Significance
Adhesion G protein-coupled receptors (GPCRs) regulate tissue development and cancer progression. Intact adhesion GPCRs are unique two-protomer receptors that undergo biosynthetic self-proteolyzation at a conserved site between the extracellular domain and the seven transmembrane (7TM) domain. We demonstrate the activation mechanism of adhesion GPCRs. The 7TM domain N-terminal stalk lies encrypted within the noncovalently bound extracellular domain. When the extracellular domain is dissociated, perhaps naturally by ligand action, the 7TM stalk is revealed. We demonstrate that the stalk acts as a tethered agonist of the 7TM to facilitate direct activation of heterotrimeric G proteins. Synthetic peptides comprising adhesion GPCR 7TM domain stalks are sufficient receptor agonists, raising the possibility for the development of adhesion GPCR synthetic peptide modulators.
Keywords: GPR56, GPR110, adhesion GPCRs, G proteins, tethered agonist
Abstract
The large class of adhesion G protein-coupled receptors (aGPCRs) bind extracellular matrix or neighboring cell-surface ligands to regulate organ and tissue development through an unknown activation mechanism. We examined aGPCR activation using two prototypical aGPCRs, GPR56 and GPR110. Active dissociation of the noncovalently bound GPR56 or GPR110 extracellular domains (ECDs) from the respective seven-transmembrane (7TM) domains relieved an inhibitory influence and permitted both receptors to activate defined G protein subtypes. After ECD displacement, the newly revealed short N-terminal stalk regions of the 7TM domains were found to be essential for G protein activation. Synthetic peptides comprising these stalks potently activated GPR56 or GPR110 in vitro or in cells, demonstrating that the stalks comprise a tethered agonist that was encrypted within the ECD. Establishment of an aGPCR activation mechanism provides a rational platform for the development of aGPCR synthetic modulators that could find clinical utility toward aGPCR-directed disease.
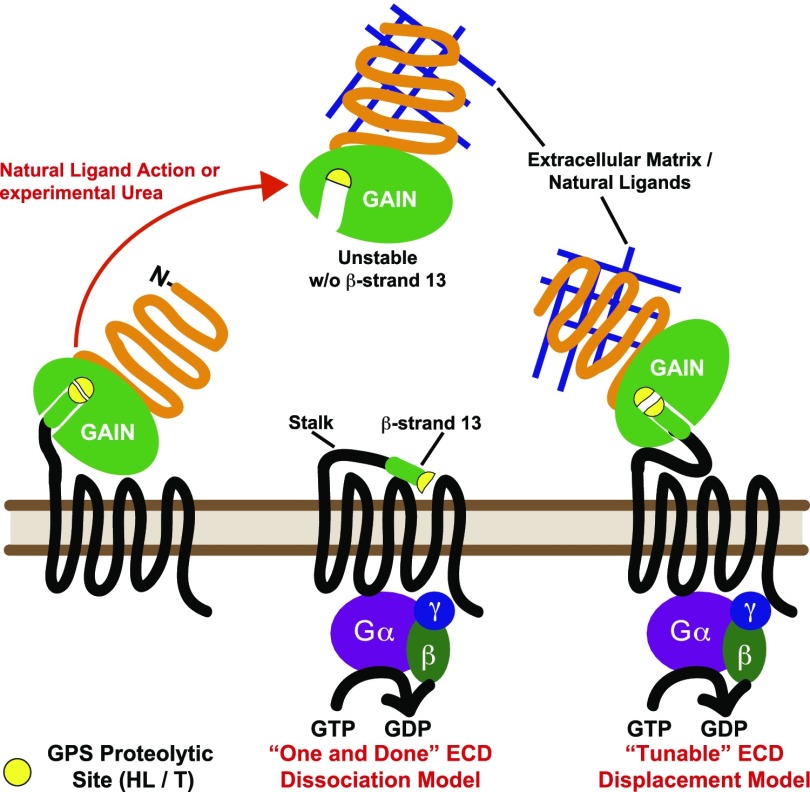
Adhesion G protein-coupled receptors (aGPCRs) are proposed to regulate tissue specification and development during embryo- and organogenesis by transmitting information received from adjacent cell-surface proteins or extracellular matrix (ECM) components to heterotrimeric G protein-directed signaling systems (1–3). aGPCRs are misregulated in developmental disorders and many cancers, and some aGPCRs are considered bona fide oncogenes (4–7). This 33-member subclass of family B GPCRs is distinguished by large extracellular N termini that harbor various adhesion modules and a ∼320-aa GPCR autoproteolysis-inducing (GAIN) domain located proximal to the seven-transmembrane spanning domain (7TM) (8, 9). GAIN-mediated receptor self-cleavage occurs constitutively during biosynthesis to produce receptors that have two protomers consisting of the extracellular domains (ECD) or N-terminal fragments (NTF) noncovalently bound to the 7TM or C-terminal fragments (CTF). The cleaved receptors traffic as one unit to the plasma membrane where the two protomers remain associated in a state poised for ligand engagement and receptor activation (Fig. 1).
Fig. 1.
Models of adhesion GPCR activation. Adhesion GPCRs are constitutively self-cleaved at the GPCR proteolytic site (GPS) within the GAIN domain. Natural ligand- or experimental urea-mediated displacement of aGPCR ECDs relieves an inhibition of the 7TM domain. The highly conserved GAIN domain β-strand-13 and stalk region could act as a tethered agonist to the 7TM domain after ECD dissociation (One and Done model) or upon displacement (Tunable model).
Most aGPCRs are orphans with little known about the identity of the downstream G protein signaling pathways they regulate. Specific aGPCR ligands include collagen subtypes and other ECM proteins that bind the adhesion motifs within the ECDs (10–13). The means of ligand action resulting in aGPCR activation is unknown. Transfection of aGPCRs with deleted N-terminal ECDs (expressed 7TM domains only) enhanced cell-based signaling outputs (11, 12). Therefore, aGPCR ECDs are suspected to impart an inhibitory influence upon the 7TM domains, and ligands are proposed to bind the ECD and alter its orientation with respect to the 7TM domain to relieve this inhibition.
We postulated that constitutive GAIN-mediated receptor self-cleavage was critical for aGPCR ligand regulation and undertook biochemical and cell-based approaches to decipher the mechanism of aGPCR activation. Human GPR56 and GPR110 aGPCRs were expressed in insect cells, and prepared receptor membranes were reconstituted with purified G protein heterotrimers of defined composition before measurement of receptor-stimulated G protein activation. The G protein coupling specificity of both receptors was determined. Dissociation of the peripherally membrane-bound ECDs from full-length GPR56 or GPR110 membranes markedly potentiated G protein activation, demonstrating that operative relief of ECD inhibition serves to activate both aGPCRs. Expressed N-terminal ECD receptor deletions constitutively activated G proteins. Progressive truncation of single amino acids from the 7TM domain N termini gradually reduced the constitutive activity to zero, showing that the aGPCR “stalk” region N-terminal to the first transmembrane domain is required for G protein activation. Synthetic peptides comprising specific portions of the GPR56 or GPR110 stalks acted as potent receptor agonists in the membrane reconstitution system and in a cell-based gene reporter assay. Given the critical role of GAIN domain receptor self-cleavage, and the high conservation of aGPCR 7TM domain stalk regions, we predict that the adhesion GPCR class is activated by stalk region tethered agonists that are revealed upon ligand-mediated ECD dissociation or displacement (Fig. 1).
Results
Biochemical Reconstitution of Adhesion GPCR G Protein Activation.
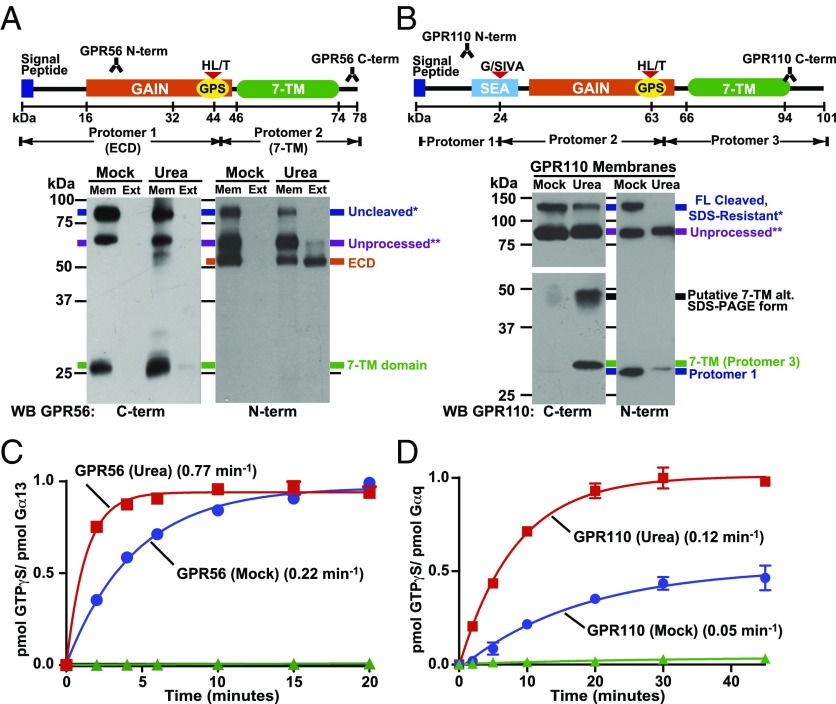
Human GPR56 and the orphan aGPCR, GPR110 were expressed in High-Five or Spodoptera frugiperda-9 (Sf9) insect cells. Prepared receptor membranes were treated with 7M urea to mimic the putative action of natural ligands: ligand-mediated ECD displacement in relation to the 7TM domain (Fig. 1). Urea treatment resulted in efficient extraction/dissociation of the glycosylated GPR56 N-terminal protomer (ECD), whereas the 7TM domain remained protected in the membrane (Fig. 2A and Fig. S1). GPR110 is an uncharacteristic three-protomer aGPCR that contains a second ECD self-cleavage site in addition to its GAIN domain, called a sperm protein/enterokinase/agrin (SEA) domain (Fig. 2B) (5). Mock-extracted GPR110 membranes had populations of self-cleaved, SDS-resistant full-length receptor (∼130 kDa) and SDS-dissociated protomer 1 (∼30 kDa). Much of the full-length SDS-resistant population was dissociated further when the GPR110 membranes were treated with urea, permitting visualization of the 7TM domain (protomer 3, ∼27 kDa) in the membrane fraction and loss of ECD protomer 1 from the membranes (Fig. 2B).
Fig. 2.
GPR56 and GPR110 ECD dissociation activates 7TM-mediated G protein activation. (A and B) Prepared insect cell membranes with expressed, full-length GPR56 or GPR110 were mock treated or extracted with urea. The presence of the GPR56 ECD and 7TM domain in the membrane (Mem) and extract (Ext) fractions was determined by Western blotting with the GPR56 N- and C-terminal antibodies. The relative levels of GPR110 ECD protomer 1 and the 7TM domain in mock and urea-extracted membranes were determined by Western blotting with the GPR110 N- and C-terminal antibodies. The GPR110 C-terminal low-molecular weight (MW) panel is a longer exposure than the high-MW panel. (C) GPR56 or (D) GPR110 urea-treated (■, red) and untreated (●, blue) membranes, or nonreceptor membranes (▲, green) were reconstituted with purified Gα13 or Gαq and Gβ1γ2, and receptor-stimulated [35S]-GTPγS binding kinetics were measured. Error bars, SEM.
Purified Gα and Gβ1γ2 subunits (Fig. S2) were used to reconstitute the GPR56 and GPR110 membranes with defined subtypes of G protein heterotrimers. Direct receptor-mediated G protein activation was measured by determining the kinetics of G protein [35S]-GTPγS binding. GPR56 activated G13 robustly (Fig. 2C), exhibited modest Gi coupling, and did not activate Gq or Gs (Fig. S3). GPR110 was defined as an exclusive Gq coupler (Fig. 2D and Fig. S3). The G protein activation kinetics mediated by both receptors were enhanced greatly when the receptor membranes were pretreated with urea to dissociate the ECDs (Fig. 2 C and D); this demonstrates that aGPCR ECD dissociation relieves an inhibitory influence that results in 7TM domain activation.
Addition of purified, recombinant GPR56 ECD (5 µM) to urea-activated GPR56 membranes had no effect to inhibit G13 activation (Fig. S4), showing that a dissociated aGPCR ECD is incapable of reinhibiting the 7TM protomer, and suggesting that an intrinsic property of the 7TM protomer renders it constitutively active.
Adhesion GPCR 7TM Domain N Termini Are Required for Receptor Activity.
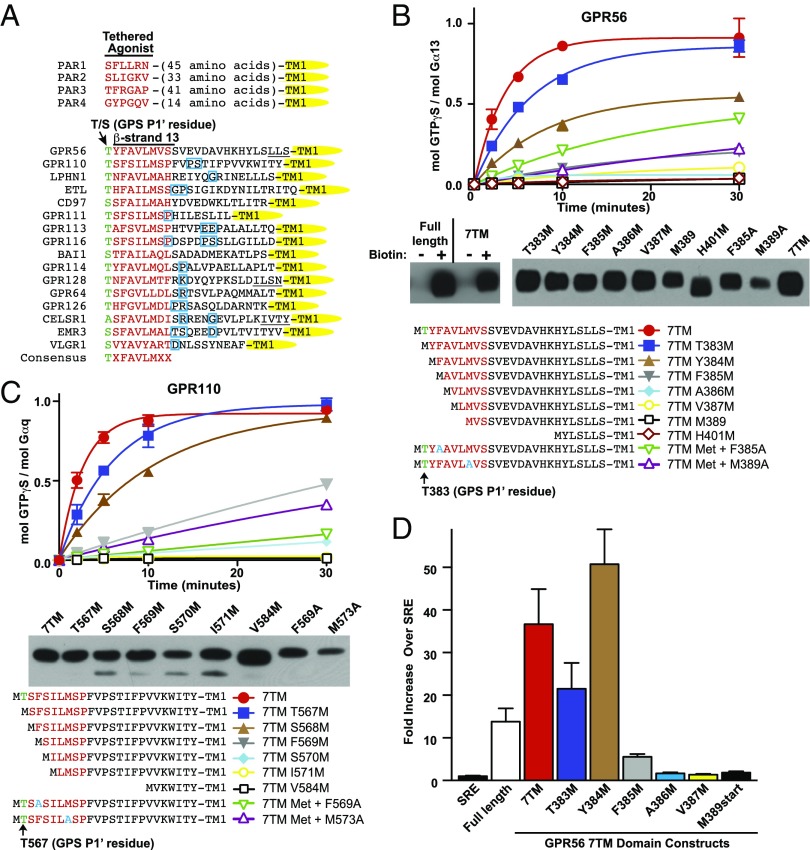
aGPCR GAIN domains are comprised predominantly of tightly packed β-strands and are sufficient to mediate autoproteolysis at the consensus site, HL/T (Fig. 1) (8, 14). β-strand-13 is buried deeply within the GAIN hydrophobic core and is an essential contributor to overall domain structural integrity despite being present C-terminal to the GAIN domain self-cleavage site. Therefore, β-strand-13 is the N terminus of the 7TM domain and would only be revealed upon ECD dissociation (Fig. 1). The β-strand-13/stalk regions of representative aGPCRs were aligned and compared with the stalk regions of protease-activated receptors (PARs; Fig. 2A). PARs have ∼6-aa cryptic tethered agonists revealed by the action of extracellular proteases including thrombin (15). The only apparent sequence conservation between PAR and aGPCR 7TM domain stalks is the N-terminal residue, Ser/Thr; however, overall stalk lengths with respect to TM1 are generally similar, with PARs being slightly longer. Among aGPCR stalks, the N-terminal 9 residues that comprise β-strand-13 share strong sequence homology, with a highly aliphatic consensus of TXFAVLMXX. Secondary structural predictions of the aGPCR β-strand-13 and adjacent stalk region suggest that many comprise a short α-helix or β-strand element followed by a predicted turn and subsequent α-helix (Fig. S5).
We investigated the necessity of β-strand-13 for aGPCR activities. GPR56 or GPR110 N-terminal truncations were engineered with initiator methionines followed by the GAIN P1′ residue, Thr, and the complete 7TM domains. GPR56 or GPR110 7TM domain membranes had high constitutive activity for G13 or Gq activation, respectively (Fig. 3 B and C), corroborating previous work and our finding that active urea-mediated ECD dissociation from full-length aGPCRs resulted in receptor activation (Fig. 2 C and D) (11, 12). Serial deletion of single amino acids from the GPR56 or GPR110 7TM domain N termini sequentially reduced the ability of the receptors to activate G proteins. Removal of the first four amino acids, TYFA or TSFS, resulted in near complete abrogation of GPR56 and GPR110 7TM domain activities, respectively. Cell-surface biotinylation pull-down assays were performed and showed that each truncated 7TM receptor was present at relatively similar levels on the plasma membrane, largely discounting the possibility that the reduced activity of the truncated mutants was due to defective trafficking (Fig. 3 B and C). Substitution (to Ala) of the highly conserved Phe residues of the GPR56 or GPR110 TXFAVLM consensus sequences also substantially reduced 7TM protomer activities. Ala substitution of the conserved Met residue of TXFAVLM consensus markedly reduced receptor activation, although cell-surface abundances of these particular GPR56 and GPR110 mutants were reduced. A direct comparison of the GPR56 7TM domain to the F385M 7TM mutant demonstrated that despite equivalent cell-surface levels of both receptors (Fig. S6A), use of up to six times more F385M membranes never approached the activity of the full 7TM domain to efficaciously stimulate G13 [35S]-GTPγS binding kinetics (Fig. S6 B and C).
Fig. 3.
Adhesion GPCR β-strand-13/stalk regions are essential for G protein activation. (A) Sequence comparison of representative aGPCR and PAR stalk regions. The β-strand-13 TXFAVLMXX consensus sequences are denoted in red, and predicted turn elements are boxed in blue (CFSSP server) (28). Underlined sequences are alternative TM1 assignments (TMPred server). (B) GPR56 or (C) GPR110 7TM domain N-terminal single amino acid truncation series stimulation of G protein [35S]-GTPγS binding kinetics. Adhesion GPCR C-terminal antibody Western blots show relative levels of the indicated receptors isolated from insect cell surfaces by biotinylation pull-down assay. (D) HEK293 SRE luciferase activity in response to expressed GPR56 full-length and 7TM domain receptor N-terminal single amino acid truncation series. Error bars, SEM.
To measure the importance of the GPR56 7TM N-terminal stalk in live cells, we used an HEK293 cell serum response element (SRE)–luciferase reporter assay shown to be responsive to GPR56-dependent G12/13 activity (16–18). Full-length GPR56 activated the reporter modestly, as expected for an inhibited receptor state, whereas the 7TM domain provided robust activation (Fig. 3D). Successive deletion of single 7TM domain N-terminal amino acids abrogated SRE reporter activity with a very similar pattern to that observed in the insect membrane receptor reconstitution assays (Fig. 3 B and C), with the exception of the “TYFAVLM…” to “MFALVM…” truncation/mutation. All 7TM truncations in both systems required use of artificial initiator codons. Methionine aminopeptidases (MetAPs) process many cellular proteins, but we do not know if authentic N termini of the 7TM truncations were produced by HEK293 and/or insect cell MetAPs (19). We suspect that the “MFAVLM…” construct (Y384M) was not processed in HEK cells because human MetAPs do not efficiently proteolyze substrates with bulky P1′ residues such as Phe. Nonetheless, these results demonstrate the necessity of the N termini of two aGPCR β-strand-13/stalk regions for receptor activity and suggest that the region comprises a cryptic element required for 7TM domain activation when revealed by ECD displacement.
Activity Screens of Synthetic Peptides Comprising aGPCR Stalk Regions.
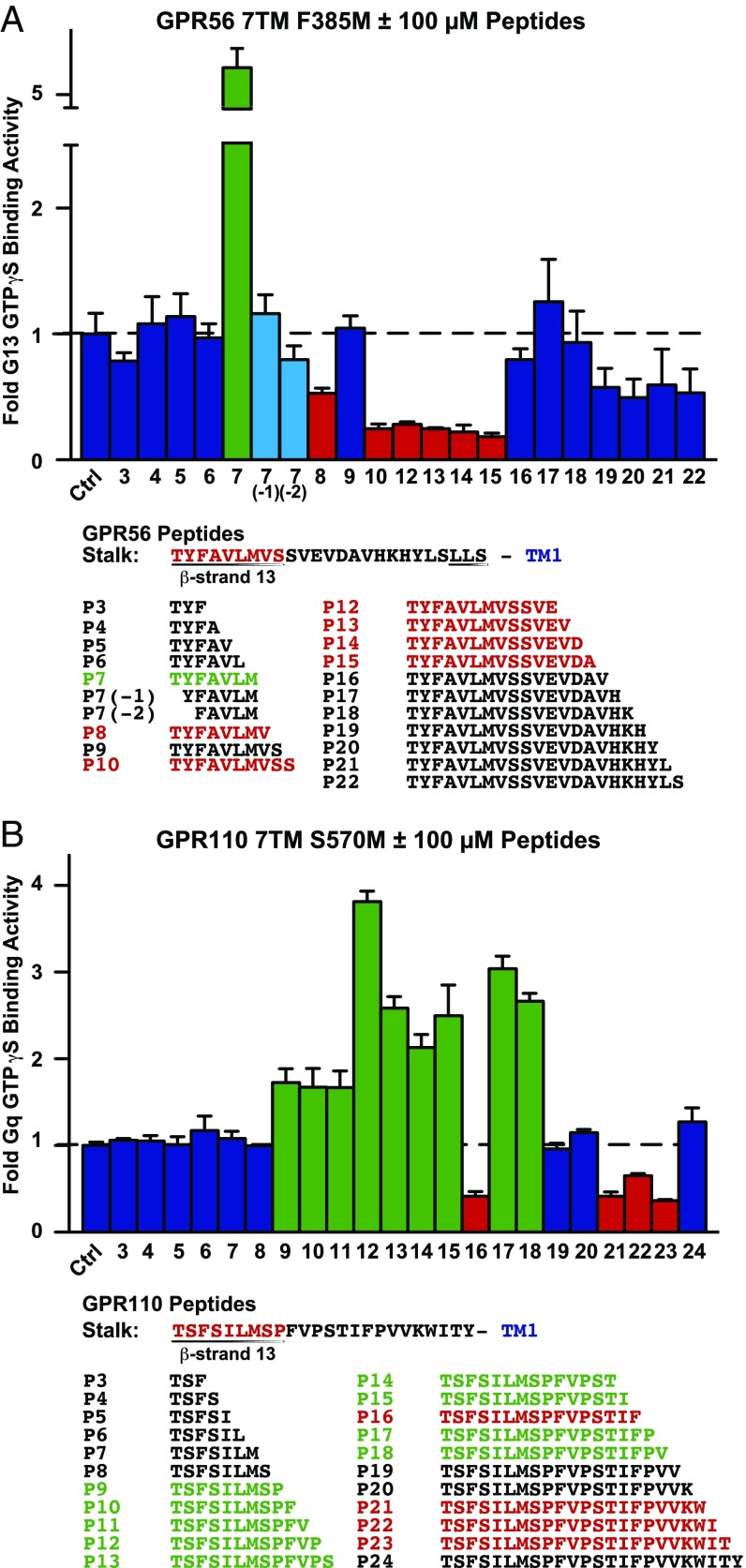
With the necessity of the GPR56 and GPR110 β-strand-13/stalk regions demonstrated, we questioned whether they harbored tethered agonists sufficient to activate the 7TM domains. Synthetic peptide libraries comprising the GPR56 and GPR110 β-strand-13/stalks were synthesized and screened for the ability to activate the truncated GPR56 7TM F385M and GPR110 7TM S570M receptors with low constitutive activities (Fig. 4). Peptide design began with the authentic P1′ residues of the GAIN cleavage site, Thr and extended C-terminally by one amino acid until the first residues of TM1. A conservative peptide concentration of 100 µM was screened based on the estimated effective concentration of PAR tethered agonists (∼400 µM) (20). A seven-residue GPR56 peptide (TYFAVLM/P7) comprising the majority of β-strand-13 substantially activated GPR56 7TM F385M (Fig. 4A). GPR56 peptides lacking the first, YFAVLM/P7(−1), or first and second, FAVLM/P7(−2), amino acids of the TYFAVLM peptide failed to stimulate the receptor, demonstrating the importance of the authentic N terminus for agonism (Fig. 4A). Nine GPR110 peptides were found to activate the GPR110 7TM S570M receptor in the peptide screen (Fig. 4B). These peptides did not stimulate Gq [35S]-GTPγS binding in the presence of control, nonreceptor membranes (Fig. S7). The most efficacious GPR110 peptide agonist was the 12-residue peptide (TSFSILMSPFVP/P12), although all nine peptides enhanced the kinetics of GPR110-mediated Gq activation (Fig. 4B and Fig. S8).
Fig. 4.
GPR56 and GPR110 synthetic stalk peptide screens for modulation of 7TM domain-mediated G protein activation. Synthetic peptides (100 µM) comprising the indicated portions of the (A) GPR56 or (B) GPR110 β-strand-13/stalk regions were tested for the ability to modulate low activity GPR56 7TM F385M domain-stimulated G13 or GPR110 7TM S570M domain-stimulated Gq [35S]-GTPγS binding. Values are expressed as fold vehicle treatment (Ctrl). Error bars, SEM.
Pharmacological and Kinetic Analyses of Adhesion GPCR Peptide Agonists.
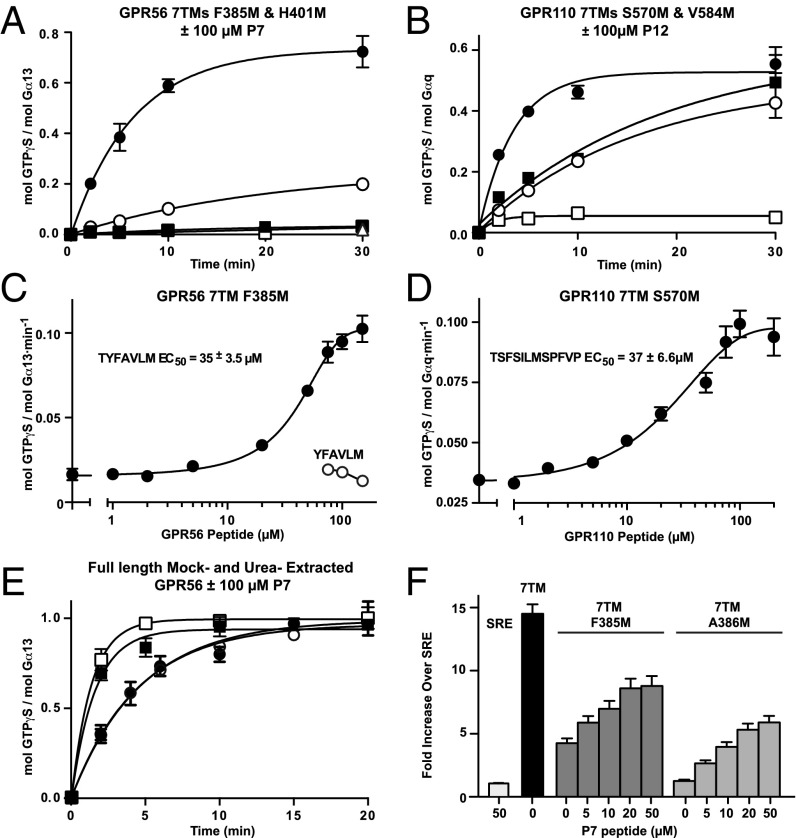
The abilities of the GPR56 and GPR110 agonist peptides to enhance receptor-stimulated G protein [35S]-GTPγS binding kinetics were measured directly. The 7TM receptors with 3–4 amino acids truncated from the N termini that had low constitutive activities (GPR56 F385M and GPR110 S570M), and 7TM receptors with the majority of the stalks truncated that had no constitutive activities (GPR56 H401M and GPR110 V584M), were tested with 100 µM each of the respective peptide agonists. GPR56 P7 dramatically enhanced F385M activity, but was incapable of activating the complete stalk truncation, H401M (Fig. 5A). In contrast, GPR110 P12 activated both equivalent GPR110 7TM receptor versions very well (Fig. 5B). The GPR56 P7 and GPR110 P12 peptide agonists enhanced the initial rates of GPR56 F385M or GPR110 S570M-stimulated G protein [35S]-GTPγS binding at EC50s of ∼35 and ∼37 µM, respectively (Fig. 5 C and D). High concentrations of GPR56 P7(−1) YFAVLM peptide had no ability to activate the GPR56 7TM receptor (Fig. 5C). The GPR56 P7 peptide elicited a steeper Hill slope than the GPR110 P12 peptide, suggesting better accessibility of the smaller peptide for its binding site.
Fig. 5.
Specific aGPCR synthetic peptides act as agonists. (A) GPR56 7TM F385M membranes (●, with 100 µM P7 peptide; ○, no peptide), 7TM H401M membranes (■, with 100 µM peptide; □, no peptide) and nonreceptor membranes (△) with 100 µM P7 peptide were reconstituted with G13 before measurement of receptor-stimulated [35S]-GTPγS binding kinetics (30-min point only, △). (B) GPR110 7TM S570M membranes (●, with 100 µM P12 peptide; ○, no peptide) and GPR110 7TM V584M membranes (■, with 100 µM P12 peptide; □, no peptide) were reconstituted with Gq before measurement of receptor-stimulated [35S]-GTPγS binding kinetics. (C) GPR56 7TM F385M membranes were incubated with the indicated concentrations of GPR56 peptides, TYFAVLM (P7) or YFAVLM (P7-1), and (D) GPR110 7TM S570M membranes were incubated with the indicated concentration of GPR110 P12 peptide before reconstitution with G13 or Gq and measurement of initial [35S]-GTPγS binding rates. Rates were plotted vs. peptide concentrations. (E) Urea- (squares) and mock-extracted (circles) full-length GPR56 membranes were incubated with 100 µM P7 peptide (closed symbols) or without peptide (open symbols) before G13 reconstitution and measurement of receptor-stimulated [35S]-GTPγS binding kinetics. (F) HEK293 SRE luciferase activity in response to empty vector (SRE) or expressed GPR56 7TM receptors and the indicated concentrations of P7 peptide. Error bars, SEM.
The GPR56 P7 peptide was tested for specificity using low-activity GPR110 F569M and S570M 7TM receptors, which are equivalent to the GPR56 F385M and A386M truncated 7TM domains, respectively. GPR56 P7 provided no activation of the GPR110 receptors and actually inhibited them modestly (Fig. S9). The GPR56 P7 peptide curiously lacked the ability to activate intact or urea-treated full-length GPR56 (Fig. 5E). We speculated that the ECD might occlude synthetic peptides from entering the agonist binding site on the intact full-length receptor and that the presence of the preferred, decrypted tethered agonist of the urea-treated full-length receptor did not allow the P7 peptide to activate this fully active receptor further.
Finally, we sought evidence for GPR56 P7 peptide agonism in live cells. A GPR56-responsive SRE–luciferase gene reporter assay was used to show that supplementation of HEK293 culture media with the P7 agonist peptide resulted in efficacious and concentration-dependent activation of low constitutive activity 7TM domain receptors (F385M and A386M) in cells, and did not activate the SRE reporter independent of transfected receptor (Fig. 5F). The agonistic activities of the GPR56 P7 peptide agonist in the reconstitution assays and cell-based assay were highly consistent.
Discussion
We have revealed an unexpected mechanism of adhesion GPCR activation. The 7TM domains of these two protomer receptors are inhibited by the noncovalently bound ECDs. The means of inhibition is to encrypt an element of the 7TM domain, the so-called β-strand-13/stalk region, that when revealed, serves as a tethered agonist to activate G protein signaling. Given the strong conservation of the agonist stalk regions among aGPCRs and the commonality of GAIN domain self-cleavage, most aGPCRs may use a tethered agonist regulatory mechanism.
Pivotal research questions remain: Do natural aGPCR ligands cause the ECD to dissociate fully or, in some cases, undergo more subtle rearrangement to decrypt the tethered agonist? And what is the precise composition of the tethered agonist(s)? We envision two possible models that are consistent with current knowledge of aGPCR activation and account for subtle differences of individual aGPCRs.
Complete ECD dissociation to reveal β-strand-13 would be a very energy-intensive process, although aGPCR ECD “shedding” from membranes was documented (21). Known aGPCR ligands include collagen subtypes and other components of the insoluble extracellular matrix (10, 11, 13). Mechanical shearing forces generated by cell movement in relation to a fixed ECM component bound to the aGPCR ECD might generate the force required to “tear” an ECD away from its 7TM. Once dissociated, it would seem to be entropically unfavorable for a GAIN domain/ECD to ever reform around β-strand-13 to reencrypt the agonist and terminate signaling. Our results with the in trans application of purified GPR56 ECD to 7TM domain membranes demonstrated that the ECD was not inhibitory. However, because β-strand-13 is an essential structural component of GAIN domains, a functional ECD could not be purified without this element (Fig. S4A) (8). Therefore, we do not have absolute confirmation that a dissociated ECD could reinhibit the 7TM domain, although the possibility seems highly unlikely given structural and energetic considerations. In this scenario, aGPCRs would be activated once, much like PARs and could be turned off only by a receptor desensitization mechanism (Fig. 1, One and Done Model).
We observed predicted turn elements in the middle of many of the ∼20- to 24-aa aGPCR stalk regions, suggesting some conformational flexibility for the agonist and, perhaps, supersecondary structure within the stalks (Fig. S5). These ideas lead to a second scenario that is also consistent with features of our data: the agonist is not exclusively defined by β-strand-13, but consists of β-strand-13 and a region of the stalk more proximal to TM1. Ligand binding to aGPCR ECDs could elicit internal force upon β-strand-13 resulting in a conformational change of the C-terminal stalk region to render it the competent agonist. This mechanism could be regulated and would not always result in receptor down-regulation, because the ligand signal could be terminated (Fig. 1, Tunable Model). This model could account for how cleavage-deficient aGPCRs, such as GPR111 and GPR115, may activate G proteins (22).
aGPCRs represent unexploited potential therapeutic targets (4–7). Our results in deciphering the activation mechanism of two aGPCRs and the identification of synthetic peptide agonists may aid rational design of synthetic aGPCR modulators.
Materials and Methods
Materials.
GPR56 N-terminal antibody was from R&D Systems. GPR56 C-terminal antibody was a gift from Randy Hall, Emory University, Atlanta, GA. GPR110 N- and C-terminal antibodies were from Sigma. Firefly luciferase reagent was from NanoLight. Streptavidin Sepharose HP, SP Sepharose, HiTrap, and Superdex 200 columns were from GE Healthcare. [35S]-GTPγS was from PerkinElmer. Sulfo-NHS-Biotin was from Thermo Scientific.mbf.
Human GPR110 (DNASU Plasmid Repository, dnasu.org; HsCD00295179) and GPR56 (accession no. NM_201524) (13) plasmids were used as PCR templates for full-length and designed truncations and subcloned into pcDNA3.1+, pFastBac1, or pDEST8. Baculoviruses were generated according to manufacturer’s instructions (Bac-to-Bac system; Invitrogen). Secreted GPR56 ECD had a gp67 signal sequence, mature GPR56 residues 26–394, and a His6-tag in the pACgp67-B plasmid. This baculovirus was generated using the BacPAK system (Clontech). Plasmids phRLuc-N1 and pGL4.33 [luc2/SRE/Hygro] were from PerkinElmer and Promega, respectively.
Peptide Synthesis and Solubilization.
GPR56 and GPR110 peptides were synthesized using solid-phase Fmoc chemistry at GenScript or Biomatik with free N termini and amidation-blocked C termini. Peptides were purified by HPLC (75–100%) with sequence verification by mass spectrometry. Peptides were suspended in DMSO and diluted to ≤4% DMSO (vol/vol) in experiments.
Insect Cell Culture and aGPCR Membrane Preparation.
Sf9 cells were grown in IPL41 containing 10% (vol/vol) heat-inactivated FBS (Gibco), and 1× Yeastolate at 27 °C. High-Five cells were grown in SF900II (Invitrogen). Baculoviruses were generated in Sf9 cells (Invitrogen; Bac-to-Bac). Amplified viruses were made by infecting Sf9 cultures with 1/100 dilutions of low-passage virus for 72–96 h. High-Five cells (2.0 × 106 cells/mL) were infected with baculovirus at a 1/50 dilution of primary-amplified viruses or a 1/100 dilution of secondary-amplified viruses for 48 h. Cells were harvested by centrifugation at 500 × g, lysed in HE buffer [10 mM Hepes (pH 7.4), 1 mM EGTA, and protease inhibitor mixture (23 µg/mL phenylmethylsulfonyl fluoride, 21 µg/mL Nα-p-tosyl-l-lysine-chloromethyl ketone, 21 µg/mL L-1-p-tosylamino-2-phenylethyl-chloro ketone, 3.3 µg/mL leupeptin, and 3.3 µg/mL lima bean trypsin inhibitor)] using a Parr disruption vessel (Parr Instrument Co.). Cell debris was cleared by centrifugation at 1,500 × g. Membranes were recovered by centrifugation at 100,000 × g and Dounce homogenized in HE buffer (mock) or HE buffer containing 7M urea and incubated on ice for 30 min. Treated membranes were recentrifuged and the HE (mock) and urea extracts were collected for analysis. Washed membranes were Dounce-homogenized into membrane storage buffer [HE buffer with 12% (vol/vol) sucrose] and stored at −80 °C.
Protein Purification.
Gα subunits were purified using the coexpressed GST-Ric-8 affinity purification method (23). Gβ1γ2 was purified as described (24, 25). His6-tagged GPR56 ECD was purified using a hybrid of methods (8, 16). Briefly, 3 L of High-Five cells (2 × 106 cells/mL) were infected with 1/50 dilution of amplified GPR56 ECD baculovirus for 72 h. The culture medium was loaded onto a 20-mL SP Sepharose FF column. The column was washed with 50 mM Na2HPO4, pH 7.0, and eluted with 50 mM Na2HPO4, pH 7.0, 500 mM NaCl, 10 mM imidazole, and protease inhibitors. The eluate was loaded onto a 5-mL His-Trap HP column using a BioRad DuoFlow FPLC. The column was washed with 97.5% (vol/vol) buffer A (50 mM Na2HPO4, pH 7.0, 10 mM imidazole, 300 mM NaCl, and protease inhibitor mixture) and 2.5% (vol/vol) buffer B (buffer A + 400 mM imidazole) and eluted with a linear imidazole gradient to 50% vol/vol buffer B. GPR56-ECD was dialyzed against 50 mM Tris, pH 7.7, 150 mM NaCl, 1 mM DTT, and 1 mM EDTA and concentrated to ∼5 mg/mL. The protein was gel filtered over a Superdex 200 HR 10/300 column.
[35S]-GTPγS Binding Assays.
Prepared aGPCR membranes (1–10 µg/per assay point) were incubated for 5 min with 200 nM Gα and 1 µM Gβ1γ2 in preincubation buffer (50 mM Hepes, pH 7.4, 1 mM DTT, 1 mM EDTA, 20 µM GDP, 3 µg/mL BSA). For GPR56 peptide assays, membranes were incubated with peptide for 5 min followed by G protein incubation. For GPR110 peptide assays, peptide and G proteins were incubated for 30 min in GDP-free preincubation buffer (actual GDP was ∼50 nM from prepared Gα). Kinetic assays were initiated by addition of an equal volume of [35S]-GTPγS binding buffer [50 mM Hepes (pH 7.4), 1 mM DTT, 1 mM EDTA, 0 or 20 µM GDP, 3 µg/mL BSA, 10 mM MgCl2, 50 mM NaCl, 2 µM GTPγS, [35S]-GTPγS (20,000–50,000 cpm/pmol)]. Reactions were performed in triplicate, quenched with 20 mM Tris, pH 7.7, 100 mM NaCl, 10 mM MgCl2, 1 mM GTP, and 0.08 (m/v) deionized polyoxyethylene 10 lauryl ether C12E10, then filtered onto Protran BA85 nitrocellulose filters (GE Healthcare). Filters were washed with 20 mM Tris, pH 7.7, 100 mM NaCl, and 2 mM MgCl2, dried, and subjected to liquid scintillation counting. Data were fit to one-phase monoexponential association functions (GraphPad Prism).
aGPCR Cell Surface Levels.
High-Five cells were washed with PBS containing protease inhibitor mixture, and suspended in PBS containing 2 mM Sulfo-NHS-LC-Biotin (Thermo Scientific) for 30 min. Cells were washed with PBS containing 100 mM glycine and lysed in 25 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 2 mM MgCl2, 1 mM EDTA, 2% glycerol. Lysates were centrifuged at 21,000 × g and precleared with G-25 Sephadex. Biotinylated proteins were isolated with Streptavidin Sepharose. Cell-surface aGPCRs were visualized by Western blotting.
Luciferase Reporter Assays.
HEK293T cells (2 × 105) were transfected in 24-well format with 25 ng of aGPCR in pcDNA3.1, 100 ng SRE–luciferase reporter, 1 ng phRLuc, and balanced with pcDNA3.1 using a polyethylenimine transfection method (26). At 24 h, cells were serum starved for 12 h, harvested in media, washed in Tyrode’s solution (137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 0.2 mM Na2HPO4, 12 mM NaHCO3, 5.5 mM d-glucose), and lysed in Firefly Luciferase Assay Reagent (NanoLight). Reactions were quenched using Renilla luciferase buffer containing 3 µM coelenterazine H (27). Firefly luciferase data were normalized to Renilla luciferase and compared with SRE–luciferase-only controls. In agonist experiments, peptide was added in one-quarter doses over the last 5 h of serum starvation (e.g., 2.5 µM added each h to achieve a final concentration of 10 µM).
Supplementary Material
Acknowledgments
We thank A. Smrcka and P. Hinkle for critical discussions and reading of the manuscript; E. Marvin and B. Gehl for technical support; and R. Hall for the gift of the GPR56 7TM antibody. Support for this study was provided by National Institutes of Health Grants R01GM088242B (to G.G.T.) and R01GM098591 (to L.X.). H.M.S is supported by a fellowship from the PhRMA Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421785112/-/DCSupplemental.
References
- 1.Bjarnadóttir TK, et al. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics. 2004;84(1):23–33. doi: 10.1016/j.ygeno.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Langenhan T, Aust G, Hamann J. Sticky signaling—adhesion class G protein-coupled receptors take the stage. Sci Signal. 2013;6(276):re3. doi: 10.1126/scisignal.2003825. [DOI] [PubMed] [Google Scholar]
- 3.Paavola KJ, Hall RA. Adhesion G protein-coupled receptors: Signaling, pharmacology, and mechanisms of activation. Mol Pharmacol. 2012;82(5):777–783. doi: 10.1124/mol.112.080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aust G. Adhesion-GPCRs in tumorigenesis. Adv Exp Med Biol. 2010;706:109–120. doi: 10.1007/978-1-4419-7913-1_9. [DOI] [PubMed] [Google Scholar]
- 5.Lum AM, et al. Orphan receptor GPR110, an oncogene overexpressed in lung and prostate cancer. BMC Cancer. 2010;10(1):40. doi: 10.1186/1471-2407-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piao X, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303(5666):2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Xu L. GPR56 in cancer progression: Current status and future perspective. Future Oncol. 2012;8(4):431–440. doi: 10.2217/fon.12.27. [DOI] [PubMed] [Google Scholar]
- 8.Araç D, et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31(6):1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prömel S, Langenhan T, Araç D. Matching structure with function: The GAIN domain of adhesion-GPCR and PKD1-like proteins. Trends Pharmacol Sci. 2013;34(8):470–478. doi: 10.1016/j.tips.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Luo R, et al. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci USA. 2011;108(31):12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci Signal. 2014;7(338):ra76. doi: 10.1126/scisignal.2005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem. 2011;286(33):28914–28921. doi: 10.1074/jbc.M111.247973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci USA. 2006;103(24):9023–9028. doi: 10.1073/pnas.0602681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prömel S, et al. Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Dev Dyn. 2012;241(10):1591–1602. doi: 10.1002/dvdy.23841. [DOI] [PubMed] [Google Scholar]
- 15.Vu T-KH, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64(6):1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 16.Iguchi T, et al. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G α 12/13 and Rho pathway. J Biol Chem. 2008;283(21):14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- 17.Shashidhar S, et al. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24(10):1673–1682. doi: 10.1038/sj.onc.1208395. [DOI] [PubMed] [Google Scholar]
- 18.Wu MP, et al. G-protein coupled receptor 56 promotes myoblast fusion through serum response factor- and nuclear factor of activated T-cell-mediated signalling but is not essential for muscle development in vivo. FEBS J. 2013;280(23):6097–6113. doi: 10.1111/febs.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradshaw RA, Brickey WW, Walker KW. N-terminal processing: The methionine aminopeptidase and N α-acetyl transferase families. Trends Biochem Sci. 1998;23(7):263–267. doi: 10.1016/s0968-0004(98)01227-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C, et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492(7429):387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Z, et al. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16(16):1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- 22.Promel S, et al. Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Dev Dyn. 2012;241(10):1591–1602. doi: 10.1002/dvdy.23841. [DOI] [PubMed] [Google Scholar]
- 23.Chan P, et al. Purification of heterotrimeric G protein alpha subunits by GST-Ric-8 association: Primary characterization of purified G alpha(olf) J Biol Chem. 2011;286(4):2625–2635. doi: 10.1074/jbc.M110.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozasa T. Purification of Recombinant G Protein α and βγ Subunits from Sf9 Cells. CRC; Boca Raton, FL: 1999. pp. 23–38. [Google Scholar]
- 25.Kozasa T, Gilman AG. Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits. Characterization of alpha 12 and inhibition of adenylyl cyclase by alpha z. J Biol Chem. 1995;270(4):1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 26.Oner SS, Blumer JB, Lanier SM. Group II activators of G-protein signaling: Monitoring the interaction of Gα with the G-protein regulatory motif in the intact cell. Methods Enzymol. 2013;522:153–167. doi: 10.1016/B978-0-12-407865-9.00009-1. [DOI] [PubMed] [Google Scholar]
- 27.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem. 2000;282(1):158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 28.Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.