Significance
In this paper, we characterize, for the first time to our knowledge, synaptic phenotypes in contactin associated protein-like 2 (Cntnap2) knockout neurons and reveal a novel role for CNTNAP2 in the correct trafficking of AMPA-type glutamate receptors. In addition, we report that cellular phenotypes emerge late in postnatal development, suggesting a mechanism for the apparent late emergence of some CNTNAP2-associated disorders. Taken together, our findings may provide insight into the mechanism underlying pathogenesis of CNTNAP2-associated neuropsychiatric disorders.
Keywords: CNTNAP2, synapse, AMPA, GluA1, schizophrenia
Abstract
Central glutamatergic synapses and the molecular pathways that control them are emerging as common substrates in the pathogenesis of mental disorders. Genetic variation in the contactin associated protein-like 2 (CNTNAP2) gene, including copy number variations, exon deletions, truncations, single nucleotide variants, and polymorphisms have been associated with intellectual disability, epilepsy, schizophrenia, language disorders, and autism. CNTNAP2, encoded by Cntnap2, is required for dendritic spine development and its absence causes disease-related phenotypes in mice. However, the mechanisms whereby CNTNAP2 regulates glutamatergic synapses are not known, and cellular phenotypes have not been investigated in Cntnap2 knockout neurons. Here we show that CNTNAP2 is present in dendritic spines, as well as axons and soma. Structured illumination superresolution microscopy reveals closer proximity to excitatory, rather than inhibitory synaptic markers. CNTNAP2 does not promote the formation of synapses and cultured neurons from Cntnap2 knockout mice do not show early defects in axon and dendrite outgrowth, suggesting that CNTNAP2 is not required at this stage. However, mature neurons from knockout mice show reduced spine density and levels of GluA1 subunits of AMPA receptors in spines. Unexpectedly, knockout neurons show large cytoplasmic aggregates of GluA1. Here we characterize, for the first time to our knowledge, synaptic phenotypes in Cntnap2 knockout neurons and reveal a novel role for CNTNAP2 in GluA1 trafficking. Taken together, our findings provide insight into the biological roles of CNTNAP2 and into the pathogenesis of CNTNAP2-associated neuropsychiatric disorders.
Abnormalities in excitatory synapses of cortical pyramidal neurons have emerged as key cellular substrates in the pathogenesis of several psychiatric disorders. Disease-specific disruptions in synaptic morphology or number and glutamate receptors accompany several neuropsychiatric disorders, particularly those that involve deficits in information processing (1). In support of this view, postmortem neuropathological studies found altered dendritic spine density on cortical pyramidal neurons in individuals with intellectual disability (2), autism spectrum disorders (3), and schizophrenia (4). Dendritic spines are the sites of the majority of excitatory glutamatergic synapses in the mammalian brain and represent the postsynaptic compartment of these synapses. Spines are rich in actin, and their morphology changes during development and in various physiological conditions (5). Spines contain glutamate receptors, of which the AMPA subtype are the ones responsible for fast neurotransmission (6). AMPA receptors, like other membrane proteins, are processed in the endoplasmic reticulum and the Golgi complex, then delivered to synapses by forward trafficking mechanisms (7). At the synapse, they undergo constitutive and regulated exo- and endocytosis, known to modulate synapse strength (6). Together, changes in spine number and morphology, along with glutamate receptor content in synapses, underlie functional connectivity in synaptic circuits.
Recent genome-wide association studies, copy number variant (CNV) analyses, and exome sequencing studies have implicated networks of genes that encode glutamatergic synaptic proteins in the etiology of neuropsychiatric disorders (8–10). One such gene, which has received considerable attention, is contactin associated protein-like 2 (CNTNAP2). Gene dosage, rare mutations, and common variations in CNTNAP2 have been associated with several neuropsychiatric disorders. Whereas all subjects with disruptions in the CNTNAP2 gene have complex phenotypes, it appears that shared common features are intellectual disability, seizures, autistic phenotype, and language impairments. Genotypes detected in subjects with intellectual disability include inversion of exons 11 and 12, large deletions, frameshift mutation, splice site mutation, exon deletions, deletions of the 3′ UTR, and a complex rearrangement (11). Subjects with autism have been shown to carry copy number variations, complex rearrangements, an intron deletion, a promoter deletion, and amino acid substitutions (12–20). Disruptions detected in schizophrenia include deletion of exons 9–24, an intron deletion, and copy number variations (21–25). Other disease-associated disruptions include a large deletion, intron deletion in attention deficit and hyperactivity disorder (26, 27), deletion of exons 2–4 in epilepsy (28, 29), and deletion of exons 2–4 in speech delay (30). In addition, common variants in CNTNAP2 have been associated with autism, dyslexia, schizophrenia, and bipolar disorder (31).
A potentially important role for CNTNAP2 in mental disorders is supported by the presence of autism-related behavioral phenotypes in Cntnap2 knockout (KO) mice (32). Sequence variation in CNTNAP2 is associated with altered functional connectivity in the human frontal lobe (33). By contrast, little is understood about the cellular and molecular mechanisms whereby CNTNAP2 controls brain function. Hence, understanding the roles of Cntnap2 gene product in synapses can provide insight into the mechanisms underlying normal and pathological synapse function.
The CNTNAP2 gene encodes a neurexin-related cell adhesion molecule, CNTNAP2 or Caspr2. CNTNAP2 was first identified in the peripheral nervous system, where it was shown in vitro to cluster K+ channels at the juxtaparanodal region (34). However, CNTNAP2 is also abundant in the brain, where it is enriched in the synaptic plasma membrane fraction (14). Notably, a recent article reported that shRNA-mediated knockdown of CNTNAP2 in young developing cultured cortical neurons causes a significant and cell-autonomous reduction of AMPA and NMDA mEPSCs (miniature excitatory postsynaptic currents) and GABA mIPSCs (miniature inhibitory postsynaptic currents), without affecting presynaptic function (35). Knockdown also resulted in reduced dendritic arborization and reduced spine head size, but not spine density. However, the mechanisms whereby CNTNAP2 affects synapses are still poorly understood.
In this study, we investigated the cellular phenotypes of neurons generated from Cntnap2 knockout mice, focusing on synapses. We compared the effects of absence of CNTNAP2 in developing and mature neurons and found late-emerging alterations in spine morphology and GluA1 AMPA receptor subunit localization. When investigating the cellular substrates of AMPA receptor abnormalities, we found an unexpected presence of cytoplasmic GluA1-containing aggregates. Taken together, our data show that CNTNAP2 is important for GluA1 localization to spines, and its loss causes the formation of GluA1-containing aggregates. These mechanisms may contribute to cognitive deficits in CNTNAP2-associated disorders and may represent a general mechanism of pathogenesis in these disorders.
Results
CNTNAP2 Is Present in Multiple Neuronal Compartments.
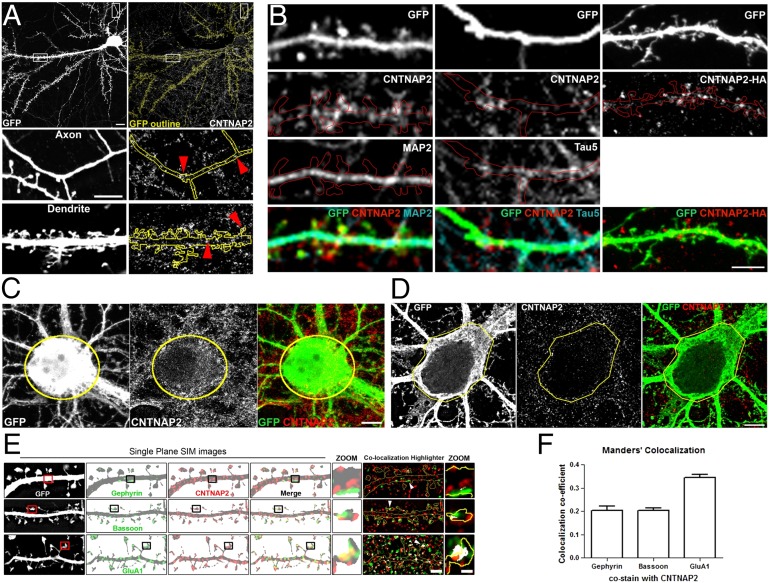
CNTNAP2 has been shown to be required for the development of spines, synapses, and dendrites, but the underlying mechanisms are not known. We therefore used immunofluorescence confocal imaging to examine the subcellular distribution of CNTNAP2 in cultured neurons. We found that CNTNAP2 is present in punctate structures throughout the neurons, including axons, dendrites, spines, and somata (Fig. 1). This antibody was specific for CNTNAP2, as it did not detect signal in Cntnap2-knockout neurons or neuronal homogenates, by immunofluorescence or Western blotting (Fig. S1; see also Fig. 3E). Filling neurons with GFP, immunostaining for CNTNAP2 and dendritic marker MAP2, and imaging with standard confocal microscopy revealed that a subset of CNTNAP2 puncta are present in dendrites and spines (Fig. 1 A and B). However, CNTNAP2 puncta were also present in structures not stained for MAP2, suggesting CNTNAP2’s axonal distribution. This is further supported by costaining with axonal marker Tau5 (Fig. 1B). Similarly, exogenously overexpressed CNTNAP2 (CNTNAP2-HA) was targeted to dendrites and spines (Fig. 1B). Endogenous CNTNAP2 can also be visualized in the soma of GFP-filled neurons (Fig. 1C).
Fig. 1.
CNTNAP2 is present in multiple neuronal compartments. (A) Confocal images show GFP-transfected wild-type mouse neurons immunostained for endogenous CNTNAP2. White boxes show axon and dendrite regions for zoom panels on Right. Red arrows indicate CNTNAP2 present in both axons and dendrites. Neuronal outline of GFP fill shown in yellow. (Scale bars: 10 μm and 5 μm, respectively.) (B) Confocal images of neurons filled with GFP and immunostained for CNTNAP2 and MAP2 or tau5, or overexpressing HA-tagged CNTNAP2. Neuronal outline of GFP fill shown in red. (Scale bar: 5 μm.) (C) Confocal images of GFP-filled neurons immunostained for CNTNAP2 to show presence within the soma. (Scale bar: 10 µm.) (D) SIM images of somatic endogenous CNTNAP2. (Scale bar: 5 µm.) (E) SIM images of GFP-filled neurons immunostained for CNTNAP2 along with gephyrin, bassoon, or GluA1. Boxed regions are shown in zoom panels. CNTNAP2 and the costained protein were analyzed with the colocalization highlighter feature in ImageJ, shown in Right (colocalization in white). (Scale bars: 1 μm and 500 nm, respectively.) White arrowheads show colocalization in spines. (F) Mander’s colocalization coefficients for SIM images of GFP-filled neurons coimmunostained for CNTNAP2, gephyrin, bassoon, and GluA1.
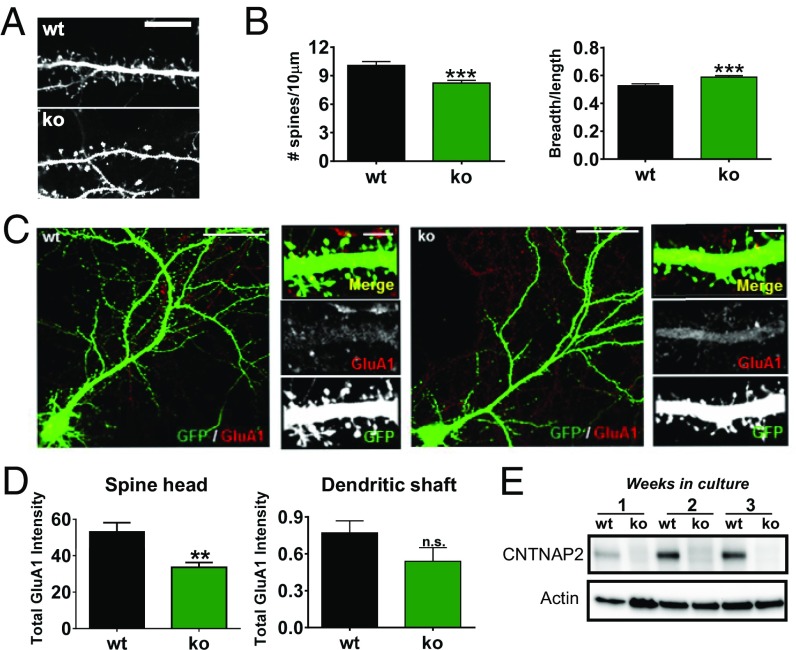
Fig. 3.
Alterations in spine morphology and GluA1 content in spines in cultured cortical pyramidal neurons from Cntnap2−/− mice. (A) Analysis of spine morphology in wild-type and Cntnap2−/− mature cultured cortical neurons (21 DIV). (B) Quantification of average spine density and breadth/length ratios. (C) Immunostaining with a GluA1-specific antibody of GFP-expressing cultured wild-type and Cntnap2−/− neurons (21 DIV). (Scale bars: 20 µm in whole neuron images and 5 µm in magnifications.) (D) Quantification of average GluA1 cluster immunofluorescence intensity/area in spines and shaft. Data shown are means ± SEM; **P < 0.01, ***P < 0.001 compared with WT by Student’s unpaired t test in number of spines, and GluA1 intensity in spine head and shaft and Mann-Whitney’s test in bread/length ratio. (E) Developmental time course of CNTNAP2 protein expression in cultured cortical neurons (1, 2, and 3 weeks in vitro) from Cntnap2-WT and KO mice.
To achieve higher imaging resolution, we used structured illumination microscopy (SIM), a superresolution method that allows a twofold increase in lateral resolution (36). SIM imaging revealed presence of CNTNAP2 within the soma (Fig. 1D), in dendrites, and at excitatory synapses, shown by partial colocalization with AMPA receptor subunit GluA1 with less colocalization evident with presynaptic marker bassoon and inhibitory marker gephyrin (Fig. 1 E and F). These data indicate that CNTNAP2 is more abundant at or near excitatory synapses and that the mechanisms of CNTNAP2-dependent regulation of excitatory and inhibitory synapses may be different. In addition, SIM imaging suggests that CNTNAP2 may be implicated in GluA1 trafficking in dendrites and spines.
CNTNAP2 Is Dispensable for Synapse Formation and Axon and Dendrite Outgrowth.
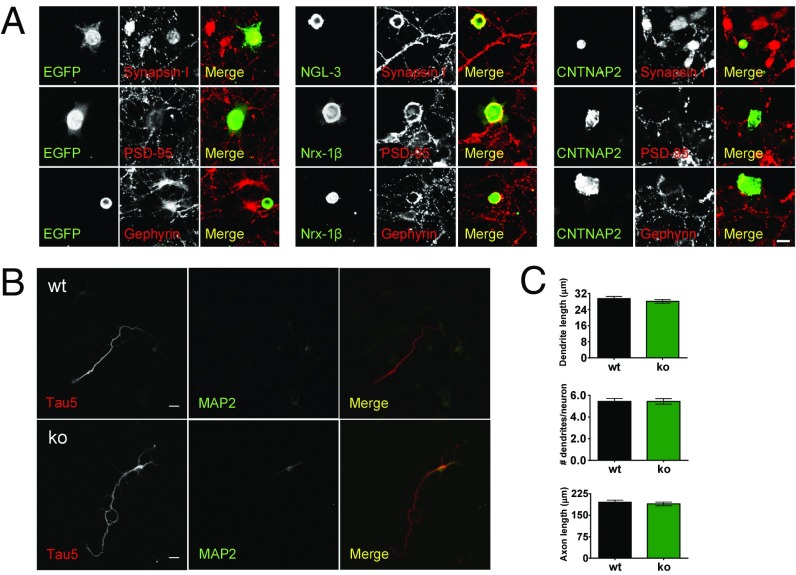
Because CNTNAP2 is at excitatory synapses, and Anderson et al. (35) have shown that it regulates synapse development, we wanted to see whether it has synaptogenic properties. We used a coculture assay in which we expressed tagged CNTNAP2 constructs in HEK293 cells, which we then cocultured with hippocampal neurons. We then assessed the ability of surface-expressed CNTNAP2 to induce formation of pre- or postsynaptic sites along the interface of neuronal processes with the HEK293 cells (Fig. 2A). We found that CNTNAP2 was not capable of inducing the differentiation of either pre- (synapsin I labeled) nor postsynaptic (PSD-95 labeled) sites in excitatory synapses, similar to GFP alone. On the other hand, netrin-G ligand 3 (NGL-3) or neurexin-1β (Nrx-1β) were readily able to induce pre- or postsynaptic differentiation, respectively. Similarly, CNTNAP2 was not able to induce the differentiation of inhibitory synapses, as labeled by gephyrin (Fig. 2A). Control experiments show that GFP alone was unable to induce pre-, post-, or inhibitory synapse development, whereas neuroligin and neurexin were able to induce synapse differentiation, as shown previously (37).
Fig. 2.
CNTNAP2 does not affect synaptogenesis and axon/dendrite outgrowth. (A) Coculture synapse formation assays show that exogenous CNTNAP2 displayed on HEK293T cells does not induce pre- or postsynaptic differentiation in contacting axons or dendrites of cocultured hippocampal neurons (10–13 DIV), as measured by the clustering of synapsin I (presynaptic) and PSD-95 or gephyrin (excitatory and inhibitory postsynaptic, respectively). (Scale bar: 20 μm.) (B) Young cultured cortical neurons (3 DIV) from wild-type and Cntnap2−/− mice do not show alterations in axon and dendrite outgrowth. (Scale bar: 10 μm.) (C) Quantification of dendrite length, number of dendrites/neuron, and axon length. Data shown are means ± SEM; Student’s unpaired t test.
We next examined the role of CNTNAP2 in the initial formation of axons and dendrites. To this end, we examined the early stages of neuronal differentiation in 3 day in vitro (DIV) cultures from wild-type and Cntnap2−/− mice. We did not detect any differences in the total length of axons [t90 = 0.5508; P = 0.5831; n = 42 (WT) and 52 (KO)] or dendrites [t440 = 1.043; P = 0.2976; n = 205 (WT) and 237 (KO)] or in the number of dendrites (t88 = 0.0; P = 1; n = 45) between wild-type and Cntnap2−/− neurons (Fig. 2 B and C). In addition, we examined the time course of CNTNAP2 protein expression in cultured cortical neurons (Fig. 3E). We found that CNTNAP2 expression was low in young (1 week old) neurons, and expression increased in the following weeks in culture. These data suggest that CNTNAP2 is not likely to play an essential role in synapse formation or outgrowth of neuronal processes.
Absence of CNTNAP2 Affects Mature Glutamatergic Synapses.
To determine whether absence of CNTNAP2 affected the morphology and glutamate receptor content of mature excitatory synapses, we examined mature cultures (21 DIV) of pyramidal neurons generated from wild-type and Cntnap2−/− mouse embryos. We detected a reduced density of spines in knockout neurons (Fig. 3 A and B) [t146 = 3.824; P = 0.0002; n = 67 (WT) and 78 (KO)]. Interestingly, spines on Cntnap2−/− neurons had an increased breadth-to-length ratio [U = 55,611; P = 0.0014; n = 483 (WT) and 286 (KO)]. Moreover, knockdown of CNTNAP2 also resulted in a decrease of spine density [t31 = 4.650; P < 0.0001; n = 13 (WT) and 20 (KO)] (Fig. S2), although overexpression (OE) of CNTNAP2 did not affect spine density [t15 = 0.564; P = 0.5811; n = 16 (WT) and 18 (KO)].
We next examined AMPA receptor localization in dendrites and spines of GFP-expressing wild-type and Cntnap2−/− neurons by imaging the immunofluorescence signal of the GluA1 subunit, along with GFP. Interestingly, total GluA1 content in spines was decreased in Cntnap2−/− neurons, but no difference could be detected in the shaft between wild-type and Cntnap2−/− neurons (Fig. 3 C and D). Also, surface staining of GluA1 showed the same pattern as total GluA1; a decrease was found in the spine head [U = 10,262; P < 0.0001; n = 7 (WT) and 12 (KO)] but no differences were found in the shaft [t17 = 1.796; P = 0.0903; n = 67 (WT) and 78 (KO)] (Fig. S3).
In addition, Western blotting of 21 DIV cortical neurons (Fig. S4) from wild-type and Cntnap2−/− mice detected no alterations in the expression levels of GluA1 (t8 = 0.02323; P = 0.9820; n = 5 per group) or GluA2 [t11 = 0.7235; P = 0.4845; n = 7 (WT) and 6 (KO)]. Similarly caspr1 was not altered [t10 = 0.3469; P = 0.7359; n = 7 (WT) and 5 (KO)], indicating that it did not compensate for the absence of CNTNAP2.
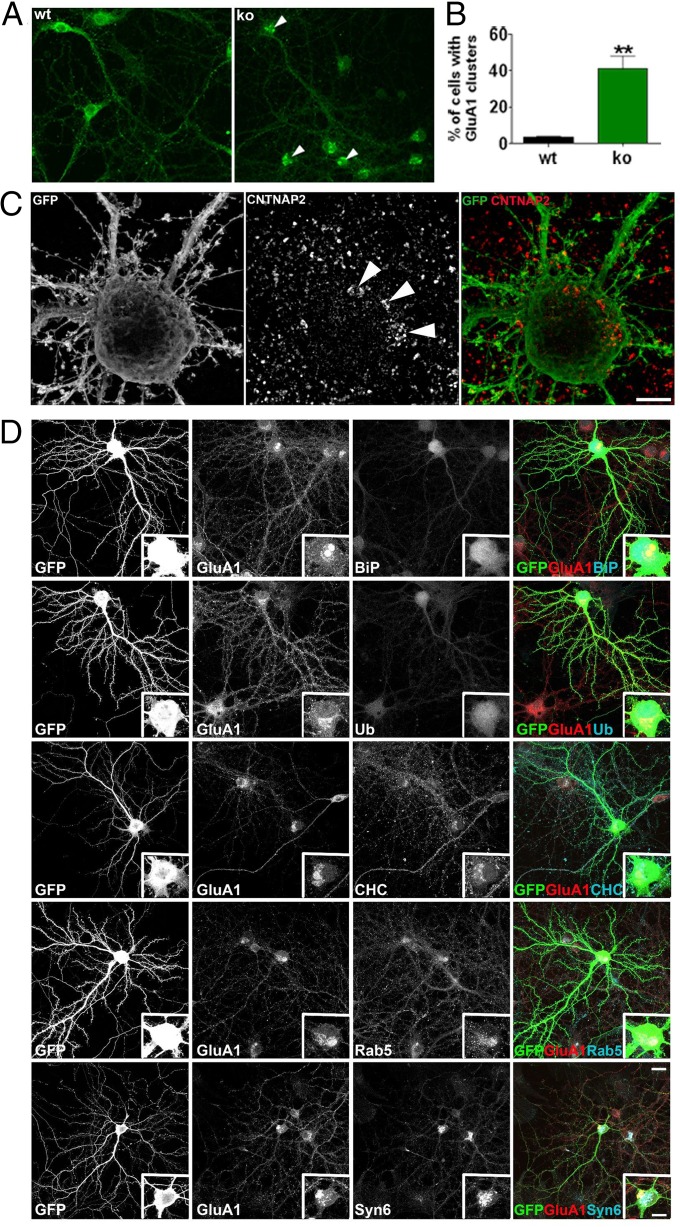
GluA1 Accumulates in Cytoplasmic Aggregates in Cntnap2−/− Neurons.
To understand the cellular mechanisms underlying the reduced AMPA receptor levels in spines, we examined images of wild-type and Cntnap2−/− neurons stained for GluA1. We observed the presence of large aggregates of GluA1 in the soma of knockout neurons (Fig. 4A). Quantification shows that a significant fraction (40%) of Cntnap2−/− neurons presented this phenotype (Fig. 4B) (t4 = 5; P = 0.0063; n = 3 per group). SIM of Cntnap2−/−neurons suggests that these somatic aggregates are composed of clusters of puncta immunopositive for GluA1 (Fig. 4C). Further characterization of the aggregates (Fig. 4D), with coimmunostaining for GluA1 and aggresome markers BiP/GRP98/HSPA5 and ubiquitin do not show extensive colocalization. Meanwhile, coimmunostaining of GluA1 with trafficking markers clathrin (CHC), rab5 and syntaxin6, show increased colocalization with the GluA1 positive aggregates, suggestive of a deficit in trafficking processes.
Fig. 4.
Cytoplasmic aggregates containing GluA1 are present in Cntnap2−/− cortical pyramidal neurons. (A) Low magnification images of GluA1 in wild-type and Cntnap2−/− cultured pyramidal neurons (21 DIV). Arrows point to GluA1 aggregates in the soma. (B) Quantification of the percent of cells containing GluA1 positive aggregates in the soma in wild-type and Cntnap2−/− neurons. (C) SIM images of GluA1 in Cntnap2−/− cultured pyramidal neurons. Arrows point to GluA1 positive puncta of aggregates. (Scale bar: 5 µm.) (D) Confocal images of neurons transfected with GFP and coimmunostained with GluA1 and BiP, ubiquitin (Ub), clathrin heavy chain (CHC), rab5, and syntaxin 6. Insets show zoom of soma. (Scale bars: 20 µm and 10 µm, respectively.)
Interestingly, these GluA1 aggregates seem to be specific to this AMPA receptor subunit, because immunostaining for GluA2 showed no apparent alteration in the distribution in the soma (Fig. S4 D and E) and furthermore, cells that presented GluA1 aggregates did not show the same pattern for GluA2 (Fig. S4E).
Discussion
In this study, we investigated synaptic phenotypes caused by the absence of CNTNAP2 in developing and mature cortical pyramidal neurons. We found late-emerging, but not early, alterations in spine morphology and AMPA receptor distribution. When investigating the cellular substrates of AMPA receptor abnormalities, we found an unexpected presence of GluA1-containing cytosolic aggregates and abnormal GluA1 trafficking. Taken together, our data show that CNTNAP2 is present in synaptic compartments in dendrites, is important for spine morphology and AMPA receptor trafficking, and its loss causes formation of GluA1-containing aggregates. These cellular mechanisms may contribute to behavioral deficits in Cntnap2−/− mice and cognitive deficits in CNTNAP2-associated disorders, and may represent a general mechanism of pathogenesis in such disorders.
Subcellular Localization.
CNTNAP2 has been originally identified as a major component of juxtaparanodes of myelinated axons. However, recent studies (35) have shown that loss of CNTNAP2 affects the development of excitatory and inhibitory synapses. However, the underlying mechanisms are not known. Here we show for the first time to our knowledge that CNTNAP2 protein is localized to multiple subcellular sites in mature pyramidal neurons. We found that CNTNAP2 was present in axons, dendrites, dendritic spines, and the soma. Superresolution microscopy revealed that CNTNAP2 is present in a subset of excitatory synapses and overall showed a higher degree of colocalization with GluA1 than with inhibitory synapse marker gephryin or presynaptic marker bassoon. This finding suggests different mechanisms whereby CNTNAP2 may regulate excitatory and inhibitory synapses. Interestingly, superresolution images also showed that CNTNAP2 colocalizes with GluA1 inside the dendritic shaft, suggesting a potential role in glutamate receptor trafficking.
Late-Emerging Cellular Phenotypes.
Whereas CNTNAP2 knockdown has been shown to affect the development of pyramidal neurons, it is not known what stage of their development requires CNTNAP2. Like other adhesion molecules, CNTNAP2 may be involved in one or several steps of development, including synaptogenesis, axon, or dendrite outgrowth, or synapse maturation. Our data show that CNTNAP2 does not have the ability to induce synapse formation in pyramidal neurons, in a coculture system, and unlike its structural relative neurexin1, is not a synaptogenic molecule. Similarly, absence of CNTNAP2 does not interfere with neurite outgrowth at early stages of neuronal differentiation in culture. These findings suggest that CNTNAP2 is dispensable for processes during early postnatal development. However, analysis of mature neurons, with developed synapses, showed that absence of CNTNAP2 caused a reduction in spine density and alterations in spine morphology. In addition, knockout neurons showed reduced levels of GluA1 AMPA receptor subunit on spines, similar to knockdown (35). These data indicate that CNTNAP2 is most likely necessary for the maturation and maintenance of spiny synapses, but not for earlier stages of postnatal development of pyramidal neurons when the expression levels of this protein are the lowest. In agreement with the idea of CNTNAP2 having a role in maturation and maintenance, Anderson et al. (35) showed a decrease in total dendritic length and branching and a decreased spine size in mature CNTNAP2 knockdown neurons. Such findings may suggest a potential mechanism for the delayed emergence of symptoms in some clinical cases associated with CNTNAP2 mutations. For example, CNTNAP2 CNVs and truncations are associated with schizophrenia, which emerges in adolescence. Interestingly, language regression is present in Amish patients carrying a CNTNAP2 truncation, suggesting a later-onset clinical phenotype.
Cytosolic GluA1 Aggregates.
One intriguing and unexpected cellular phenotype in knockout neurons is the presence of large aggregates containing GluA1. Presence of GluA1 in cytoplasmic aggregates could be caused by a number of mechanisms, including defective protein folding, defective assembly of subunits, abnormal proteolytic degradation, or defective trafficking. Immunostaining for trafficking markers such as rab5 and syntaxin6 showed the highest degree of colocalization; therefore, it seems that the presence of aggregates is at least in part due to a trafficking problem. The exact mechanisms will be the subject of future studies. Whereas AMPA receptor-containing aggresomes have not previously been reported in pyramidal neurons, GluA1-containing aggresomes have been reported in normally developing spinal cord neurons (38). These aggresomes appeared transiently between embryonic day 17 and postnatal day 19 and did not contain other AMPA receptor subunits.
Our findings could provide a molecular and cellular substrate for some of the behavioral phenotypes reported in Cntnap2−/− mice (32). Interestingly, protein aggregates have been reported in postmortem studies of brain tissue from patients with age-related neurodegenerative diseases, including Parkinson's disease, Alzheimer's disease, Huntington's disease, and amyotrophic lateral sclerosis (39). However, presence of aggregates, and specifically AMPA receptor-containing aggregates, have not yet been reported in autism, schizophrenia, intellectual disability, or in animal models of these disorders. Our data suggest that similar mechanisms may contribute to the pathogenesis of CNTNAP2-associated neuropsychiatric disorders, and are therefore worthy of further investigation. Furthermore, our data suggest that abnormalities in synaptic protein trafficking are a mechanism of potential interest in psychiatric disorders.
Materials and Methods
Antibodies and Plasmids.
The following antibodies were purchased: CNTNAP2 (rabbit polyclonal; Millipore), CNTNAP2 (mouse monoclonal; NeuroMab), actin (mouse monoclonal; Abcam), GluA1 (rabbit polyclonal, intracellular epitope; Millipore), GluA1 (mouse monoclonal, extracellular epitope; Millipore), bassoon (mouse monoclonal; NeuroMab), PSD95 (rabbit polyclonal; Cell Signaling), PSD-95 (mouse monoclonal; NeuroMab), gephyrin (mouse monoclonal; Connex), GluA2 (mouse monoclonal; NeuroMab), MAP2 (mouse monoclonal; Millipore), HA (mouse monoclonal; Sigma), GFP (mouse monoclonal; Millipore), GFP (chicken polyclonal; Abcam), clathrin (rabbit monoclonal; Cell Signaling), Rab5 (rabbit monoclonal; Cell Signaling), Syntaxin6 (rabbit monoclonal; Cell Signaling), BiP (rabbit polyclonal; Enzo Life), and ubiquitin (rabbit polyclonal; Sigma). For mixed-culture assays the following antibodies were used: Guinea pig polyclonal EGFP antibodies (#1997) were raised against H6-EGFP (aa 1–240); other antibodies were purchased, HA (mouse monoclonal; Covance and rabbit polyclonal; Santa Cruz), Synapsin I (rabbit polyclonal; Millipore), PSD-95 (mouse monoclonal; NeuroMab), and gephyrin (mouse monoclonal; Synaptic Systems). Plasmids used in this study were: pEGFP-N1 (Clontech) and WT CNTNAP2-HA (kindly provided by Elior Peles, Weizmann Institute of Science, Rehovot, Israel). Interference plasmids specific for Cntnap2 (shRNA) and scrambled control (Sc) were purchased from OriGene. NGL-3 EGFP and EGFP pDisplay constructs have been described (40). Neurexin-1β CFP was a kind gift from Ann Marie Craig, University of British Columbia, Vancouver.
Neuronal Culture and Transfections.
Medium (50,000 cells/cm2) and high density (100,000 cells/cm2) cortical neuron cultures were prepared from Sprague-Dawley rat E18 embryos as described previously (41). Briefly, neurons were plated onto coverslips coated with poly-D-lysine (0.2 mg/mL; Sigma), in feeding medium (neurobasal medium supplemented with B27 (Invitrogen) and 0.5 mM glutamine). A total of 200 μM DL-aminophosphonovalerate (APV) (Ascent Scientific, now Abcam) was added to the medium 4 d later. Cortical neurons were transfected at DIV 23 using Lipofectamine 2000 following the manufacturer's recommendations and the neurons were maintained in the feeding media for 3 d after the transfection (unless stated otherwise). Neurons were then fixed in 4% (wt/vol) formaldehyde + 4% (wt/vol) sucrose in PBS for 10 min. Coverslips were then processed for immunostaining. Only neurons that exhibited a pyramidal asymmetric morphology, with a single long, highly branching protrusion, likely to be the apical dendrite, and many shorter dendrites radiating from the soma, likely to be the basal dendrites, were selected for further analysis as it has been previously described (41). Any signs of poor neuronal health meant the exclusion of the cell from quantification. Rats and mice were used in accordance with Northwestern University Institutional Animal Care and Use Committee and national guidelines and regulations.
Mixed-Culture Assays.
Mixed-culture/coculture assays were performed as described previously (40). In brief, primary hippocampal neuron cultures at 10 DIV prepared from E18–19 rat brains were cocultured for 3 d with HEK293T cells expressing CNTNAP2 (CNTNAP2-HA), NGL-3, neurexin-1β, or EGFP in the presence of 10 μM 5-fluoro-2′-deoxyuridine (Sigma; F-0503) and 10 μM uridine (Sigma; U-3003) to suppress HEK293T cell proliferation, followed by immunostaining for EGFP, HA, synapsin I, or PSD-95 at 13 DIV.
Cntnap2 Knockout Mice.
The Cntnap2−/− mice were generated by Elior Peles (Weizmann Institute of Science). The colony was genotyped following the primers and conditions previously described (34), and it was maintained in their original strain, CD1. Mice cortical neurons were obtained from newborn mice (P0), either wild type or Cntnap2−/−, following the same method used for the rat neurons described above. The transfection of mouse neurons was identical to the rat cortical neuron protocol and conditions. For the comparison between wild-type and Cntnap2−/− neurons, the experiments were performed in parallel. The immunocytochemistry of wild type and Cntnap2−/− was done using the same antibody preparation and the same conditions were applied for acquisition of images.
Immunocytochemistry.
For the staining of endogenous proteins, medium density 24 DIV neurons were first washed in PBS and then fixed in 4% formaldehyde 4% (wt/vol) sucrose in PBS for 10 min. Fixed neurons were then permeabilized and blocked simultaneously [NGS 2% (vol/vol), Triton X-100 0.1% in PBS, 1 h at room temperature], followed by incubation of primary antibodies (overnight, 4 °C). The coverslips were washed three times with PBS and incubated with the correspondent fluorophore-secondary antibodies (25 °C, 1 h) (Alexa Fluor 488, Alexa Fluor 596; Life Technologies), as described previously (42). The coverslips were then mounted onto slides using ProLong Antifade reagent (Invitrogen) and stored at 4 °C until the acquisition of the images. All images were acquired in the linear range of fluorescence intensity.
To analyze the localization of GluA1, EGFP-transfected neurons were labeled as indicated above. The resulting immunofluorescence images were quantified using MetaMorph software (Universal Imagining) (42). Images were acquired using a Zeiss LSM 5 Pascal confocal microscope. The background corresponding to areas without cells was subtracted to generate a “background-subtracted” image. Dendritic spines were designated as “regions” using the EGFP-filled neuron image, and both area and the average intensity of each AMPA receptor cluster immunofluorescence were measured automatically. Cultures that were directly compared were stained simultaneously and imaged with the same acquisition parameters. Experiments were carried out blind to condition and on sister cultures. All statistical analysis was performed as described below.
Quantitative Analysis of Spine Morphology and Density in Immunofluorescence Staining.
Confocal images of double-stained neurons were obtained as described previously (41). Briefly, images were acquired with a Zeiss LSM 5 Pascal confocal microscope using a 63× oil immersion objective [Zeiss; numerical aperture (N.A.) = 1.4] as a z series (z sectioning: 0.37 µm). The acquisition parameters were kept the same for all conditions. Two dimensional maximum projection reconstructions of images were generated, and morphometric analysis (spine number, area, length, and breadth) was done using MetaMorph software. To examine the morphologies of dendritic spines, individual spines on dendrites were manually traced, and spine dimensions were measured by MetaMorph. Cultures that were directly compared were stained simultaneously and imaged with the same acquisition parameters. For each condition, 11–12 neurons from at least three separate experiments were used. For overall dendritic spine linear density measurements (represented as number of spines/10 μm of dendrite length), at least two dendrites (secondary or tertiary branches), totaling 100 μm, were analyzed per neuron. Experiments were done blind to conditions and on sister cultures.
SIM Imaging.
Neurons were fixed after 24 DIV, and immunocytochemistry was performed as described above. Three-channel images were taken using a Nikon SIM microscope with a 100× 1.4 N.A. objective (Cell Imaging Facility and Nikon Imaging Center, Northwestern University, Feinberg School of Medicine) that allows a resolution range of 110–130 nm. Images were analyzed with ImageJ with the McMaster Biophotonics Facility set of plugins.
Statistical Analysis.
For quantitative immunofluorescence experiments, Western blots, and dendrite length or number measurements, differences between condition means were identified by Student's unpaired t tests or analyses of variance performed in GraphPad. Error bars represent SEM.
Supplementary Material
Acknowledgments
We thank Drs. Teng-Leong Chew and Joshua Zachary Rappoport for help with SIM imaging. This work was supported by Grant MH097216 from the NIH National Institute of Mental Health (to P.P.), IBS-R002-D1 from Institute for Basic Science (to E.K.), German Research Foundation Research Fellowship SCHU2710/1-1 (to B.S.), and NIH Grant NS50220 (to E.P.). SIM imaging work was performed at the Northwestern University Center for Advanced Microscopy, which is generously supported by National Cancer Institute Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. SIM imaging was performed on a Nikon N-SIM system, purchased through the support of NIH Grant 1S10OD016342-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423205112/-/DCSupplemental.
References
- 1.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10(10):981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 3.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 4.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57(1):65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 5.Woolfrey KM, et al. Epac2 induces synapse remodeling and depression and its disease-associated forms alter spines. Nat Neurosci. 2009;12(10):1275–1284. doi: 10.1038/nn.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin DT, et al. Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat Neurosci. 2009;12(7):879–887. doi: 10.1038/nn.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeyifous O, et al. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci. 2009;12(8):1011–1019. doi: 10.1038/nn.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nurnberger JI, Jr, et al. Psychiatric Genomics Consortium Bipolar Group Identification of pathways for bipolar disorder: A meta-analysis. JAMA Psychiatry. 2014;71(6):657–664. doi: 10.1001/jamapsychiatry.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zweier C, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85(5):655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alarcón M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregor A, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med Genet. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi E, et al. A 12Mb deletion at 7q33-q35 associated with autism spectrum disorders and primary amenorrhea. Eur J Med Genet. 2008;51(6):631–638. doi: 10.1016/j.ejmg.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Ballarati L, et al. Cytogenetic, FISH and array-CGH characterization of a complex chromosomal rearrangement carried by a mentally and language impaired patient. Eur J Med Genet. 2009;52(4):218–223. doi: 10.1016/j.ejmg.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Burbach JP, van der Zwaag B. Contact in the genetics of autism and schizophrenia. Trends Neurosci. 2009;32(2):69–72. doi: 10.1016/j.tins.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Jackman C, Horn ND, Molleston JP, Sokol DK. Gene associated with seizures, autism, and hepatomegaly in an Amish girl. Pediatr Neurol. 2009;40(4):310–313. doi: 10.1016/j.pediatrneurol.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Li X, et al. Association analysis of CNTNAP2 polymorphisms with autism in the Chinese Han population. Psychiatr Genet. 2010;20(3):113–117. doi: 10.1097/YPG.0b013e32833a216f. [DOI] [PubMed] [Google Scholar]
- 21.Friedman JI, et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13(3):261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 22.Corvin AP. Neuronal cell adhesion genes: Key players in risk for schizophrenia, bipolar disorder and other neurodevelopmental brain disorders? Cell Adhes Migr. 2010;4(4):511–514. doi: 10.4161/cam.4.4.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KS, Liu XF, Aragam N. A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124(1-3):192–199. doi: 10.1016/j.schres.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 24.O’Dushlaine C, et al. International Schizophrenia Consortium Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16(3):286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 25.Levinson DF, et al. Schizophrenia Psychiatric GWAS Consortium Genome-wide association study of multiplex schizophrenia pedigrees. Am J Psychiatry. 2012;169(9):963–973. doi: 10.1176/appi.ajp.2012.11091423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elia J, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15(6):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sizoo B, et al. Do candidate genes discriminate patients with an autism spectrum disorder from those with attention deficit/hyperactivity disorder and is there an effect of lifetime substance use disorders? World J Biol Psychiatry. 2010;11(5):699–708. doi: 10.3109/15622975.2010.480985. [DOI] [PubMed] [Google Scholar]
- 28.Strauss KA, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354(13):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 29.Mefford HC, et al. Genome-wide copy number variation in epilepsy: Novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6(5):e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poot M, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010;11(1):81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- 31.Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Brain Res Rev. 2006;52(2):293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Peñagarikano O, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147(1):235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott-Van Zeeland AA, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2(56):56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poliak S, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162(6):1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson GR, et al. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci USA. 2012;109(44):18120–18125. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustafsson MG. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102(37):13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulopoulos A, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63(5):628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 38.Serrando M, Casanovas A, Esquerda JE. Occurrence of glutamate receptor subunit 1-containing aggresome-like structures during normal development of rat spinal cord interneurons. J Comp Neurol. 2002;442(1):23–34. doi: 10.1002/cne.1419. [DOI] [PubMed] [Google Scholar]
- 39.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 40.Woo J, et al. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci. 2009;12(4):428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava DP, Woolfrey KM, Penzes P. Analysis of dendritic spine morphology in cultured CNS neurons. J Vis Exp. 2011;(53):e2794. doi: 10.3791/2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava DP, et al. An autism-associated variant of Epac2 reveals a role for Ras/Epac2 signaling in controlling basal dendrite maintenance in mice. PLoS Biol. 2012;10(6):e1001350. doi: 10.1371/journal.pbio.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.