Significance
RNA-binding proteins use diverse mechanisms for generating specificity toward distinct RNA molecules. Different subfamilies of bacterial dihydrouridine synthases (Dus) modify specific uridines in tRNA, but the mechanism for selection of the target nucleotide is unknown. We solved crystal structures of the U16-specific Dus from Escherichia coli complexed with two different tRNAs. These structures reveal that the tRNA is bound in a completely different orientation from that observed in a U20-specific enzyme. The major reorientation of the substrate tRNA, driven by unique amino acid “binding signatures” and plasticity in the position of the C-terminal recognition domain, appears to be an evolutionary innovation to the known strategies that define specificity of enzymes toward tRNA.
Keywords: dihydrouridine synthase, tRNA modification, protein–RNA interaction, substrate specificity, X-ray crystallography
Abstract
The reduction of specific uridines to dihydrouridine is one of the most common modifications in tRNA. Increased levels of the dihydrouridine modification are associated with cancer. Dihydrouridine synthases (Dus) from different subfamilies selectively reduce distinct uridines, located at spatially unique positions of folded tRNA, into dihydrouridine. Because the catalytic center of all Dus enzymes is conserved, it is unclear how the same protein fold can be reprogrammed to ensure that nucleotides exposed at spatially distinct faces of tRNA can be accommodated in the same active site. We show that the Escherichia coli DusC is specific toward U16 of tRNA. Unexpectedly, crystal structures of DusC complexes with tRNAPhe and tRNATrp show that Dus subfamilies that selectively modify U16 or U20 in tRNA adopt identical folds but bind their respective tRNA substrates in an almost reverse orientation that differs by a 160° rotation. The tRNA docking orientation appears to be guided by subfamily-specific clusters of amino acids (“binding signatures”) together with differences in the shape of the positively charged tRNA-binding surfaces. tRNA orientations are further constrained by positional differences between the C-terminal “recognition” domains. The exquisite substrate specificity of Dus enzymes is therefore controlled by a relatively simple mechanism involving major reorientation of the whole tRNA molecule. Such reprogramming of the enzymatic specificity appears to be a unique evolutionary solution for altering tRNA recognition by the same protein fold.
During the posttranscriptional maturation of tRNA, about 10% of its nucleosides are enzymatically modified at specific positions (1). Altered levels of tRNA modification have been linked to several disorders including cancers (2–8). One of the most common modified nucleosides, dihydrouridine, is produced by reduction of the C5–C6 double bond in uridine. The resulting nonplanar base cannot form stabilizing stacking interactions with neighboring nucleotides and favors the C2′-endo ribose conformation (9). In Escherichia coli, dihydrouridine is commonly found at positions 16, 17, 20, and 20a of the D loop of tRNA (Fig. S1A). The formation of dihydrouridine is catalyzed by dihydrouridine synthases (Dus) (10–12). Different Dus subfamilies display specificity toward distinct subsets of target uridines in tRNA. Whereas the specificity of the four Saccharomyces cerevisiae Dus enzymes has been established (13), little is known about the three E. coli Dus proteins (DusA, DusB, and DusC), except that the specificities are nonoverlapping and that DusA modifies U20 (10). The mechanistic basis of the exquisite substrate specificity of Dus is an intriguing problem because the target uridines are exposed at spatially distinct faces of folded tRNAs, and yet all Dus subfamilies are predicted to adopt the same fold with highly conserved active-site residues (14–16).
A structure of the DusA-like U20-specific Thermus thermophilus Dus (TtDus) in complex with tRNAPhe showed that tRNA is bound with the surface containing U20 facing the protein and U20 covalently bound in the active site (16). Although the crystal structure of E. coli DusC is also available (17), how it binds RNA remained unknown. We show that in solution DusC is specific toward U16, which is located on the opposite side of the D loop to U20 (Fig. S1B). To understand how specificity toward different uridines in tRNA is generated using the same fold, we determined the crystal structures of DusC in complex with two substrate tRNAs: tRNAPhe and tRNATrp. Unexpectedly, we found that Dus enzymes that modify uridines at positions 16 and 20 bind their tRNA substrates in completely different orientations. The binding modes of the two Dus subfamilies differ by a major (∼160°) rotation of the whole tRNA molecule, providing the basis for their distinct specificities.
Results
Structural Basis for U16 Specificity.
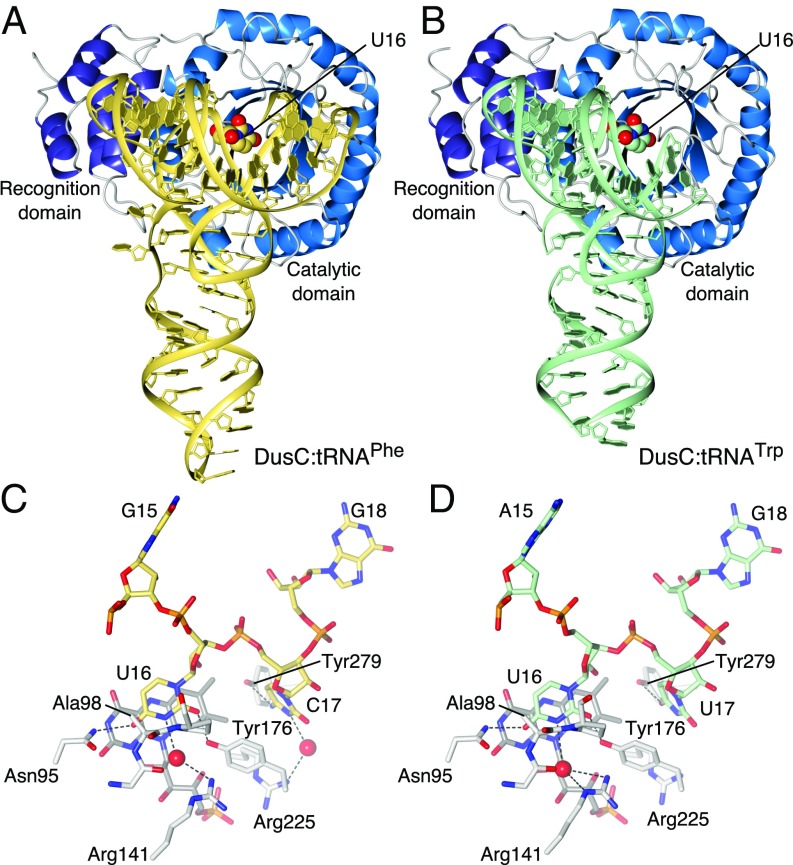
DusC is a bipartite molecule with an N-terminal triosephosphate isomerase barrel catalytic domain (residues 1–242) and a C-terminal α-helical “recognition” domain (residues 245–309) (Fig. S2A). Substitution of Cys98 by Ala in the active site allowed formation of stable complexes of DusCC98A with tRNAPhe or tRNATrp, and structures of these complexes were determined at 2.1 Å and 2.55 Å resolution, respectively. In both complexes (Fig. 1 A and B), the catalytic domain interacts with the D stem loop of the tRNA (Fig. 1 C and D), whereas the C-terminal recognition domain interacts with the D and TΨC stem loops. Recognition of tRNA involves direct hydrogen bonds to bases of specific nucleotides: U16, which is inserted into the active site, the adjacent nucleotide (C17 in tRNAPhe, U17 in tRNATrp), and C56 (Fig. 1 C and D). In the two complexes, virtually identical networks of direct and water-mediated hydrogen-bonding interactions form between the enzyme and sugar-phosphate backbone of tRNA (Fig. S3). The interactions occur mainly between both domains of the protein and the D loop of the tRNA, but several hydrogen-bonding contacts also exist between the recognition domain and the TΨC loop. The only base-specific interaction with the TΨC loop is between the side chain of Lys274 and the O2 carbonyl group of the universally conserved C56.
Fig. 1.
Structures of DusCC98A complexes with tRNAPhe and tRNATrp. (A) tRNAPhe complex. (B) tRNATrp complex. (C) Recognition of U16 and C17 in the tRNAPhe complex. (D) Recognition of U16 and U17 in the tRNATrp complex.
Upon binding to tRNA, DusC undergoes only small rearrangements: The backbone rmsds (residues 5–300) with tRNA-free DusC are 0.53 Å and 0.42 Å for the complex with tRNAPhe and tRNATrp, respectively. The most prominent conformational changes occur within the active-site loop (residues 97–108) that projects toward the D stem of tRNA (Fig. S2 A and B). In contrast to most tRNA–enzyme complexes (18–20), the overall fold of tRNA does not change upon binding, except for minor adjustments in the D loop that facilitate insertion of the U16 base into the active site of DusC.
The Watson–Crick edge of U16 is bound through hydrogen bonds between its O2 and O4 carbonyl groups and the side chains of Tyr176 (a residue that is conserved in the DusC subfamily only) and Asn95, respectively (Fig. 1 C and D). An additional water-mediated hydrogen bond between N3 and the highly conserved Arg141 also contributes to U16 recognition (Fig. 1 C and D). Residues equivalent to Asn95 and Arg141 are essential for the activity of the U20-specific DusA and TtDus (16, 21). Notably, the water molecules that mediate the hydrogen bonds with Arg141 in both complexes (Fig. S4C) occupy a region of electron density that was previously ascribed to an unknown cofactor in TtDus:tRNAPhe and in E. coli DusC structures (16, 17). We show, however, that the presence of such a cofactor is inconsistent with the electron density observed for both DusC and for DusC:tRNA complexes (Fig. S4). E. coli tRNAs containing dihydrouridine at position 16 typically contain a cytidine or uridine at position 17 (Fig. S1A) (1). Accommodation of either C17 or U17 in the same binding pocket is possible due to an interaction with the side chain hydroxyl of Tyr279, which forms a hydrogen bond with N4 of C17 in tRNAPhe or with O4 of U17 in tRNATrp (Fig. 1 C and D).
The crystal structures of DusC–tRNA complexes show that U16 is inserted into the active site. To validate that DusC specifically modifies U16, we used an assay that detects dihydrouridine position based on reverse transcriptase primer extension termination after alkaline hydrolysis (13) (Fig. S5 A and B). Furthermore, analysis of DusC and DusCC98A binding to several tRNA transcripts by EMSA and analytical size-exclusion chromatography (SEC) (Fig. S5 C–G) indicated that uridine in position 16 is essential not only for modification but also for recognition of tRNA by DusC.
Structural Comparison of U16- and U20-Specific Dus.
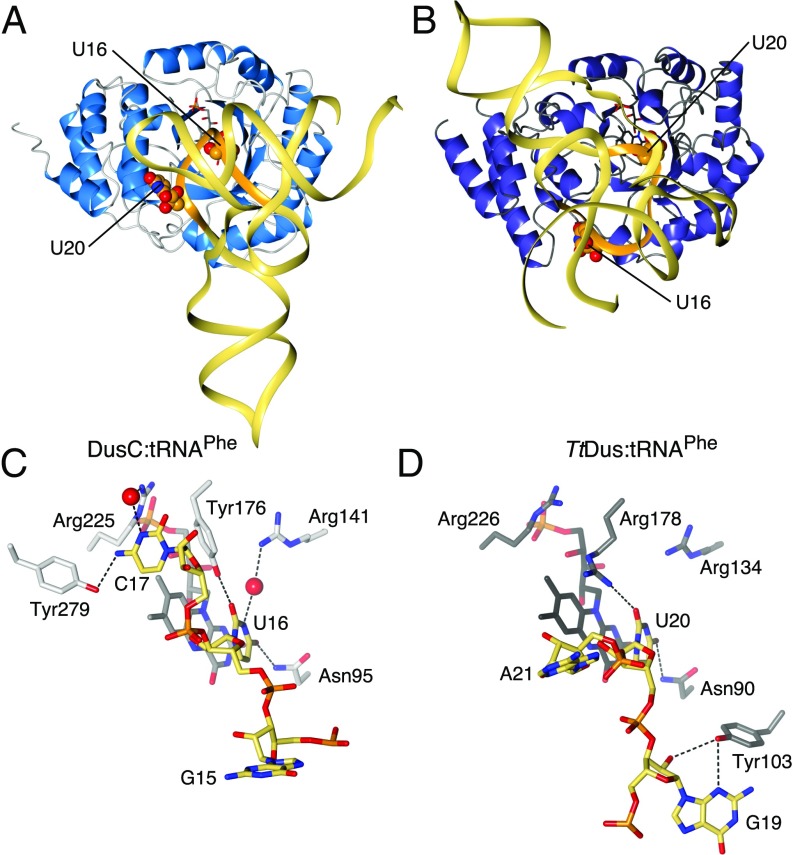
The U16-specific DusC adopts the same overall fold as the U20-specific TtDus (16) (Fig. 2 A and B), with the catalytic and recognition domains superposing with main chain rmsds of 2.0 Å (213 residues) and 2.4 Å (42 residues), respectively. The basis for the targeting of different nucleotides by the two Dus becomes immediately apparent when the structures of DusC:tRNAPhe and DusC:tRNATrp complexes are compared with that of TtDus:tRNAPhe (Figs. 1 A and B and 2 A and B). The tRNA substrate is rotated as a rigid body by ∼160°, such that U16 is placed in the active site in DusC, whereas the active site in TtDus accommodates U20 (Fig. 2 C and D). The nucleosides 5′ and 3′ of the target uridine are, therefore, in substantially different positions. In DusC, C17 of tRNAPhe is specifically recognized by Tyr279 and Arg225 (Fig. 2C). In contrast, in TtDus G19 (the nucleoside 5′ of U20) is specifically recognized by Tyr103 (Fig. 2D). The counterparts of DusC Tyr279 and TtDus Tyr103 (TtDus Asn281 and DusC Gly108, respectively) do not interact directly with the tRNA substrate. Therefore, these residues constitute subfamily-specific selection determinants.
Fig. 2.
Comparison of tRNAPhe binding by U16- and U20-specific Dus. (A) Structure of DusC:tRNAPhe complex. (B) Structure of TtDus:tRNAPhe shown with the protein in the same orientation as in A. (C) Recognition of U16 and C17 in the DusC:tRNAPhe complex. (D) Recognition of U20 and G19 in the TtDus:tRNAPhe complex. D loop is highlighted in orange in A and B. Protein orientation is the same as in A–D.
“Binding Signatures” Define tRNA Orientation.
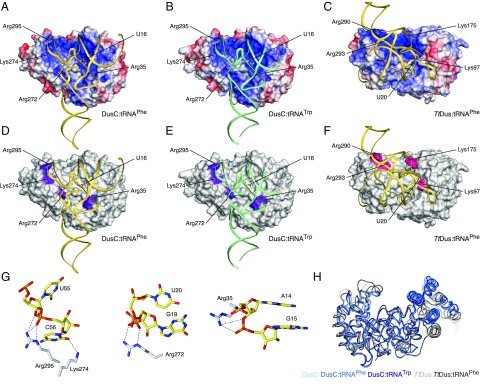
Analysis of the superposition of the DusC and TtDus enzymes reveals differences in the distribution of charges over the tRNA-binding surfaces of the two enzymes. In the DusC complex structures, tRNA binds in an L-shaped positively charged groove that accommodates the D and TΨC loops (Fig. 3 A and B). By contrast, in the TtDus complex, the D stem loop lies in a cylindrical groove formed between the recognition and catalytic domains of the protein, with the TΨC loop not held in a clearly defined binding site (Fig. 3C). Comparison of the sequences of diverse members of the U16-specific DusC subfamily (Fig. S6) and the U20-specific DusA subfamily (Fig. S7) suggests subfamily-specific binding signatures (Fig. S8) define the orientation of tRNA through direct hydrogen bonding and ionic interactions (Fig. 3 D–F). In U16-specific enzymes, the binding signatures comprise residues Lys274/Arg295, Arg272, and Arg35 (Fig. 3 D and E), whereas in U20-specific enzymes the binding signatures are Arg290/Arg293, Lys175, and Lys97 (Fig. 3F). Intriguingly, Lys175 (DusA) is present also in DusB and DusC subfamilies. However, its location within the 3D structure is unique due to an eight-amino-acid insertion in sequences of the DusA subfamily (Fig. S8), bringing this residue into close contact with tRNA (Fig. 3F). Lys274 and Arg295 of the DusC binding signatures are located in the recognition domain and interact with the TΨC loop of tRNA; Arg272 makes a salt bridge with the phosphodiester connecting G19 and U20; and Arg35 binds A14 and G15 at the 5′ of the D loop (Fig. 3G).
Fig. 3.
Determinants of Dus modification specificity. (A) DusC:tRNAPhe. (B) DusC:tRNATrp. (C) TtDus:tRNAPhe. DusC and TtDus orientations are the same as in Fig. 2. tRNA is shown as a ribbon. Protein electrostatic potentials are contoured at ±5 kTe−1 (blue, positive; red, negative; white, neutral); binding signature positions and target uridine are labeled. (D–F) Same as A–C but with protein surfaces in white and binding signatures in color (U16-specific in purple and U20-specific in pink). (G) Interactions of DusC binding signature residues with tRNAPhe. (H) Difference in the relative position of the recognition domain between DusC and TtDus when proteins are superimposed using only catalytic domains (catalytic domain on the Left; recognition domain on the Right). tRNA-free and tRNA-bound DusCs are in blue. tRNA-free and tRNA-bound TtDus are in gray.
In addition to the binding signatures, the relative position of the catalytic and recognition domains differs in the U16- and U20-specific Dus subfamilies. The cylindrical groove between the two domains is narrower in DusC than in TtDus (Fig. 3H), thereby obstructing the binding of the substrate tRNA in the U20-specific orientation and thus contributing to the specificity of DusC toward U16.
When electrostatic surfaces are calculated for free DusC and TtDus, the positive isosurfaces have shapes that are complementary to the face of the tRNA bound in the respective complex (Fig. S9). This complementarity may contribute to defining the dramatically different orientations through long-range electrostatic steering as the negatively charged tRNA approaches the enzymes. The binding signatures then fine-tune docking of the substrate tRNA to insert the target uridine into the active site.
Discussion
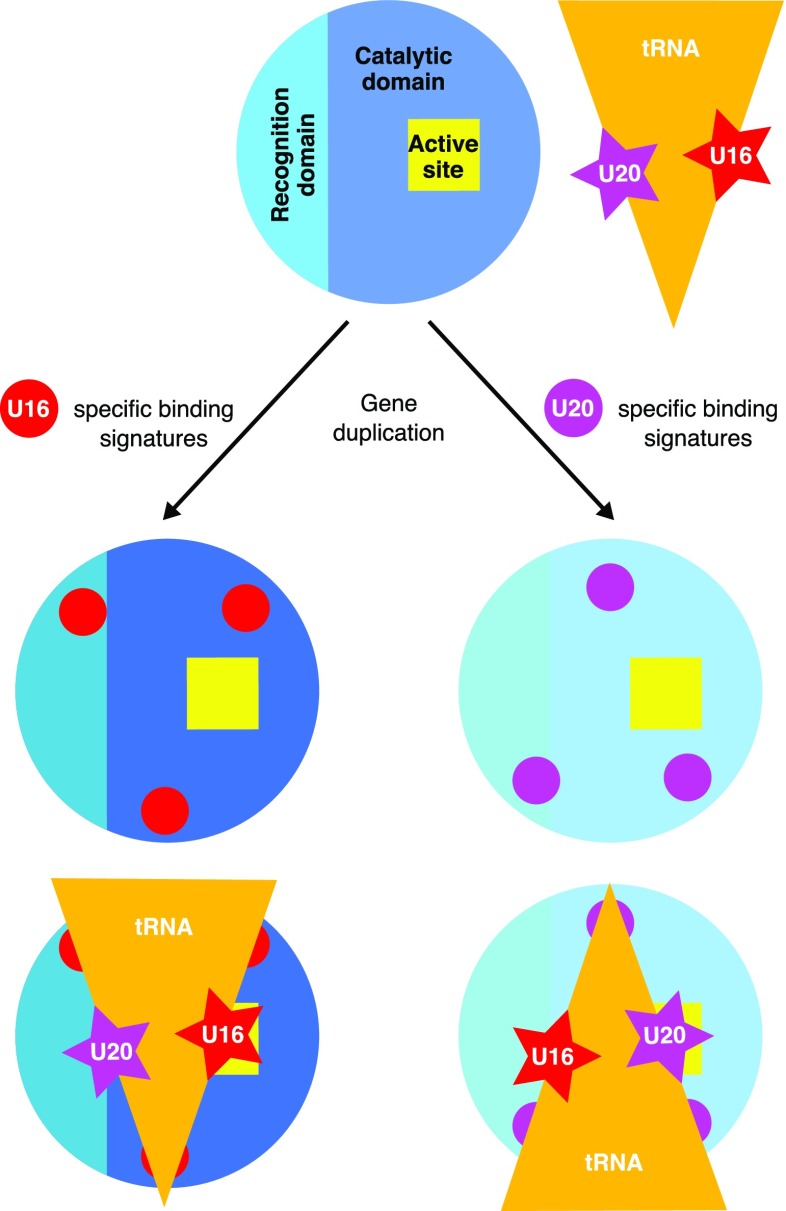
Dus that modify uridines at different positions emerged via gene duplication (14). Our findings indicate that the specificity of Dus has evolved by tuning of three putative binding signatures containing charged residues that guide the docking of the tRNA in a specific orientation (Fig. 4). The selection of binding orientation is further defined by a slight repositioning of the C-terminal recognition domain in the U16- or U20-specific families.
Fig. 4.
The role of binding signatures in generating U16 and U20 specificity.
The specificity and structure of DusB, the third Dus enzyme found in E. coli, are unknown. Dihydrouridine modification, in addition to U16 and U20 positions, occurs also at U17 and U20a of E. coli tRNAs (1). DusB might be specific toward at least one of these two uridines. Although half of the binding signature residues that are conserved in either DusA or DusC are also present in the DusB subfamily, several additional residues are uniquely conserved in the DusB sequences (Fig. S8). Thus, DusB might bind tRNA in yet another orientation using a unique set of binding signatures.
DusC is represented only in a limited range of Proteobacteria and is thought to have evolved as a result of a duplication that occurred at the branch leading to beta- and gamma-Proteobacteria (14). Thus, the emergence of the binding signatures that cause a major reorientation of tRNA and hence modification of U16 appears to be an evolutionary innovation. In contrast, in other tRNA modification enzymes, such as pseudouridine synthases (18, 22, 23), guanine transglycosylases (24, 25), and S-adenosyl-L-methionine–dependent methyltransferases (26–28), specificity depends either on the presence of additional domains or on change in the oligomerisation state of the protein.
In summary, structural data on U16-specific Dus (this report) and comparison with the U20-specific Dus (16) reveal an elegant mechanism to endow closely related tRNA-modifying enzymes with distinctly different positional specificities (Fig. 4). In these two Dus subfamilies, specific clusters of amino acid binding signatures appear to guide docking of the substrate tRNA in completely different orientations, and orientation of the tRNA is further constrained by slight positional differences between the C-terminal recognition domains. This unexpectedly simple mechanism underlying the specificity of Dus appears to be a previously unidentified addition to the recognition strategies used by tRNA-modifying enzymes.
Methods
Expression and purification of wild-type and Cys98Ala DusC and crystallization of native and SeMet DusC and DusCC98A–tRNAPhe and DusCC98A–tRNATrp complex procedures are described in SI Methods. The structure of DusC was solved by single-wavelength anomalous diffraction (Table 1) using SHELX (29) and Phaser (30), followed by density modification with Parrot (31) and autobuilding with Buccaneer (32). The structures of the DusCC98A–tRNAPhe and DusCC98A–tRNATrp complexes (Table 1) were determined by molecular replacement with Phaser (30). In all cases, the models were further improved through alternate cycles of manual rebuilding with Coot (33) and refinement with Refmac5 (34). tRNA geometry was improved using RCrane (35) and ERRASER (36). Detailed descriptions of the in vitro tRNAPhe dihydrouridylation and dihydrouridine detection, electrophoretic mobility shift assays, and analytical SEC are provided in SI Methods.
Table 1.
Data collection and refinement statistics
| Native DusC | SeMet DusC | DusC:tRNAPhe | DusC:tRNATrp | |
| Data collection | ||||
| Space group | P212121 | P43212 | C2221 | P6522 |
| Cell dimensions | ||||
| a, b, c, Å | 68.5, 99.5, 119.9 | 94.1, 94.1, 119.7 | 100.6, 176.9, 238.4 | 98.5, 98.5, 231.2 |
| Resolution, Å* | 60.0–1.65 (1.74–1.65) | 59.8–2.60 (2.74–2.60) | 49.2–2.10 (2.14–2.10) | 49.3–2.55 (2.66–2.55) |
| Rmerge | 0.105 (0.708) | 0.117 (0.843) | 0.061 (0.999) | 0.141 (1.099) |
| I/σI | 11.0 (2.6) | 12.2 (2.2) | 15.7 (1.7) | 8.8 (1.5) |
| Completeness,% | 100.0 (100.0) | 99.9 (100.0) | 99.9 (100.0) | 99.6 (99.9) |
| Redundancy | 7.3 (6.6) | 7.7 (7.9) | 4.5 (4.7) | 5.3 (5.5) |
| Refinement | ||||
| Resolution, Å | 51.1–1.65 | 58.2–2.60 | 49.3–2.10 | 49.3–2.55 |
| No. reflections | 99,520 | 17,086 | 123,501 | 22,329 |
| Rwork/Rfree | 13.4/17.7 | 17.7/21.4 | 19.2/22.3 | 20.8/23.5 |
| No. atoms | ||||
| Protein | 4,970 | 2,495 | 7,397 | 2,398 |
| RNA | — | — | 4,533 | 1,287 |
| Ligand/ion | 86 | 31 | 121 | 42 |
| Water | 611 | 104 | 614 | 52 |
| B factors | ||||
| Protein | 26.1 | 53.8 | 40.1 | 47.8 |
| RNA | — | — | 65.1 | 61.3 |
| Ligand/ion | 17.4 | 40.3 | 48.1 | 46.3 |
| Water | 41.8 | 41.1 | 44.6 | 38.7 |
| rmsd | ||||
| Bond lengths, Å | 0.009 | 0.011 | 0.008 | 0.006 |
| Bond angles, ° | 1.2 | 1.4 | 1.3 | 1.0 |
Values in parentheses are for the highest resolution shell.
Supplementary Material
Acknowledgments
The authors thank Ralf Flaig (Diamond Light Source), Gideon Grogan, Sam Hart, Johan Turkenburg, Celina Whalley (University of York), Robert Nicholls (Medical Research Council Laboratory of Molecular Biology), and David Waterman (Science and Technology Facilities Council) for assistance and useful discussions. We thank Diamond Light Source for access to beamlines I02, I04, and I24 (under Proposals MX1221, MX7864, and MX9948), which contributed to the results presented here. This work was funded by Wellcome Trust Fellowship 098230 (to A.A.A.), a Biotechnology and Biological Sciences Research Council PhD studentship (to R.T.B.), and Russian Science Foundation Grant 14-34-00023 (to A.L.K.). E.V.K. is supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine). M.V.R. is supported by funding from the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4bfa, 4bf9, 4yco, and 4ycp).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500161112/-/DCSupplemental.
References
- 1.Jühling F, et al. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, Tuck S, Byström AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5(7):e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakovich T, et al. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J Biol Chem. 2011;286(22):19354–19363. doi: 10.1074/jbc.M111.219576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9(8):e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato T, et al. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005;65(13):5638–5646. doi: 10.1158/0008-5472.CAN-05-0600. [DOI] [PubMed] [Google Scholar]
- 6.Kuchino Y, Borek E. Tumour-specific phenylalanine tRNA contains two supernumerary methylated bases. Nature. 1978;271(5641):126–129. doi: 10.1038/271126a0. [DOI] [PubMed] [Google Scholar]
- 7.Begley U, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol Med. 2013;5(3):366–383. doi: 10.1002/emmm.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spinola M, et al. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene. 2005;24(35):5502–5509. doi: 10.1038/sj.onc.1208687. [DOI] [PubMed] [Google Scholar]
- 9.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. Conformational flexibility in RNA: The role of dihydrouridine. Nucleic Acids Res. 1996;24(6):1073–1079. doi: 10.1093/nar/24.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AC, Xu J, Johnson RC, Schimmel P, de Crécy-Lagard V. Identification of the tRNA-dihydrouridine synthase family. J Biol Chem. 2002;277(28):25090–25095. doi: 10.1074/jbc.M203208200. [DOI] [PubMed] [Google Scholar]
- 11.Xing F, Martzen MR, Phizicky EM. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA. 2002;8(3):370–381. doi: 10.1017/s1355838202029825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rider LW, Ottosen MB, Gattis SG, Palfey BA. Mechanism of dihydrouridine synthase 2 from yeast and the importance of modifications for efficient tRNA reduction. J Biol Chem. 2009;284(16):10324–10333. doi: 10.1074/jbc.M806137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing F, Hiley SL, Hughes TR, Phizicky EM. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J Biol Chem. 2004;279(17):17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]
- 14.Kasprzak JM, Czerwoniec A, Bujnicki JM. Molecular evolution of dihydrouridine synthases. BMC Bioinformatics. 2012;13:153. doi: 10.1186/1471-2105-13-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park F, et al. The 1.59 A resolution crystal structure of TM0096, a flavin mononucleotide binding protein from Thermotoga maritima. Proteins. 2004;55(3):772–774. doi: 10.1002/prot.20086. [DOI] [PubMed] [Google Scholar]
- 16.Yu F, et al. Molecular basis of dihydrouridine formation on tRNA. Proc Natl Acad Sci USA. 2011;108(49):19593–19598. doi: 10.1073/pnas.1112352108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, et al. Structure of dihydrouridine synthase C (DusC) from Escherichia coli. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 8):834–838. doi: 10.1107/S1744309113019489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoang C, Ferré-D’Amaré AR. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107(7):929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 19.Ishitani R, et al. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113(3):383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 20.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Natl Acad Sci USA. 2003;100(22):12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savage DF, de Crécy-Lagard V, Bishop AC. Molecular determinants of dihydrouridine synthase activity. FEBS Lett. 2006;580(22):5198–5202. doi: 10.1016/j.febslet.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 22.Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell. 2007;26(2):189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol. 2007;373(5):1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 24.Ishitani R, et al. Crystal structure of archaeosine tRNA-guanine transglycosylase. J Mol Biol. 2002;318(3):665–677. doi: 10.1016/S0022-2836(02)00090-6. [DOI] [PubMed] [Google Scholar]
- 25.Romier C, Reuter K, Suck D, Ficner R. Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. EMBO J. 1996;15(11):2850–2857. [PMC free article] [PubMed] [Google Scholar]
- 26.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci USA. 2008;105(19):6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto-Ito S, et al. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins. 2008;72(4):1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 28.Walbott H, Leulliot N, Grosjean H, Golinelli-Pimpaneau B. The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic Acids Res. 2008;36(15):4929–4940. doi: 10.1093/nar/gkn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 30.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Keating KS, Pyle AM. RCrane: Semi-automated RNA model building. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 8):985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou FC, Sripakdeevong P, Dibrov SM, Hermann T, Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat Methods. 2013;10(1):74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.