). In contrast, genetic studies suggest that assembly of Drosophila NMJ type A and type B glutamate receptors requires four subunits: either GluRIIA or GluRIIB, plus GluRIIC, GluRIID, and GluRIIE (6–9); both subtypes desensitize rapidly, with time constants of 18.8 and 2.0 ms, respectively (6). In flies, lack of GluRIIA and GluRIIB or any other single subunit induces embryonic paralysis. Furthermore, in the absence of GluRIIA and GluRIIB, or any other subunit, none of the remaining iGluR subunits cluster at nascent synapses, suggesting that recruitment and stabilization of iGluRs at synaptic sites requires heterotetramers. Despite a decade of work, the molecular basis for this unique profile has remained obscure. In part, this is because the reconstitution of functional glutamate receptors (iGluRs) in heterologous systems, which has been a powerful tool for analysis of receptor function in other species, has not been achieved for the Drosophila NMJ (10).
We recently demonstrated that the synaptic distribution of Drosophila NMJ iGluRs requires Neto (Neuropillin and Tolloid-like), a protein essential for NMJ function (11). Neto belongs to a family of highly conserved auxiliary proteins that modulate the function of vertebrate kainate receptors (12) and C. elegans AMPA receptors (13, 14). In these species Neto plays a minor role in delivery and synaptic targeting, and instead modulates receptor gating properties. In contrast, in flies Neto is absolutely required for clustering of iGluRs at the NMJ and lack of Neto induces embryonic paralysis (11). This finding may reflect a role for Neto in receptor assembly, surface expression, synaptic trafficking and stabilization, or modulation of iGluR gating. To distinguish among these possibilities, a recombinant expression system for Drosophila NMJ iGluRs is required. We have recently found that Drosophila Neto encodes two isoforms, Neto α and Neto β, which differ only in their cytoplasmic C terminus, and that Neto β is the predominant isoform at the larval NMJ. Using heterologous expression we now show that Neto β has a modest effect on the surface delivery of Drosophila NMJ iGluRs in Xenopus oocytes and that both Neto α and Neto β increase glutamate activated currents by several orders of magnitude.
Results
Neto Modulates but Is Not Required for Cell Surface Expression of Drosophila iGluRs.
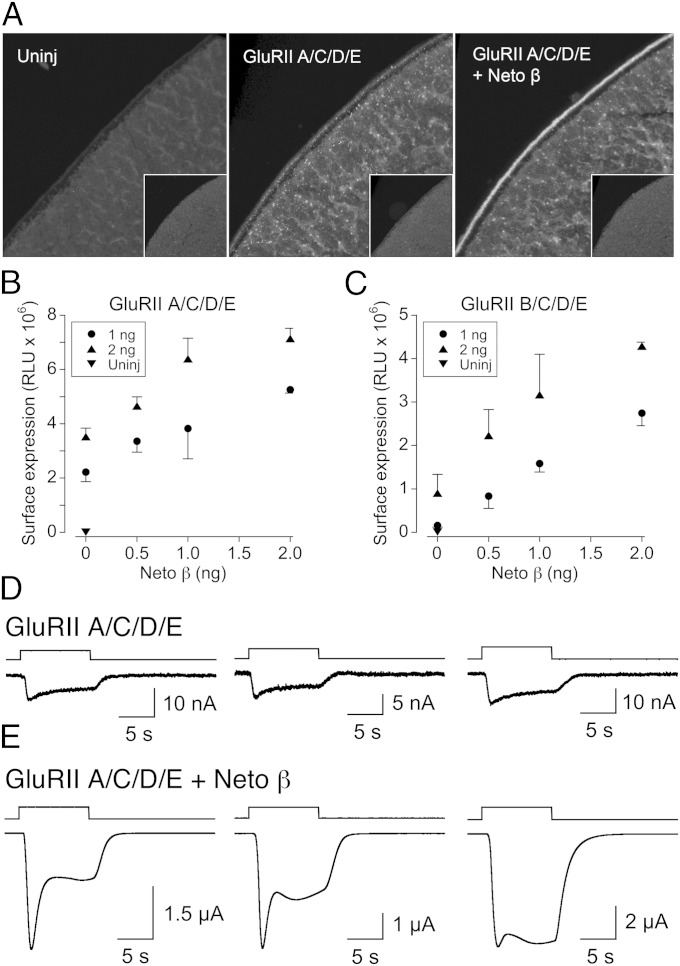
To facilitate cell surface targeting and detection of Drosophila NMJ iGluRs, we replaced endogenous signal peptides with an optimized sequence and added a C-terminal RGSH6 epitope to all iGluR constructs. Microinjection of Xenopus laevis oocytes with 2 ng each of GluRIIA, GluRIIC, GluRIID, and GluRIIE subunit cRNAs (hereafter referred to as GluRIIA/C/D/E) induced accumulation of RGSH6-positive signals on the surface of oocytes (Fig. 1A). The RGSH6 signals were not detected in uninjected oocytes and appeared to increase when iGluR cRNAs were coexpressed with Neto β. Similar but less intense immunoreactivity was observed in oocytes injected with cRNAs for GluRIIB/C/D/E with or without Neto β (Fig. S1A). To quantify the effect of Neto β on receptor surface expression, we added an extracellular HA-epitope to each iGluR subunit and measured surface expression by chemiluminescence (Fig. 1 B and C). We found that Neto β cRNA (0.5–2.0 ng) enhanced surface expression for both HA-GluRIIA/C/D/E and HA-GluRIIB/C/D/E complexes by up to fourfold. However, signals were lower than for oocytes injected with 0.5 ng of HA-GluK2 cRNA (12), which exceeded the linear range of the assay optimized for Drosophila NMJ iGluRs. Similarly, HA-positive immunoreactivity was more intense in oocytes injected with 0.5 ng HA-GluK2 cRNA than for 2 ng of each HA-GluRIIA/C/D/E cRNAs and Neto β (Fig. S1B).
Fig. 1.
Drosophila Neto modulates NMJ glutamate receptor cell surface expression and function. (A) Confocal images of oocytes injected as indicated with cRNA for RGSH6-tagged GluRIIA/C/D/E with or without Neto β, and immunolabeled for anti-RGSH6; the box size for each image is 170 × 170 μm. Surface receptors were detected at the animal but not vegetal pole (Insets) and were increased by coexpression of Neto β. (B) Cell surface expression for HA-tagged GluRIIA/C/D/E measured by chemiluminescence is facilitated by but does not require Neto β; error bars show SEM. (C) Cell surface expression for HA-tagged GluRIIB/C/D/E reveals a similar effect, but with approximately twofold lower signals; error bars show SEM. (D) Small amplitude responses to 3 mM glutamate at −60 mV for three representative oocytes injected with cRNA for GluRIIA/C/D/E and treated with Con A. (E) Large amplitude responses to 3 mM glutamate at −60 mV for three representative oocytes injected with cRNA for GluRIIA/C/D/E and Neto β and treated with Con A, illustrating variability in transient inward current responses. Responses in D and E were recorded on the same day from the same batch of oocytes.
In pilot voltage-clamp experiments we observed no response to glutamate in oocytes injected with GluRIIA/C/D/E, either with or without Neto β. Because native Drosophila NMJ iGluRs show rapid desensitization (6, 15), we tested whether the lectin Concanavalin A (Con A), which attenuates desensitization of iGluRs from a wide variety of species—including locust NMJ iGluRs (16), vertebrate kainate receptors (17), and AvGluR1 (18)—would have a similar effect on Drosophila NMJ iGluRs. After application of Con A (0.6 mg/mL for 4 min), 3 mM glutamate consistently evoked small currents (15.6 ± 0.9 nA, n = 8) for oocytes injected with GluRIIA/C/D/E alone (Fig. 1D); however, when GluRIIA/C/D/E was coexpressed with Drosophila Neto β, glutamate-evoked currents (12 ± 3.1 µA, n = 11) recorded on the same day and from the same batch of oocytes were on average 770-fold larger (Fig. 1E). A similar profile was obtained for GluRIIB/C/D/E (Fig. S1C); small currents, typically 1 nA or less (n = 10), were evoked by 3 mM glutamate in the absence of Neto β, whereas coexpression with Neto β gave large responses (5.6 ± 1.3 µA, n = 8). Similarly, large amplitude responses for both GluRIIA/C/D/E (6.0 ± 1.1 µA, n = 14) and GluRIIB/C/D/E (0.63 ± 0.12 µA, n = 15), recorded on the same day, were obtained following coexpression with Neto α (Fig. S1 D and E). Treatment with Con A and coexpression with Neto β, as described above, was used in all subsequent voltage-clamp experiments reported in the following sections.
Efficient Receptor Expression and Function Requires Multiple iGluR Subunits.
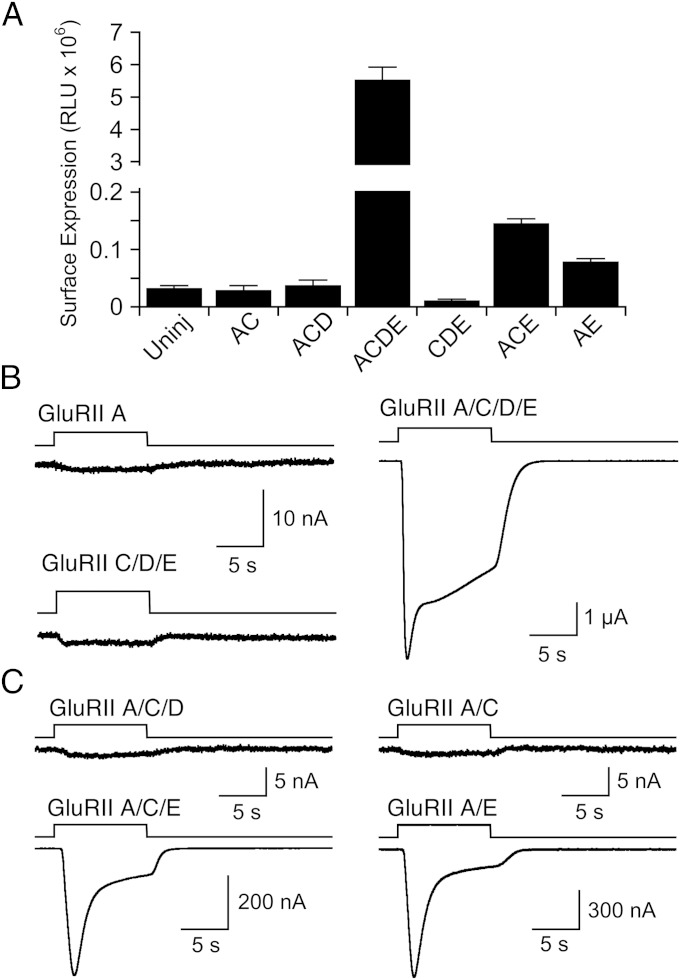
Studies on Drosophila mutants have established that multiple iGluR subunits are required for NMJ function (6, 8, 9, 11). It was previously unclear if this reflects control of receptor assembly, receptor function, cell surface expression, clustering at the NMJ, or a combination of these processes. To begin to address this we used chemiluminescence assays to examine the level of receptor surface expression in oocytes injected with Neto β and various iGluR subunits, combined with functional assays for receptor activity. We found no detectable surface expression in oocytes injected with either HA-GluRIIA, HA-GluRIIA/C, HA-GluRIIA/C/D, or HA-GluRIIC/D/E cRNAs; in contrast, responses for HA-GluRIIA/C/D/E were 170 ± 12-fold above the background signal recorded in uninjected oocytes (Fig. 2A). A similar requirement for surface expression was observed for HA-GluRIIB–containing heterotetramers (Fig. S2A). We then examined additional subunit combinations and found that for HA-GluRIIA/C/E or HA-GluRIIA/E the signals were 4.5 ± 0.2- and 2.4 ± 0.2-fold above background, indicating weak cell surface expression, but much less than for GluRIIA/C/D/E (Fig. 2A).
Fig. 2.
Subunit dependence of Drosophila NMJ glutamate receptor cell surface expression and function. (A) Cell surface expression measured by chemiluminescence for different combinations of HA-tagged GluRII subunits and Neto β; error bars show SEM. (B) Responses to 3 mM glutamate at −60 mV for representative oocytes injected with cRNA for GluRIIA, GluRIIC/D/E, and GluRIIA/C/D/E and Neto β, all treated with Con A, and all recorded on the same day. (C) Responses to 3 mM glutamate at −60 mV for representative oocytes injected with cRNA for Neto β and GluRIIA/C/D, GluRIIA/C, GluRIIA/C/E, or GluRIIA/E, all treated with Con A and recorded on the same day from the same batch of oocytes.
Because with Xenopus oocyte two-electrode voltage-clamp recordings we can in principle detect the activation of as few as 200 Drosophila wild-type NMJ iGluRs of conductance 100 pS (6, 19), we used this higher level of sensitivity to test whether any of these subunit combinations, including those that did not yield detectable cell surface expression, could generate functional receptors. For GluRIIA, GluRIIC/D/E, and as a positive control a mixture of the same cRNAs, we tested for responses to 3 mM glutamate on the same day from the same batch of oocytes. Both GluRIIA (n = 10) and GluRIIC/D/E (n = 10) failed to generate functional receptors, with responses to 3 mM glutamate less than 1 nA in amplitude (Fig. 2B), whereas the mixture GluRIIA/C/D/E gave robust responses of amplitude 7.9 ± 2.4 µA (n = 9). In a second experiment, a similar profile was obtained for GluRIIB (Fig. S2B), with responses to glutamate of less than 1 nA (n = 6) for GluRIIB expressed alone, and robust responses for GluRIIB/C/D/E (5.9 ± 0.8 µA, n = 6); however, in this experiment GluRIIC/D/E also gave small but consistent responses to glutamate (8.8 ± 1.6 nA, n = 6). We then tested whether GluRIIA in combination with just one or two additional Drosophila NMJ iGluR subunits could generate functional receptors (Fig. 2C). In this experiment, the combinations GluRIIA/C (n = 10), GluRIIA/B (n = 5), and GluRIIA/C/D (n = 10) were nonfunctional, with responses to 3 mM glutamate of around 1 nA or less. In contrast, both GluRIIA/E and GluRIIA/C/E gave substantial responses to 3 mM glutamate, 872 ± 300 nA (n = 9) and 608 ± 300 nA (n = 10); however, these responses were on average still only 2–3% of the amplitude of those recorded for GluRIIA/C/D/E recorded on the same day (39 ± 4.6 µA, n = 6), consistent with the results of cell surface expression assays.
Drosophila iGluRs Are Ca2+-Permeable and Blocked by Polyamines.
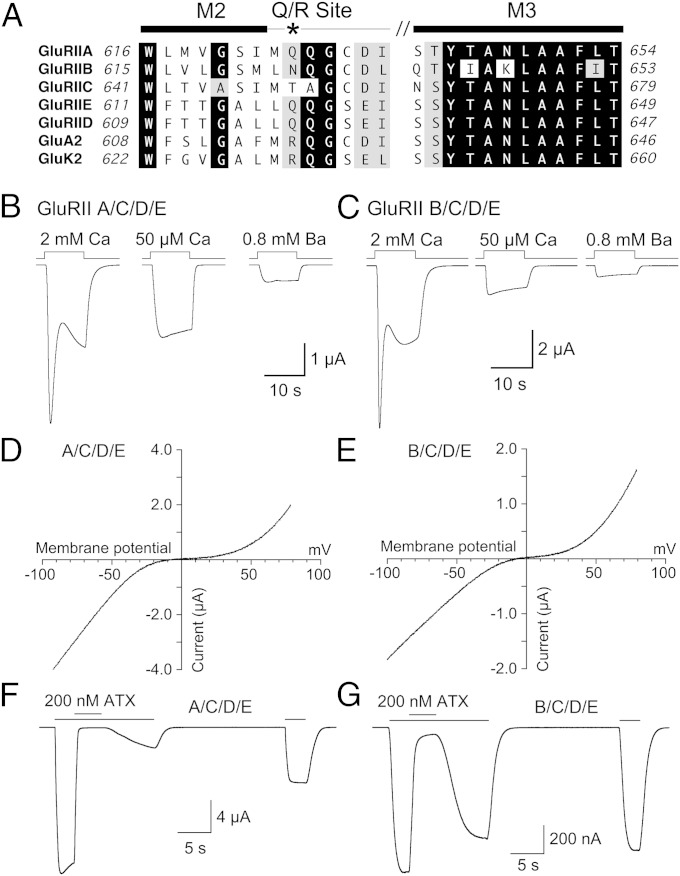
Amino acid sequence alignments for Drosophila NMJ iGluRs derived from cDNA sequences reveals a glutamine, asparagine, or threonine residue at the Q/R site in the pore loop (Fig. 3A), a position that in vertebrate GluA2 and GluK2 AMPA and kainate receptors is converted to an arginine by RNA editing (20–22). In the M3 helix, which forms the entrance to the ion channel pore in glutamate receptors (23–26), Drosophila GluRIIB has three amino acid substitutions; most notable is the replacement of asparagine by lysine at position 647 (Fig. 3A).
Fig. 3.
Calcium permeability and voltage dependence of Drosophila NMJ glutamate receptor currents. (A) Sequence alignments for the M2, pore loop, and M3 ion channel segments of the five Drosophila NMJ iGluRs, a vertebrate AMPA (GluA2), and a vertebrate kainate (GluK2) receptor; the asterisk indicates the Q/R RNA editing site. (B) Responses to 3 mM glutamate at −60 mV for GluRIIA/C/D/E and Neto β recorded from a single oocyte with either 2 mM Ca2+, 50 µM Ca2+, or 0.8 mM Ba2+ in the extracellular solution; transient inward Ca2+-activated chloride currents are a result of activation of Xenopus TMEM16A channels. (C) Responses to 3 mM glutamate at −60 mV for GluRIIB/C/D/E and Neto β recorded from a single oocyte with either 2 mM Ca2+, 50 µM Ca2+, or 0.8 mM Ba2+ in the extracellular solution. (D and E) Current-voltage plots for glutamate activated responses for Neto β and GluRIIA/C/D/E (D) and GluRIIB/C/D/E (E) recorded with 0.8 mM extracellular Ba2+; the smaller outward current response and extended region of near zero slope conductance for GluRIIA/C/D/E versus GluRIIB/C/D/E was observed consistently. (F and G) Block of responses to 3 mM glutamate at −60 mV by 200 nM argiotoxin for Neto β and GluRIIA/C/D/E (F) or GluRIIB/C/D/E (G), illustrating much faster recovery for GluRIIB/C/D/E.
In vertebrate AMPA and kainate receptors a glutamine at the Q/R site confers Ca2+ permeability (20, 22), voltage-dependent block by cytoplasmic polyamines (27, 28), and sensitivity to polyamine toxins, such as argiotoxin (ATX) (29, 30). It is wellestablished that in Xenopus oocytes Ca2+ influx through recombinant cation permeable ion channels can trigger transient Ca2+-dependent chloride currents because of activation of endogenous TMEM16A channels (31, 32). Consistent with the predicted Ca2+ permeability of Drosophila NMJ iGluRs, the transient inward current for both GluRIIA/C/D/E (Fig. 3B) and GluRIIB/C/D/E (Fig. 3C) was abolished when the extracellular Ca2+ concentration was reduced to 50 µM, or when Ca2+ was replaced by Ba2+. Responses recorded in the presence 0.8 mM Ba2+ were only 24 ± 0.4% (n = 10) of those recorded with 50 µM Ca2+ for GluRIIA/C/D/E (Fig. 3B) and 45 ± 1% (n = 8) for GluRIIB/C/D/E (Fig. 3C), indicating that Ba2+ blocks Na+ flux through both channel types, similar to vertebrate kainate receptors (33). The large amplitude of the Ca2+-dependent chloride current compared with those recorded previously for GluK2 suggests that both types of Drosophila NMJ iGluRs have a high permeability to Ca2+.
Current-voltage plots for both GluRIIA/C/D/E (Fig. 3D) and GluRIIB/C/D/E (Fig. 3E) responses to glutamate, recorded with either 50 µM extracellular Ca2+ or when Ca2+ was replaced by Ba2+, exhibited a characteristic biphasic rectification exactly like that observed for vertebrate Ca2+-permeable AMPA and kainate receptors (22, 34, 35), with slightly greater relief from block at positive membrane potentials for GluRIIB/C/D/E (Fig. 3E and Fig. S3). It is well established that such biphasic rectification is produced by voltage-dependent ion channel block by endogenous cytoplasmic spermine and spermidine, with polyamine permeation on depolarization to positive membrane potentials (28, 36). Analysis of conductance voltage plots revealed a 13-mV depolarizing shift in the half-block potential for GluRIIB/C/D/E −31.9 ± 1.3 mV (n = 6) compared with GluRIIA/C/D/E −44.8 ± 0.9 mV (n = 9), suggesting a lower polyamine affinity for channels containing the GluRIIB subunit (Fig. S3), perhaps because of the amino acid substitution at the Q/R site or the lysine substitution in M3 (Fig. 3A).
ATX produced a block of responses to 3 mM glutamate at −60 mV for both GluRIIA/C/D/E and GluRIIB/C/D/E (Fig. 3 F and G). At a concentration of 200 nM ATX produced 99 ± 0.6% block (n = 6) for GluRIIA/C/D/E, with slow recovery after correction for rundown (Fig. S4), to 15.4 ± 1 and 41 ± 2% of the initial response to glutamate at 8 and 30 s after removal of toxin (Fig. 3F). For GluRIIB/C/D/E 200 nM ATX produced 94 ± 0.3% block (n = 10), but with much faster recovery after correction for rundown (Fig. S4), to 88 ± 1 and 96 ± 1% of the initial response to glutamate at 8 and 30 s after removal of toxin (Fig. 3G). Block was weaker for the synthetic toxin 1-naphthyl acetyl spermine (NASPM; 1 µM), 86 ± 1% (n = 6) for GluRIIA/C/D/E, and 61 ± 2% (n = 10) for GluRIIB/C/D/E, with faster recovery, to 102 ± 3% and 102 ± 3% of control at 8 s after removal of toxin (Fig. S4). The difference in toxin sensitivity for GluRIIA/C/D/E and GluRIIB/C/D/E agrees well with that for Drosophila NMJ type A and type B glutamate receptors (6).
Drosophila iGluRs Have Low Affinity for Glutamate.
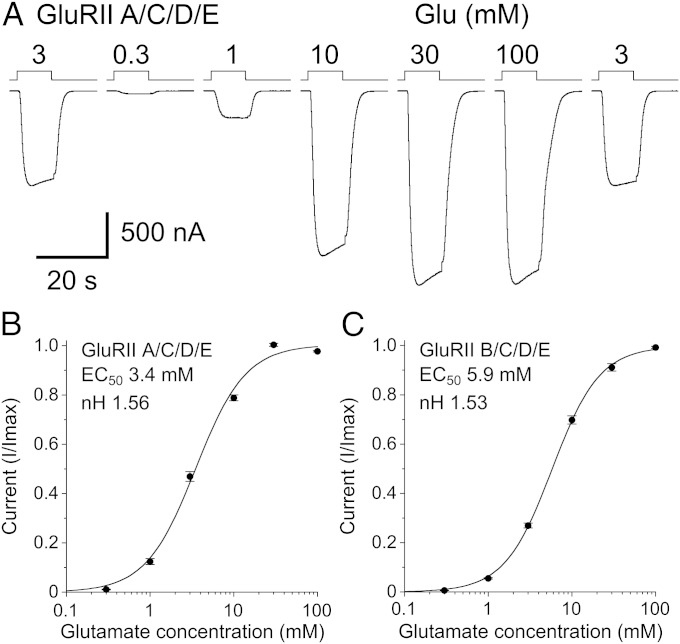
Pilot experiments revealed that at the high amino acid concentrations required to saturate activation of Drosophila NMJ iGluRs, the accompanying change in Na+ concentration distorted the concentration response relationship. To circumvent this we used an extracellular solution containing 20 mM NaCl, to which the sodium salts of gluconic and glutamic acids was added to maintain a constant extracellular Na+ concentration, with glutamate varied from 0.3 to 100 mM (Fig. 4A and Fig. S5A). This approach, combined with substitution of extracellular Ca2+ by 0.8 mM Ba2+, allowed analysis of the concentration response relationship for both GluRIIA/C/D/E (Fig. 4B) and GluRIIB/C/D/E (Fig. 4C), yielding EC50s, Hill coefficients (nH), and maximum currents of 3.4 ± 0.2 mM, nH 1.6 ± 0.04, and 5.8 ± 1.7 µA (n = 9) for GluRIIA/C/D/E, and 5.9 ± 0.3 mM, nH 1.5 ± 0.04, and 2.0 ± 0.4 µA (n = 10) for GluRIIB/C/D/E.
Fig. 4.
Drosophila NMJ iGluRs have low affinity for glutamate. (A) Responses at −60 mV to glutamate at the indicated concentrations with 0.8 mM extracellular Ba2+ used to prevent activation of TMEM16A currents and Na+ gluconate used to maintain a constant extracellular sodium concentration. (B and C) Concentration-response plots for glutamate-activated currents for Neto β and GluRIIA/C/D/E (B) or GluRIIB/C/D/E (C) fit with the Hill equation; data points show the mean for 9 and 10 cells, respectively, normalized to the peak current recorded in individual cells; error bars show ± SEM.
Drosophila iGluRs Are Not AMPA or Kainate Receptors.
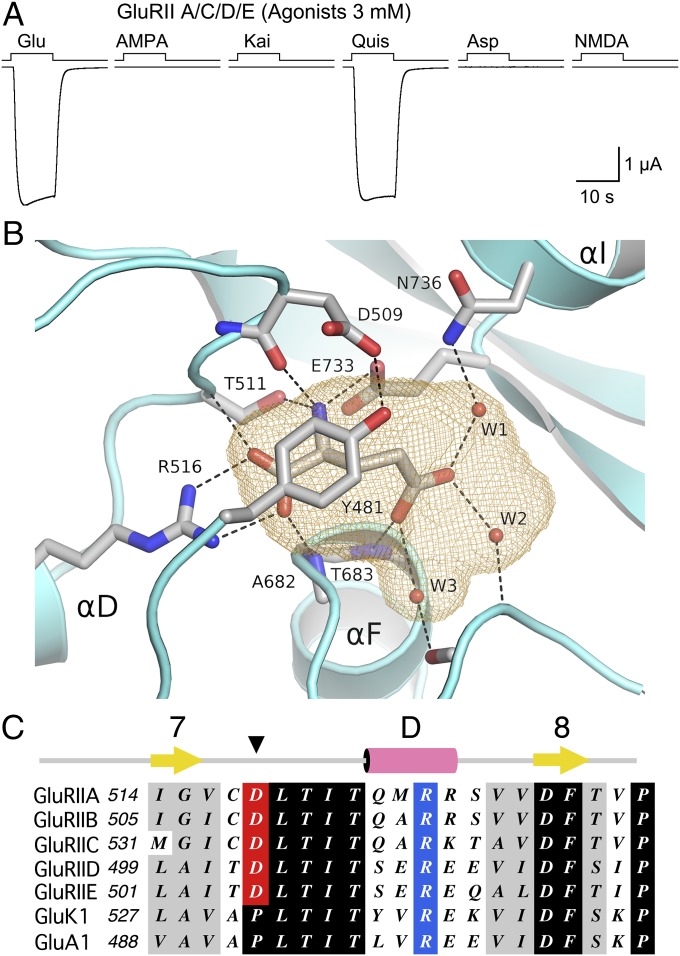
We applied AMPA, kainate, and NMDA, the canonical ligands used to classify vertebrate iGluRs (37), and compared in the same cell responses to those evoked by glutamate, aspartate, and quisqualate. All agonists were applied at a concentration of 3 mM, close to the EC50 for glutamate, with substitution of extracellular Ca2+ by 0.8 mM Ba2+ used to prevent activation of TMEM16A channels (Fig. 5A). For GluRIIA/C/D/E responses to glutamate and quisqualate, 3.3 ± 0.4 and 3.4 ± 0.4 µA (n = 6) were of similar amplitude, whereas for AMPA, kainate, aspartate, and NMDA there was no detectable response. A similar profile was obtained for GluRIIB/C/D/E, with glutamate and quisqualate responses of 0.38 ± 0.13 and 0.36 ± 0.13 µA (n = 8), and no detectable response for AMPA, kainate, aspartate, and NMDA (Fig. S5B).
Fig. 5.
Structural basis for ligand selectivity of Drosophila NMJ iGluRs. (A) Responses at −60 mV to sequential applications of six glutamate receptor agonists, all at 3 mM, recorded from a single oocyte expressing Neto β and GluRIIA/C/D/E. (B) Ribbon diagram of the GluRIIB ligand binding domain crystal structure showing a bound glutamate molecule trapped in a cavity (orange mesh) together with three water molecules. The cavity is capped by Tyr481 which is held in place by a hydrogen bond formed with the side chain of Asp509. (C) Sequence alignments for part of domain 1 for five Drosophila NMJ iGluRs, and representative vertebrate kainate (GluK1) and AMPA (GluA1) receptors, showing the exchange of a conserved proline residue for an aspartate in the Drosophila NMJ iGluRs.
To investigate the structural basis for this unique profile we screened ligand binding domain (LBD) S1S2 constructs of the five Drosophila NMJ iGluRs for expression as soluble proteins in Escherichia coli, and identified GluRIIB as a promising candidate for crystallization. X-ray diffraction data for the GluRIIB S1S2 complex with glutamate, at a resolution of 2 Å (Table S1), revealed the classic back-to-back LBD dimer assembly (Fig. S6A), as first reported for the GluA2 AMPA receptor (38). In both subunits glutamate was bound in a cavity of volume 208 Å3 together with three trapped water molecules (Fig. 5B and Fig. S6B). The glutamate ligand α-carboxyl and α-amino groups make ion pair and hydrogen bond contacts with conserved arginine and glutamate residues, identical to the binding mechanism for AMPA and kainate receptors (38, 39), with the γ-carboxyl group forming a series of solvent mediated interactions with main-chain and side-chain groups in domain 2 (Fig. 5B and Fig. S6C). The cavity volume for GluRIIB is similar to that for GluA2 (218 Å3), which binds both AMPA and kainate, as well as quisqualate (38, 40), but smaller than that for GluK1, GluK2, and GluK3 volume 305, 255, and 299 Å3, respectively (39, 41), suggesting that structural features unique to Drosophila NMJ iGluRs control ligand selectivity.
To gain insight into why quisqualate but not AMPA or kainate can activate Drosophila NMJ iGluRs, we superimposed crystal structures for vertebrate GluA2 and GluK2 LBD complexes with these ligands on the GluRIIB LBD crystal structure. This process revealed that, as is the case for GluA2 and GluK2 (39, 40), the quisqualate ligand is easily accommodated in the GluRIIB binding site by displacement of water molecule W1 (Fig. S7A). Within domain 1 of the GluRIIB LBD structure, in the loop between β-strand 7 and α-helix D, the side chain of Asp509 forms a hydrogen bond with the hydroxyl group of Tyr481, a conserved aromatic residue that caps the entrance to the ligand binding cavity, sealing it from extracellular solvent. Stacked above Tyr481, the side chain of Arg429 forms a cation π interaction with the aromatic ring, further stabilizing the conformation of Tyr481 (Fig. S6C). Amino acid sequence alignments (Fig. 5C) reveal that Asp509 is conserved in all Drosophila NMJ iGluRs, whereas in all vertebrate AMPA and kainate receptor subunits there is a proline at this position; similarly, cation π stacking by Arg429 is unique to GluRIIA, GluRIIB, and GluRIIC, because vertebrate AMPA and kainate receptor subunits have an Ile residue at this position. As a result, because of the different conformation of the isoxazazole group, AMPA is unable to bind to GluRIIB because the ligand 5-methyl group makes steric clashes with Asp509 and Asn736 (Fig. S7B). Similarly, although the ligand α-carboxyl, α-amino, and γ-carboxyl groups of kainate are isosteric with those of glutamate, the isopropenyl group makes steric clashes with the Asp509 and Tyr481 side chains (Fig. S7C).
Discussion
The Drosophila NMJ has been used extensively for genetic analysis of synapse assembly and regulation. However, lack of a recombinant system for expression of functional receptors has made it difficult to differentiate between defects in the subcellular targeting and distribution of receptors and their activity. Here we show that Neto is required for the function of Drosophila NMJ iGluRs in Xenopus oocytes, and use this system to examine the subunit dependence and function of these receptors. Because trafficking and synaptic stabilization of NMJ iGluRs depend on receptor activity (10), our findings pave the way toward obtaining an understanding of changes in receptor function before the studying the compound phenotypes in vivo.
Assembly and Function of NMJ iGluRs.
Our results indicate that only iGluR heterotetramers are efficiently delivered at the cell surface (Fig. 2 and Fig. S2). Coexpression of three or fewer iGluR subunits with Neto β induced very small currents primarily because of diminished surface delivery (Fig. 2 and Fig. S2). Because the assembly of iGluR heterotetramers is mediated via a dimer of dimers organization (25, 26, 42) and coexpression of GluRIIA/E and Neto β induced small but significant currents, we speculate that the order of subunit arrangement in the Drosophila NMJ heterotetramers entails the formation of GluRIIA/E and the reciprocal C/D dimers. In Xenopus oocytes, surface expression of iGluR heterotetramers is much lower than that of vertebrate GluK2 (Fig. S1). This finding suggests that additional auxiliary subunits may aid in targeting the NMJ heterotetramers to the cell surface. Indeed, the Drosophila genome codes for Stargazin-like (Stg1) and two Cornichon-type proteins that may regulate the surface expression of NMJ receptors (4, 43). Our results suggest that the function of Drosophila Neto is conserved with that in vertebrates and C. elegans: modulation of receptor function is the major role of Neto, with much less of an effect on surface delivery (12, 13). Because trafficking and stabilization of iGluRs at the postsynaptic density depend on receptor activity (6, 10), Drosophila Neto may have a major effect on iGluR clustering partly because of its role on receptor function.
Functional Properties of Recombinant Drosophila NMJ iGluRs.
The expression of fly NMJ iGluRs in Xenopus oocytes, and structural analysis of the GluRIIB ligand binding domain, has uncovered similarities to vertebrate Ca2+ permeable AMPA and kainate receptors and unique features of Drosophila receptors. The rectification properties of Drosophila larval NMJ iGluRs has not been analyzed in detail before, but biphasic responses were observed previously using either microelectrode or whole-cell recordings (2, 44, 45), although not for single-channel recording using isolated membrane patches (45, 46). This difference mimics the behavior of vertebrate Ca2+ permeable AMPA and kainate receptors, and is caused by washout of cytoplasmic polyamines in isolated patches (27, 28).
We find a modest difference in half-block potential for GluRIIA and GluRIIB (Fig. S3), indicating a slightly lower affinity of GluRIIB for polyamines; this small difference is likely caused by incorporation of only a single GluRIIB subunit in a heterotetramer. Because of the Asn and Lys substitutions at the Q/R site and in the M3 helix, it is possible that the Ca2+ permeability for GluRIIB will also be less than for GluRIIA, although in the experiments reported here both types produced robust activation of Ca-dependent Cl currents. Experiments on unidentified iGluRs in cultured Drosophila myotubes revealed a high Ca2+ permeability, PCa/PNa 9.6 (46), comparable to the value for NMDA receptors, PCa/PNa 10.6 (47), and much higher than that for vertebrate AMPA and kainate receptors, PCa/PNa 1.2–1.4 (22, 48). It is thus noteworthy that although RNA editing occurs in Drosophila and modifies Shab potassium channel inactivation (49), a genome-wide analysis by single-molecule sequencing found no evidence of editing for Drosophila NMJ iGluRs (50). As a consequence, activity-dependent Ca2+ influx at the Drosophila NMJ is likely to play an unusually prominent role in synapse development and regulation.
The ligand-binding properties of Drosophila NMJ iGluRs are strikingly different from those for vertebrate glutamate receptors. Both GluRIIA and GluRIIB subtypes have a similar, low sensitivity to glutamate, EC50 3 and 6 mM, respectively, which agrees well with the EC50 of 2 mM obtained using single-channel analysis for native Drosophila NMJ iGluRs (19). Similarly, in larval NMJ preparations, only l-glutamate mimicked the action of the natural neurotransmitter among many agonists tested, including aspartic acid (2), which also produced no response in our experiments. However, despite high structural conservation in the LBDs of vertebrate AMPA and kainate receptors and Drosophila NMJ iGluRs, sequence identity and similarity of 34–44% and 51–62%, respectively, Drosophila NMJ iGluRs are not activated by any of the classic ligands used to classify vertebrate iGluR subtypes. By solving the structure of the GluRIIB LBD complex with glutamate we established that the volume of the ligand binding cavity is similar to that of AMPA receptors, and that steric factors prevent the binding of AMPA and kainate. Most notable is the locking into position of Tyr481 both by Asp509 and by a stacking interaction with Arg429 (Fig. S6C) that, in combination, prevent binding of AMPA and kainate by steric hindrance. In domain 2 Asn736 plays a similar role to Asn690 in GluK2 in preventing binding of AMPA, which in the AMPA-sensitive GluK1 subtype Asn690 is replaced by Ser (39).
Prior studies on the Drosophila NMJ have relied extensively on the analysis of genetically modified flies. This powerful approach has one drawback for the study of the molecular mechanisms of ion-channel function, namely the complex compound phenotypes that ion-channel mutants can produce. The ability to functionally characterize mutant iGluRs before introducing them into flies will greatly facilitate future analysis of this important model for synaptic development.
Materials and Methods
Construct Design, Immunohistochemical Detection, and Chemiluminescence Assays.
Drosophila iGluR coding sequences were PCR-amplified from full-length cDNAs and cloned into a pSP64 expression vector using standard techniques for recombinant cDNA manipulation, which were used to add tags for immunohistochemical detection and analysis of cell surface expression by chemiluminescence using commercially available antibodies, as described in detail in SI Materials and Methods.
Expression in Xenopus Oocytes and Functional Analysis.
Defolliculated stage 5–6 oocytes were injected with 0.5–10 ng cRNA generated from linearized plasmids using the Ambion, mMessage mMachine transcription kit. Two-electrode voltage-clamp recordings were performed as described in detail in SI Materials and Methods, essentially as reported previously (33).
Protein Expression and X-Ray Crystallography.
The GluRIIB LBD S1S2 construct, residues D416-K537 and D660-D802, was expressed as a soluble protein in E. coli, purified to homogeneity using metal affinity and ion-exchange chromatography, and crystallized. X-ray diffraction data collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, was used to solve the structure by molecular replacement to a resolution of 2.0 Å, as described in detail in SI Materials and Methods. The coordinates and structure factors have been deposited to the protein data bank with PDB ID code 4WXJ.
Supplementary Material
Acknowledgments
We thank Carla Glasser and Peter Nguyen for technical assistance; Aaron DiAntonio (Washington University) for the GluRIIB plasmid; Susumu Tomita (Yale University) for the HA-tagged GluK2 plasmid; and Prof. Koji Nakanishi (Columbia University) for the gift of argiotoxin. Data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. This work was supported by the Intramural Research Program of The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4WXJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500458112/-/DCSupplemental.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol Rev. 2010;62(3):405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jan LY, Jan YN. l-glutamate as an excitatory transmitter at the Drosophila larval neuromuscular junction. J Physiol. 1976;262(1):215–236. doi: 10.1113/jphysiol.1976.sp011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi A, Takeuchi N. The effect on crayfish muscle of iontophoretically applied glutamate. J Physiol. 1964;170:296–317. doi: 10.1113/jphysiol.1964.sp007332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker CS, et al. Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc Natl Acad Sci USA. 2006;103(28):10781–10786. doi: 10.1073/pnas.0604482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janovjak H, Sandoz G, Isacoff EY. A modern ionotropic glutamate receptor with a K(+) selectivity signature sequence. Nat Commun. 2011;2:232. doi: 10.1038/ncomms1231. [DOI] [PubMed] [Google Scholar]
- 6.DiAntonio A, Petersen SA, Heckmann M, Goodman CS. Glutamate receptor expression regulates quantal size and quantal content at the Drosophila neuromuscular junction. J Neurosci. 1999;19(8):3023–3032. doi: 10.1523/JNEUROSCI.19-08-03023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J Neurosci. 2004;24(6):1406–1415. doi: 10.1523/JNEUROSCI.1575-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Featherstone DE, et al. An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J Neurosci. 2005;25(12):3199–3208. doi: 10.1523/JNEUROSCI.4201-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin G, et al. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J Neurosci. 2005;25(12):3209–3218. doi: 10.1523/JNEUROSCI.4194-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petzoldt AG, et al. Gating characteristics control glutamate receptor distribution and trafficking in vivo. Curr Biol. 2014;24(17):2059–2065. doi: 10.1016/j.cub.2014.07.051. [DOI] [PubMed] [Google Scholar]
- 11.Kim YJ, Bao H, Bonanno L, Zhang B, Serpe M. Drosophila Neto is essential for clustering glutamate receptors at the neuromuscular junction. Genes Dev. 2012;26(9):974–987. doi: 10.1101/gad.185165.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, et al. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61(3):385–396. doi: 10.1016/j.neuron.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, et al. The SOL-2/Neto auxiliary protein modulates the function of AMPA-subtype ionotropic glutamate receptors. Neuron. 2012;75(5):838–850. doi: 10.1016/j.neuron.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Mellem JE, Brockie PJ, Madsen DM, Maricq AV. SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature. 2004;427(6973):451–457. doi: 10.1038/nature02244. [DOI] [PubMed] [Google Scholar]
- 15.Heckmann M, Dudel J. Desensitization and resensitization kinetics of glutamate receptor channels from Drosophila larval muscle. Biophys J. 1997;72(5):2160–2169. doi: 10.1016/S0006-3495(97)78859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathers DA, Usherwood PN. Concanavalin A blocks desensitisation of glutamate receptors on insect muscle fibres. Nature. 1976;259(5542):409–411. doi: 10.1038/259409a0. [DOI] [PubMed] [Google Scholar]
- 17.Wong LA, Mayer ML. Differential modulation by cyclothiazide and concanavalin A of desensitization at native alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid- and kainate-preferring glutamate receptors. Mol Pharmacol. 1993;44(3):504–510. [PubMed] [Google Scholar]
- 18.Lomash S, Chittori S, Brown P, Mayer ML. Anions mediate ligand binding in Adineta vaga glutamate receptor ion channels. Structure. 2013;21(3):414–425. doi: 10.1016/j.str.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heckmann M, Parzefall F, Dudel J. Activation kinetics of glutamate receptor channels from wild-type Drosophila muscle. Pflugers Arch. 1996;432(6):1023–1029. doi: 10.1007/s004240050230. [DOI] [PubMed] [Google Scholar]
- 20.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell. 1993;75(7):1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 22.Köhler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: Diversity by RNA editing. Neuron. 1993;10(3):491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 23.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462(7274):745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dürr KL, et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158(4):778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344(6187):992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CH, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511(7508):191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15(2):453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 28.Kamboj SK, Swanson GT, Cull-Candy SG. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol. 1995;486(Pt 2):297–303. doi: 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herlitze S, et al. Argiotoxin detects molecular differences in AMPA receptor channels. Neuron. 1993;10(6):1131–1140. doi: 10.1016/0896-6273(93)90061-u. [DOI] [PubMed] [Google Scholar]
- 30.Brackley PT, Bell DR, Choi SK, Nakanishi K, Usherwood PN. Selective antagonism of native and cloned kainate and NMDA receptors by polyamine-containing toxins. J Pharmacol Exp Ther. 1993;266(3):1573–1580. [PubMed] [Google Scholar]
- 31.Boton R, Dascal N, Gillo B, Lass Y. Two calcium-activated chloride conductances in Xenopus laevis oocytes permeabilized with the ionophore A23187. J Physiol. 1989;408:511–534. doi: 10.1113/jphysiol.1989.sp017473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134(6):1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panchenko VA, Glasser CR, Partin KM, Mayer ML. Amino acid substitutions in the pore of rat glutamate receptors at sites influencing block by polyamines. J Physiol. 1999;520(Pt 2):337–357. doi: 10.1111/j.1469-7793.1999.t01-1-00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253(5023):1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- 35.Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991;252(5013):1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- 36.Bähring R, Bowie D, Benveniste M, Mayer ML. Permeation and block of rat GluR6 glutamate receptor channels by internal and external polyamines. J Physiol. 1997;502(Pt 3):575–589. doi: 10.1111/j.1469-7793.1997.575bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watkins JC, Evans RH. Excitatory amino acid transmitters. Annu Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- 38.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28(1):165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 39.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: Molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45(4):539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 40.Jin R, Horning M, Mayer ML, Gouaux E. Mechanism of activation and selectivity in a ligand-gated ion channel: Structural and functional studies of GluR2 and quisqualate. Biochemistry. 2002;41(52):15635–15643. doi: 10.1021/bi020583k. [DOI] [PubMed] [Google Scholar]
- 41.Veran J, et al. Zinc potentiates GluK3 glutamate receptor function by stabilizing the ligand binding domain dimer interface. Neuron. 2012;76(3):565–578. doi: 10.1016/j.neuron.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71(2):319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YJ, Serpe M. Building a synapse: A complex matter. Fly (Austin) 2013;7(3):146–152. doi: 10.4161/fly.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broadie KS, Bate M. Development of the embryonic neuromuscular synapse of Drosophila melanogaster. J Neurosci. 1993;13(1):144–166. doi: 10.1523/JNEUROSCI.13-01-00144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishikawa K, Kidokoro Y. Junctional and extrajunctional glutamate receptor channels in Drosophila embryos and larvae. J Neurosci. 1995;15(12):7905–7915. doi: 10.1523/JNEUROSCI.15-12-07905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang H, Ciani S, Kidokoro Y. Ion permeation properties of the glutamate receptor channel in cultured embryonic Drosophila myotubes. J Physiol. 1994;476(1):1–16. [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer ML, Westbrook GL. Permeation and block of N-methyl-d-aspartic acid receptor channels by divalent cations in mouse cultured central neurones. J Physiol. 1987;394:501–527. doi: 10.1113/jphysiol.1987.sp016883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of ampa receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8(1):189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- 49.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11(10):950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 50.St Laurent G, et al. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 2013;20(11):1333–1339. doi: 10.1038/nsmb.2675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.