Significance
Colorectal cancer results from genetic lesions in epithelial cells. However, the tumor microenvironment, which is formed by nonepithelial stromal cells, also plays an important role in this disease. The influence of the microenvironment on tumorigenesis is mediated by paracrine signals between tumor epithelial cells and neighboring stromal cells. We found that expression of interleukin 33 (IL-33), an important mediator of type 2 immunity and wound repair, is induced in epithelial cells of human and mouse intestinal tumors. IL-33 promoted intestinal tumorigenesis in ApcMin/+ mice and activated two stromal cell types, subepithelial myofibroblasts and mast cells, known to mediate intestinal dysplasia. Tumor epithelial cells are proposed to coopt IL-33–mediated immune and wound-healing responses to create a microenvironment favorable to tumorigenesis.
Keywords: IL-33, colorectal cancer, ApcMin, wound healing, Th2 immunity
Abstract
Tumor epithelial cells develop within a microenvironment consisting of extracellular matrix, growth factors, and cytokines produced by nonepithelial stromal cells. In response to paracrine signals from tumor epithelia, stromal cells modify the microenvironment to promote tumor growth and metastasis. Here, we identify interleukin 33 (IL-33) as a regulator of tumor stromal cell activation and mediator of intestinal polyposis. In human colorectal cancer, IL-33 expression was induced in the tumor epithelium of adenomas and carcinomas, and expression of the IL-33 receptor, IL1RL1 (also referred to as IL1-R4 or ST2), localized predominantly to the stroma of adenoma and both the stroma and epithelium of carcinoma. Genetic and antibody abrogation of responsiveness to IL-33 in the ApcMin/+ mouse model of intestinal tumorigenesis inhibited proliferation, induced apoptosis, and suppressed angiogenesis in adenomatous polyps, which reduced both tumor number and size. Similar to human adenomas, IL-33 expression localized to tumor epithelial cells and expression of IL1RL1 associated with two stromal cell types, subepithelial myofibroblasts and mast cells, in ApcMin/+ polyps. In vitro, IL-33 stimulation of human subepithelial myofibroblasts induced the expression of extracellular matrix components and growth factors associated with intestinal tumor progression. IL-33 deficiency reduced mast cell accumulation in ApcMin/+ polyps and suppressed the expression of mast cell-derived proteases and cytokines known to promote polyposis. Based on these findings, we propose that IL-33 derived from the tumor epithelium promotes polyposis through the coordinated activation of stromal cells and the formation of a protumorigenic microenvironment.
Colorectal cancer (CRC) is a leading cause of cancer-related deaths worldwide. The origins of hereditary colon cancer (familial adenomatous polyposis) as well as sporadic CRC are closely associated with mutations in the adenomatous polyposis coli (APC) tumor suppressor gene (1, 2). Subsequent loss of heterozygosity for APC in intestinal epithelial cells (IECs) activates Wnt signaling through stabilization of β-catenin, which is sufficient to initiate polyp formation (3). Although genetic alterations in IECs are a driving force of dysplasia, intestinal tumors are rarely, if ever, entirely composed of a genetically altered epithelium. Rather, a host of ancillary cells including gut mesenchymal cells [e.g., smooth muscle cells, endothelial cells, and subepithelial myofibroblasts (SEMFs)] as well as mucosal immune cells are intermingled with the tumor epithelial cells. Importantly, these stromal cells regulate the tumor microenvironment to influence CRC initiation and progression (4, 5).
Stromal cells of the normal intestinal mucosa have an inherent ability to rapidly react to changes in epithelial cell homeostasis. In response to tissue damage such as infection, the stromal compartment produces cytokines and chemokines, extracellular matrix (ECM) proteins, ECM remodeling molecules, and growth factors to coordinate immune responses and mediate tissue repair through epithelial restitution and proliferation (6, 7). The CRC stroma acquires a similar activated phenotype and produces the same soluble factors and ECM components associated with inflammation and wound healing to promote proliferation and survival of transformed epithelia, tumor immune evasion, angiogenesis, and tissue invasion and metastasis (5, 8, 9). Importantly, the tumor epithelium directly activates stromal cells through the release of paracrine factors and cytokines, such as transforming growth factor-β (TGFβ), to establish a microenvironment favorable to tumor growth and metastasis (4, 9–11). Thus, tumor epithelial cell-derived paracrine factors that modulate stromal cell function are potential biomarkers of disease prognosis as well as targets for anticancer therapy (9, 12).
Interleukin 33 (IL-33) is a member of the IL-1 family of cytokines and is expressed in several organ systems including the gastrointestinal tract (13). Nonhematopoietic cells, including epithelial cells, myofibroblasts, fibroblasts, adipocytes, smooth muscle cells, and endothelial cells, are the primary sources of IL-33 production (14–16), but professional antigen-presenting cells such as macrophages also express IL-33 (16). Similar to IL-1α, IL-33 is a dual-function protein with roles as a nuclear factor and a classical cytokine (17). IL-33 functions as a prototypic “alarmin,” passively released by stressed, damaged, or necrotic cells to alert the immune system of a local threat such as trauma or infection (18, 19). As a cytokine, IL-33 activates a heterodimeric receptor complex comprised of IL-1 receptor-like 1 (IL1RL1; also referred to as IL1-R4 or ST2L) and its coreceptor, IL-1 receptor accessory protein (IL-1RAcp), which regulates inflammatory gene expression through MAPK and NF-κB signaling cascades (16, 20). A splice variant of IL1RL1 exists as a soluble isoform (commonly referred to as sST2) and is proposed to act as an antagonistic decoy receptor for IL-33 (21). Several immune cells, including mast cells and T helper (Th) 2 lymphocytes, as well as nonhematopoietic cells, including epithelial cells, myofibroblasts, and endothelial cells, express IL1RL1 (22).
IL-33 is an important mediator of inflammation and wound-healing responses in several tissues. In the gastrointestinal tract, IEC-derived IL-33 enhances mucosal barrier defense against helminth parasites by augmenting type 2 immune responses and Th2-associated interleukin production (e.g., IL-4 and IL-13), which mediate parasite expulsion, activate wound-healing responses in SEMFs, and promote IEC proliferation and mucus production for epithelial restitution and repair (6, 16, 23). In inflammatory bowel disease (IBD), in particular active ulcerative colitis, IL-33 expression is induced in IECs and infiltrating lamina propria mononuclear cells of ulcerative lesions and SEMFs beneath active lesions (24–28). Importantly, two recent studies have established a positive correlation between IL-33/IL1RL1 expression levels and human CRC progression and metastasis (29, 30). However, the contribution of IL-33 to overall tumor risk remains to be directly assessed, and the mechanisms by which IL-33 contributes to tumor initiation and progression remain unexplored.
In the present study, we demonstrate that IL-33 is an important cytokine mediator of intestinal tumorigenesis. In human and mouse adenomas, IL-33 and IL1RL1 expression are shown to localize primarily to the tumor epithelium and stroma, respectively. Using both genetic and antibody suppression of IL-33 signaling in ApcMin/+ mice, cytokine IL-33 is shown to promote polyposis and influence adenomatous cell (tumor epithelial cell) proliferation and apoptosis and tumor-associated angiogenesis. We demonstrate that IL-33 expression is induced in adenomatous cells of ApcMin/+ polyps and that two stromal cell types previously associated with intestinal tumorigenesis, SEMFs and mast cells, express IL1RL1. In response to IL-33, SEMFs and mast cells express ECM components and remodeling proteins, growth factors, and angiogenesis regulators, and cytokines characteristic of activated stroma and involved in tumorigenesis. Taken together, our observations suggest that tumor epithelial cells coopt IL-33–induced tissue repair responses to form a tumor microenvironment favorable to polyposis.
Results
IL-33 Expression Is Induced in Human CRC.
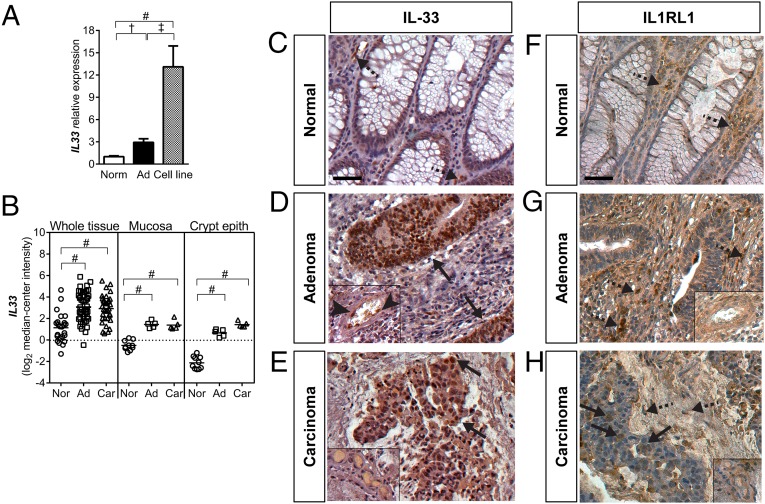
We initially tested whether IL-33 signaling is associated with CRC in humans. Using quantitative PCR (qPCR), IL33 expression was found to be significantly increased in adenocarcinomas and patient-derived CRC cell lines compared with expression in adjacent normal tissues (Fig. 1A). IL33 expression in cell lines was significantly higher than that in both normal intestinal tissues and tumors, suggesting that IL33 is enriched in the tumor epithelium. Reanalysis of IL33 expression in the CRC Affymetrix datasets published by Skrzypczak et al. (31) produced similar results, with IL33 expression significantly increased in CRC whole tissue biopsies and in laser-microdissected CRC mucosa and crypt epithelium (Fig. 1B). Reanalysis of IL-33 expression in the CRC Affymetrix datasets published by Sabates-Bellver et al. (32) produced similar results (Fig. S1A). Importantly, circulating levels of IL-33 were significantly increased in CRC patients compared with healthy individuals (Fig. S1B).
Fig. 1.
IL-33 and IL1RL1 expression in human CRC. (A) qPCR analysis of IL33 expression in adjacent normal mucosa (Norm), adenocarcinoma (Ad), and cell lines derived from adenocarcinomas (Cell line). Data plotted relative to norm ± SEM (n = 20–25). (B) IL33 expression in human whole mucosa or tumor tissue biopsies and laser-microdissected mucosa and crypt epithelium. Ad, colon adenoma or adenocarcinoma; Car, colon carcinoma; Norm, adjacent normal tissue. Data obtained from the Skrzypczak and Skrzypczak 2 colorectal oncomine datasets. Bars indicate the mean. †P ≤ 0.01; ‡P ≤ 0.001; #P ≤ 0.0001. (C–E) Immunohistochemistry for IL-33 in normal adjacent mucosa, adenoma, and carcinoma. Insets show endothelial cells associated with vessels. Arrows indicate IL-33–positive epithelium; arrowheads indicate IL-33–positive endothelial cells. (F–H) Immunohistochemistry for IL1RL1 in normal adjacent mucosa (Normal), adenoma, and carcinoma. Insets show endothelial cells associated with vessels. Arrows indicate IL1RL1-positive epithelium; dashed arrows indicate stromal cells. (Scale bars, 50 μm.)
Next, we used immunohistochemistry to examine the distribution of IL-33 and IL1RL1 in CRC and adjacent normal tissue. In normal tissue, IL-33 immunolabeling was observed in stromal cells within the lamina propria but rarely in the epithelium (Fig. 1C). In adenoma and carcinoma, the tumor epithelium represented the major source of IL-33 expression (Fig. 1 D and E). Tumor stromal cells, including endothelial cells, positive for IL-33 were rare. IL1RL1 expression in normal tissue was observed in some stromal cells of the lamina propria and rarely within the epithelium (Fig. 1F). In adenomas, IL1RL1 expression localized predominately to stromal cells. Endothelial cell and adenomatous cell immunostaining was rarely observed (Fig. 1G). In carcinoma, IL1RL1 immunolabeling of both carcinoma cells and stromal cells was observed, and endothelial cell staining was infrequent (Fig. 1H). In summary, our analysis of IL-33 and IL1RL1 expression in CRC suggests that the epithelium is a major source of IL-33 expression and that tumor-associated stromal cells predominately express IL1RL1. Thus, IL-33 appears to be an epithelial cell-derived signal that modulates stromal cell function. However, IL1RL1 expression in carcinoma cells suggests that as CRC progresses the transformed epithelium also acquires the capacity to respond to IL-33 autocrine/paracrine signals.
IL-33 Signaling Deficiency Reduces Polyp Burden in ApcMin/+ Mice.
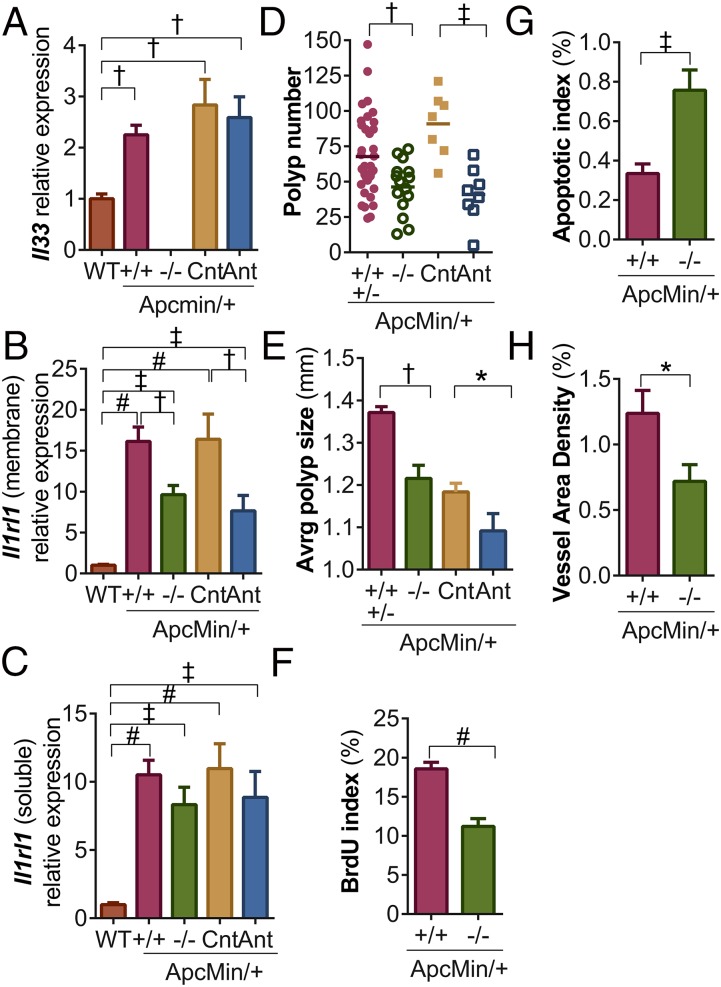
To confirm our findings that IL-33 and IL1RL1 are expressed during intestinal tumorigenesis, we next tested whether Il33 and Il1rl1 are induced in adenomatous polyps of C57BL/6J (B6) mice heterozygous for the ApcMin mutation. These mice (ApcMin/+) form multiple intestinal adenomas following loss of heterozygosity for Apc, similar to the initiation of most sporadic human CRC and familial adenomatous polyposis (33). Il33 expression increased twofold in the polyps of ApcMin/+ mice compared with normal intestinal mucosa of wild-type (WT) littermates (Fig. 2A). In addition, we measured expression of Ilrl1 in ApcMin/+ polyps. Expression of membrane-associated and -soluble Il1rl1 isoforms were increased 10- and 17-fold, respectively, in ApcMin/+ polyps compared with normal intestinal mucosa (Fig. 2 B and C). Thus, Il33 and Il1rl1 expression are also induced during polyposis in mice.
Fig. 2.
IL-33 signaling deficiency attenuates polyp growth, induces apoptosis, and suppresses angiogenesis. (A–C) qPCR analysis of Il33, membrane-bound Il1rl1, and soluble Il1rl1 expression in normal mucosa of WT mice and in adenomas of ApcMin/+ mice WT (+/+) or deficient (−/−) for Il33, or treated with an IgG control (Cnt) or Il1rl1 antagonist (Ant) antibody. Expression is plotted relative to WT ± SEM (n = 5–10). Average polyp number (D) and size (E) in the small intestines and colons of ApcMin/+ mice WT and heterozygous for the Il33 deletion (+/+ and +/−), homozygous for the Il33 deletion (−/−), treated with a control (Cnt) antibody, or treated with the Il1rl1 antagonist antibody (Ant). Bars indicate the mean in panel A. (F) Proliferative indices (BrdU incorporation), (G) apoptotic (TUNEL labeling) indices, and (H) angiogenesis assessment (size vWF-positive area/size of tumor area) in polyps from ApcMin/+ mice WT or deficient for Il33. For panels F–H, data are plotted as the percentage of positive cells or total vessel area per section ± SEM (n =16). *P ≤ 0.05; †P ≤ 0.01; ‡P ≤ 0.001; #P ≤ 0.0001.
Given the increased expression of Il33 and Il1rl1 in polyps of ApcMin/+ mice, we next tested whether IL-33 signaling deficiency blocks polyposis. B6 mice heterozygous for a null allele of Il33 (Il33+/−) (Fig. S2 A–C) were first crossed with B6-ApcMin/+ mice to produce B6-ApcMin/+,Il33+/− offspring. B6-ApcMin/+,Il33+/− mice were then crossed to B6-Il33+/− mice to produce experimental ApcMin/+ mice deficient for Il33 (ApcMin/+,Il33−/−) and littermate controls (ApcMin/+,Il33+/+ and ApcMin/+,Il33+/−). Il33 transcripts were not detected in Il33−/− mice, verifying that the engineered mutation is a null allele (Fig. 2A).
First we tested whether heterozygosity for the Il33 deletion significantly affects polyposis. At 90 d of age, polyp initiation (number) and progression (size) were indistinguishable between control ApcMin/+,Il33+/+ and ApcMin/+,Il33+/− siblings (Fig. S3 A and B). Thus, ApcMin/+,Il33+/+ and ApcMin/+,Il33+/− data were pooled in further analyses. Next, we investigated whether Il33 deficiency inhibited polyposis. At 90 d of age, a 33% and 11% reduction in polyp number and size, respectively, was observed in experimental ApcMin/+,Il33−/− mice compared with ApcMin/+,Il33+/+ and ApcMin/+,Il33+/− control siblings (Fig. 2 D and E and Fig. S3C). Because IL-33 is proposed to have dual roles as a chromatin-associated nuclear factor and a cytokine (17), we used an IL1RL1 antagonist antibody (34) to test whether the cytokine function of IL-33 promotes polyposis. B6-ApcMin/+ mice were treated with 150 μg of an anti-IL1RL1 antibody or IgG control twice a week from 21 to 90 d of age. At this time point, anti-IL1RL1 treatment significantly reduced polyp number by 55% and polyp size by 8% (Fig. 2 D and E and Fig. S3C). Thus, based on the results of two independent strategies for blocking IL-33 signaling, we find that the cytokine function of IL-33 is an important mediator of intestinal polyposis.
Various cytokines associated with polyposis stimulate IEC proliferation and survival (4, 35). Therefore, we used BrdU incorporation and TUNEL assays to compare cell proliferation and apoptosis, respectively, in ApcMin/+,Il33+/+ control and ApcMin/+,Il33−/− test polyps. Il33 deficiency caused a 56% reduction in the number of mitotic cells (Fig. 2F and Fig. S4A) and a 46% increase in apoptotic cell number in polyps (Fig. 2G and Fig. S4B). Previous studies have suggested that IL-33 signaling induces angiogenesis (36), which facilitates tumor growth and metastasis. Thus, the surface area of von Willebrand Factor (vWF) immunostained blood vessels in polyp sections was calculated for ApcMin/+,Il33+/+ control and ApcMin/+,Il33−/− test mice. Importantly, IL-33 deficiency significantly reduced mean vessel surface area by 42% in polyps (Fig. 2H and Fig. S4C). Together these results suggest that IL-33 signaling promotes tumor growth and associated angiogenesis.
Adenomatous Cells Express IL-33 in ApcMin/+ Polyps.
Expression of IL-33 in the gut mucosa has been localized to IECs, endothelial cells, SEMFs, and lamina propria mononuclear cells (25, 26). In IBD, in particular ulcerative colitis lesions, previous studies revealed that IECs and SEMFs express IL-33 (25, 26, 28). Therefore, we investigated whether IL-33 expression is also induced in IECs and SEMFs during polyposis. In the normal small intestine mucosa of WT mice, IL-33 expression localized to stromal cells (mononuclear immune cells and fibroblast-like cells) of the lamina propria but was minimally detectable in IECs, which were identified by membranous-associated β-catenin (CTNNB1) (Fig. 3A). Similarly, normal epithelial cells in tissues surrounding polyps and in deep crypts under polyp lesions remained IL-33–negative (Fig. 3A). In contrast, adenomatous cells, which were identified by cytoplasmic and nuclear accumulation of β-catenin, showed robust nuclear and cytoplasmic expression of IL-33 (Fig. 3A).
Fig. 3.
IL-33 localizes to transformed epithelial cells in polyps and SEMFs in normal mucosa. (A) Immunofluorescence for IL-33 (green) and CTNNB1 (red) in WT (Normal) mucosa and an ApcMin/+ polyp. Adenomatous cells (nuclear and cytoplasmic CTNNB1-positive) colabel for IL-33 (above the dashed line). Normal IECs (membranous CTNNB1-positive) in deep crypts beneath a polyp lesion are IL-33–negative (below the dashed line). (B) IL-33 (green) colocalized with the SEMF marker ACTA2 (red) in WT mucosa and normal mucosa underlying ApcMin/+ polyp lesion (below the dashed line). SEMFs within the lesion (above the dashed line) are IL-33–negative. For A and B, nuclei are counterstained with DAPI and images are oriented with the intestinal lumen at the top. (Scale bars, 50 μm.)
Next we tested whether polyp-associated SEMFs are a source of IL-33 expression. SEMFs exhibit features of both fibroblasts and smooth muscle cells and are characterized by immunoreactivity for smooth muscle actin (ACTA2) and vimentin (VIM) but not desmin (DES) (37). Together these markers distinguish SEMFs from smooth muscle cells (ACTA2-positive, VIM-negative, and DES-positive) and fibroblasts (ACTA2-negative, VIM-positive, and DES-negative). In sections of normal gut mucosa, IL-33 immunolabeling predominately localized to the nuclei of pericryptal ACTA2- and VIM-positive SEMFs (Fig. 3B and Fig. S5 A and B). Similarly, SEMFs surrounding deep crypts under polyp lesions were positive for nuclear IL-33 (Fig. 3B and Fig. S5 A and B). In contrast, SEMFs within polyps and surrounding IL-33–expressing adenomatous cells did not express detectable levels of IL-33 (Fig. 3B and Fig. S5B). We also assessed the expression of IL-33 in smooth muscle and endothelial cells in normal intestinal mucosa and polyps to exclude these cell types as major contributors of IL-33 signaling. Importantly, IL-33 staining was notably absent in all but a few DES-positive smooth muscle cells or platelet/endothelial cell adhesion (CD31)-positive endothelial cells in either tissue (Fig. S6 A and B). To test whether tumor-associated stromal cells expressed IL-33 but were negative for IL-33 immunolabeling due to constitutive cytokine release, we used the beta-galactosidase (LacZ) reporter in the IL-33 knockout allele (Fig. S2) to identify cells within polyps transcriptionally active for IL-33. Immunolabeling for LacZ in ApcMin/+ mice heterozygous for the IL-33 knockout allele confirmed that CTNNB1-positive adenomatous cells express LacZ (Fig. S5 C and D). Few if any stromal cells, including ACTA2-positve SEMFs, expressed LacZ (Fig. S5 C and E). In summary, these results demonstrate that adenomatous cells represent the vast majority of cells expressing IL-33 in ApcMin/+polyps.
SEMFs Express IL1RL1 in ApcMin/+ Polyps.
Previous studies have established that myofibroblasts in chronic pancreatitis tissues express either IL-33 or IL1RL1 in response to different combinations of cytokines (38). Based on evidence that myofibroblasts have the capacity to express IL-33 or IL1RL1, we tested whether polyposis in ApcMin/+ mice causes SEMFs to shift from an IL-33– to an IL1RL1-positive cell population. In sections of normal mucosa, IL1RL1 immunolabeling was weak to undetectable in all cell types (Fig. 4 A and B). In contrast immunolabeling of ApcMin/+ polyp sections revealed strong IL1RL1 localization to stromal cells, with the highest levels of immunostaining localizing to cells just beneath the surface epithelia (Fig. S7A). Coimmunolabeling for IL1RL1 and SEMF markers ACTA2 and VIM showed that IL1RL1-positive, periluminal stromal cells are SEMFs (Fig. 4 A and B and Fig. S8). DES-positive smooth muscle cells and CD31-positive endothelial cells were rarely positive for IL1RL1 immunolabeling (Fig. S6 C and D). Importantly, Il33 deficiency reduced, but did not suppress, SEMF immunolabeling for IL1RL1 in polyps, suggesting that IL-33 directly or indirectly regulates the expression of its own receptor in SEMFs (Fig. S7 A and B). Together these results demonstrate that SEMFs, in particular those in periluminal regions, are the primary nonhematopoietic cell type expressing IL1RL1 in polyps.
Fig. 4.
Activated periluminal SEMFs in polyps express IL1RL1. Immunofluorescence for IL1RL1 (green) and the SEMF marker (A) ACTA2 or (B) VIM (red) in WT (Normal) mucosa and an ApcMin/+ polyp. Polyp images on the Right are higher magnification images of the Middle panels. SEMFs within the polyps are IL1RL1-positive. (C) Immunofluorescence of polyp serial sections for the activated stroma marker TNC (green) and ACTA2, IL1RL1, or IL-33 (red). Periluminal SEMFs adjacent to IL-33–positive epithelium coexpress TNC and IL1RL1. In all images nuclei are counterstained with DAPI, and arrowheads indicate regions of colabeling. All images are oriented with the intestinal lumen at the top. (Scale bars, 50 μm).
SEMFs Express Genes Associated with Activated Stroma in Response to IL-33.
In the normal intestinal mucosa, SEMFs are positioned subadjacent to the basement membrane and have a profound influence on stem cell maintenance and IEC homeostasis (39). During infection, trauma, and neoplasia, SEMFs are induced to proliferate and release cytokines, growth factors, ECM components, and angiogenesis factors that mediate IEC proliferation, survival, differentiation, and repair (5, 39–41). Therefore, to test whether IL-33 can also induce a gene expression profile indicative of activated SEMFs, the human SEMF cell line (CCD-18Co) (42) was cultured with recombinant human IL-33 or vehicle control and then subjected to whole transcriptome sequencing (RNA-Seq). Comparison of mRNA expression between IL-33– and control-treated cells identified 1,357 transcripts that were differentially expressed more than 1.5-fold (P < 0.05) (Dataset S1). Gene Ontology (GO) Biological Processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of the 699 transcripts up-regulated by IL-33 enriched for several annotations associated with activation of stromal fibroblasts during tissue repair and tumorigenesis, including ECM processing and remodeling, collagen metabolism, angiogenesis, and growth factor signaling (Table 1 and Dataset S2) (9, 10, 43). Curiously, the 658 transcripts down-regulated by IL-33 annotated into several pathways associated with DNA repair (Table 1 and Dataset S3).
Table 1.
Pathway enrichment analysis of genes differentially expressed by SEMFs in response to IL-33
| Representative pathways enriched by up- or down-regulated genes* | P score |
| Up-regulated | |
| Angiogenesis (BP) | 9.68E-07 |
| ECM–receptor interaction (Kegg) | 7.60E-06 |
| TGF-beta signaling pathway (Kegg) | 3.33E-05 |
| MAPK signaling pathway (Kegg) | 4.17E-05 |
| Wound healing (BP) | 4.64E-05 |
| CRC (Kegg) | 6.92E-04 |
| Positive regulation of inflammatory response (BP) | 1.62E-03 |
| ECM organization (BP) | 6.74E-03 |
| Fibroblast growth factor receptor signaling pathway (BP) | 4.16E-02 |
| Positive regulation of collagen biosynthetic process (BP) | 1.77E-02 |
| Collagen catabolic process (BP) | 2.38E-02 |
| I-kappaB kinase/NF-kappaB cascade (BP) | 3.89E-02 |
| Down-regulated genes | |
| DNA repair (BP) | 6.52E-11 |
| Mismatch repair (Kegg) | 3.39E-04 |
| Nucleotide excision repair (Kegg) | 6.92E-04 |
| Base-excision repair (BP) | 2.37E-03 |
Representative pathways enriched by genes significantly increased or decreased 1.5-fold in response to IL-33. Full lists of annotated pathways and their composing genes can be found in Datasets S2 and S3. BP, Go Biological Processes; Kegg, Kegg Pathways.
Interestingly, IL-33 treatment also induced SEMF expression of transforming growth factor, beta 1 (TGFB1) (Dataset S1) and the expression of several genes that annotated for TGFβ signaling (Datasets S1 and S2). Several cell types in CRC, including IECs and SEMFs, express TGFβ (9). TGFβ release into the tumor microenvironment induces SEMF differentiation, migration, and expression of genes (e.g., ANGPTL4, PTHLH, HBEGF, CTGF, IL11, and TNC) known to promote metastasis of the tumor epithelium (5, 11). Curiously, treatment of SEMFs with IL-33 increased the expression of these same TGFβ-regulated genes by 2–15-fold (P < 0.05). Importantly, IL-33 signaling also associated with TGFβ-induced gene expression in vivo. Immunolabeling ApcMin/+ polyp sections for tenacin-C (TNC), a downstream target of TGFβ in intestinal SEMFs (44), revealed robust localization of TNC to ACTA2-positive, periluminal SEMFs (Fig. 4C). Serial sections demonstrated that the TNC-positive SEMFs also express IL1RL1 and are adjacent to IL-33–positive tumor epithelia (Fig. 4C).
IL-33 Signaling Modulates Cytokine Expression in ApcMin/+ Polyps.
Because IL-33 is an important mediator of intestinal immunity and IBD-associated inflammation (22), we next tested whether tumor-elicited cytokine production is altered in the polyps of Il33-deficient ApcMin/+ mice. IL-33 deficiency did not alter the expression of the innate immune response cytokines tumor necrosis factor (Tnf) and interleukin 1 beta (Il1b), Th1 immune response cytokine interferon gamma (Ifng), or Th17 immune response interleukins (Il17a, Il17f, Il22, and Il23) (Fig. S9 A–G). However, expression of interleukins associated with Th2 immune responses (Il4 and Il13) and chronic and tumor-elicited intestinal inflammation (Il6) was significantly reduced in IL-33–deficient polyps (Fig. 5 A–C). Interestingly, IL-33 signaling deficiency also reduced expression of membrane-bound, but not -soluble, Il1rl1 (Fig. 1 D and E), suggesting that IL-33 regulates the isoform-specific expression of its own receptor in target cells or that IL-33 influences the recruitment or proliferation of cells that express the membrane-associated isoform. Similar changes in cytokine gene expression were observed in IL1RL1 antagonist antibody-treated ApcMin/+ polyps (Fig. 5 A–C and Fig. S9 A–G). Thus, abrogation of responsiveness to IL-33 appears to primarily influence Th2-associated cytokine production during polyposis.
Fig. 5.
Abrogation of responsiveness to IL-33 signaling attenuates Th2-associated cytokine expression in polyps. qPCR analysis of (A) Il4, (B) Il13, and (C) Il6 expression in normal mucosa of WT mice and in polyps of ApcMin/+ mice WT (+/+) or deficient (−/−) for Il33 or treated with an IgG control (Cnt) or Il1rl1 antagonist antibody (Ant). Expression is plotted relative to WT ± SEM (n = 5–10). †P ≤ 0.01; ‡P ≤ 0.001; #P ≤ 0.0001.
IL-33 Signaling Contributes to Polyp-Associated Mastocytosis.
Because IL-33 signaling deficiency suppressed Th2-associated cytokine expression during polyposis, we sought to identify a cell type associated with Th2 immune responses through which IL-33 signaling may promote polyposis. IL1RL1 is expressed in the mast cell lineage starting in committed progenitor cells and promotes mast cell maturation, survival, granule accumulation, tryptase production, and expression of Th2-associated cytokines (45, 46). Previous studies showed that mast cells accumulate in human and mouse intestinal adenomas and promote polyposis in Apc mutant mouse models by regulating IEC homeostasis, stimulating angiogenesis, and modulating immune responses within the tumor microenvironment (47–49). Thus, we investigated the effect of IL-33 signaling deficiency on mast cell activation and recruitment in ApcMin/+ polyps.
In mice, mast cells are historically divided into two populations, connective tissue mast cells and mucosal mast cells, which are distinguished by the differential expression of several chymases, tryptases, and proteases (50). Both cell types are found within intestinal mucosa (47). To test whether IL-33 signaling influences tumor-associated mast cell gene expression, we measured the expression of connective tissue mast cell-associated chymase and tryptase (Cma1 and Tpsb2) and mucosal mast cell-associated proteases (Mcpt4 and Mcpt2) in the polyps of Il33-deficient, IL1RL1 antibody-treated, and control ApcMin/+ mice (Fig. 6 A and B and Fig. S9 H and I). In the polyps of ApcMin/+,Il33+/+ and control antibody-treated ApcMin/+ mice, expression of connective tissue and mucosal mast cell-related genes was substantially increased 50–800-fold compared with normal, WT mucosa. Importantly, both genetic and antibody abrogation of responsiveness to IL-33 signaling caused a 10–70-fold reduction in mast cell-associated gene expression in polyps. To test whether IL-33 signaling directly induces mast cell gene expression, we coimmunolabeled polyp sections for IL1RL1 and either tryptase, a connective tissue mast cell marker, or mast cell protease 1 (MCPT1), a mucosal mast cell marker. Both tumor-associated tryptase-positive and MCPT1-positive mast cells expressed IL1RL1 (Fig. 6 C and D). Curiously, however, mast cells within normal intestinal mucosa and polyps of Il33-deficient ApcMin/+ mice did not express detectable levels of IL1RL1 (Fig. S10), suggesting that IL-33 induces the expression of its own receptor in mast cells during polyposis.
Fig. 6.
IL-33 signaling deficiency suppresses mast cell number and gene expression in polyps. (A and B) qPCR expression analysis of mast cell genes Cma1 and Mcpt4 in normal mucosa of WT mice and in polyps of 90-d-old ApcMin/+ mice WT (+/+) or deficient (−/−) for Il33, or treated with an IgG control (Cnt) or Il1rl1 antagonist (Ant) antibody. Expression is plotted relative to WT ± SEM (n = 5–10). (C and D) Colocalization of IL1RL1 (green) with the connective tissue mast cell markers tryptase and mucosal mast cell marker MCPT1 (red) in ApcMin/+ polyps. Nuclei were counterstained with DAPI. (Scale bars, 25 μm.) (E) Representative images of CAE- and hematoxylin-stained ApcMin/+ polyps WT or deficient for IL-33. Mast cells are identified as cells with intensely red-stained granules. (Scale bars, 50 μm.) (F) Mast cells per field were counted in high-magnification (400×) images of normal WT mucosa (n = 9) and IL-33 WT (+/+) (n = 24) and deficient (−/−) (n = 18) polyp sections stained for CAE. Bars indicate the mean.
The significant reduction of mast cell gene expression in Il33-deficient and antibody-treated polyps suggests that IL-33 influences mast cell activation, infiltration/expansion, or both. Therefore, we used chloroacetate esterase (CAE) staining (51) to identify and enumerate mast cells in polyps of Il33-deficient and control ApcMin/+ mice (Fig. 6E). Importantly, polyps from ApcMin/+,Il33−/− mice had fewer mast cells (fourfold reduction) compared with ApcMin/+,Il33+/+ controls (Fig. 6F). Curiously, the fourfold decrease in mast cell numbers resulting from IL-33 deficiency is substantially less than the 10–70-fold decrease in mast cell gene expression observed in the same polyps (Fig. 6 A and B and Fig. S9 H and I). IL-33 signaling, therefore, appears to influence both mast cell accumulation and activation during intestinal polyposis.
Discussion
Carcinogenesis commonly associates with suppression of cell-mediated cytotoxic (Th1) immunity and enhanced humoral (Th2) immunity (52). Deviation toward Th1 immunity promotes tumor rejection by targeting and eliminating neoplastic cells (53). Conversely, deviation toward Th2 immunity suppresses antitumor immunity and promotes wound-healing responses that enhance tumorigenesis (54–56). In patients with CRC, total numbers of Th1 lymphocytes are decreased, Th1-associated cytokine production is reduced, and Th2-associated cytokine production is elevated (57–59). As CRC progresses, skewing toward Th2 immunity becomes more significant with Th2 cytokine levels, predictive of tumor reoccurrence and decreased disease-free survival (59, 60). A similar imbalance between Th1- and Th2-mediated immunity has been observed in the polyps of ApcMin/+ mice (61). Compared with normal mucosa, Th1 cytokine expression is lower or unchanged and Th2 cytokine expression is increased in polyps. The mechanisms by which Th2 immunity is preferentially activated during intestinal tumorigenesis are not well defined. However, induction of prostaglandin-endoperoxide synthase 2 (COX-2) expression appears to play a role. COX-2–mediated production of prostaglandin E2 (PGE2) suppresses Th1 responses and promotes Th2 immunity in both human CRC and ApcMin/+ polyps (61–63). However, the mechanisms through which PGE2 enhances Th2 immunity remain unresolved. Curiously, PGE2 has been shown to induce IL-33 expression in dendritic cells (64). Thus, enhancement of IL-33 expression within the tumor microenvironment may be one mechanism through which PGE2 induces Th2-immune and wound-repair responses that promote intestinal tumorigenesis.
A small but expanding body of evidence connects IL-33 signaling and its enhancement of Th2 immunity and wound-healing responses to tumorigenesis in several tissues (29, 30, 65–69). In the present study, we demonstrate for the first time, to our knowledge, that the IL-33 signaling axis is also an important modulator of tumor initiation and progression in the intestine. In human and ApcMin/+ adenomas, adenomatous cells were the primary source of IL-33 expression, and stromal cells expressed IL1RL1. This cell type distribution of IL-33 and its receptor mirrors the distribution observed during mucosal type 2 immune and wound-healing responses to parasite infection (6) and fits a model in which tumor epithelial cells coopt mucosal immune and tissue repair responses to promote tumorigenesis (4, 5). However, other cell types in normal mucosa surrounding intestinal tumors (e.g., SEMFs) and, in rare instances, within adenomas (e.g., endothelial cells) expressed IL-33. Thus, we cannot exclude the possibility that these cells are an additional/alternative source of cytokine IL-33 in adenomas. A conditional knockout allele of IL-33 will be necessary to evaluate the relative contribution of IL-33 expression by various cell types to tumor initiation and progression in the ApcMin/+ mouse model.
The extrinsic or intrinsic factors that induce IL-33 expression in adenomatous cells remain to be identified. Several studies previously showed that cytokines and eicosanoids associated with polyposis, such as TNF, IL-1β, TGFβ, PGE2, and pathogen-associated molecular patterns, such as lipopolysaccharide (LPS), induce IL-33 expression and in some instances release a variety of cell types (25, 26, 28, 38, 64). However, the capacity of inflammatory molecules to induce CRC cell line expression and release of IL-33 has not been consistent among studies (25–27). Alternatively, intrinsic properties of tumor epithelial cells, alone or in combination with inflammatory signals, may induce IL-33 expression. In ApcMin/+ polyps, IL-33 expression was localized to adenomatous cells with abundant nuclear localization of β-catenin. Thus, loss of heterozygosity for Apc and the constitutive activation of Wnt signaling may significantly contribute to IEC expression of Il33 expression. In support of this observation, IL-33 expression is significantly reduced following RNAi-mediated knockdown of β-catenin in CRC cell lines DLD1 and LS174T (70).
Interestingly, we found that immunolabeling for IL-33 in the epithelium of human intestinal tumors decreased as tumors progressed to the carcinoma stage. Conversely, we observed little to no immunolabeling for IL1RL1 in adenomatous cells in human or mouse. However, some carcinoma cells were IL1RL1-positive. These results suggest that additional genetic alterations or changes in the microenvironment associated with carcinoma cause the epithelium to become responsive to IL-33. In fact, studies of head and neck squamous cell carcinoma (HNSCC), pancreatic carcinoma, and colon carcinoma cell lines suggest that IL-33 is an autocrine/paracrine mediator of carcinoma cell invasiveness and metastasis (30, 67, 68). Thus, at later stages of CRC progression, IL-33 may directly influence the tumorigenic properties of both the epithelium and stroma.
Polyposis also significantly altered SEMF expression of IL-33 and IL1RL1. SEMFs are the primary mesenchymal cell type in the stroma of adenomas, and their activation leads to the production and release of several cytokines, growth factors, ECM components, and angiogenesis factors to form a microenvironment favorable to tumor initiation, progression, and metastasis (5, 9). In our study, SEMFs surrounding crypts in normal mucosa and deep crypts under polyp lesions expressed IL-33. However, within polyp lesions, SEMFs expressed neither IL-33 nor the LacZ reporter of the IL-33 knockout allele. Instead, polyp-associated SEMFs expressed IL1RL1, which was surprising because previous studies of normal and chronically inflamed intestinal mucosa did not report IL1RL1 expression by SEMFs (24, 26). Previous studies of human chronic pancreatitis tissues showed subpopulations of myofibroblasts expressing either IL-33 or IL1RL1. In vitro, pancreatic myofibroblasts expressed IL-33 or IL1RL1 in response to different cytokines: IL-1β, TNF, and LPS induced IL-33 expression and IL-4 and IFN-γ induced IL1RL1 expression (38). Thus, based on these observations, it is feasible that a cytokine milieu specific to polyposis activates SEMF expression of IL1RL1.
Transcriptional profiling of the human SEMF cell line CCD-18Co demonstrated that IL-33 activates SEMF expression of factors that contribute to tumorigenesis (Dataset S1). In particular, IL-33 induced the expression of several growth factors, including fibroblast growth factors (FGF2 and FGF7), nerve growth factor (NGF), and amphiregulin (AREG), known to promote epithelial cell proliferation and tumor initiation and progression (9). Additionally, IL-33 stimulated SEMF expression of matrix metalloproteinases previously associated with CRC, including MMP1 and MMP3, which degrade collagens, activate growth factors, promote tumor invasion, and mediate angiogenesis (43). IL-33 induced the expression of several additional proangiogenic factors including VEGFC and IL8 (71, 72). Thus, the decreased proliferation, increased apoptosis, and reduced vascularization observed in Il33-deficient ApcMin/+ polyps may be attributed, in part, to a reduction in SEMF activation. Curiously, IL-33 also stimulates SEMF expression of TGFB1. TGFB1 is a known paracrine/autocrine mediator of SEMF differentiation and contributor to stromal cell expression of secreted and membrane-bound factors that promote carcinoma cell metastasis (11). SEMF expression of these same TGFβ-induced genes increased in response to IL-33, suggesting that IL-33 promotes metastasis through a TGFβ transcriptional program in the tumor stroma. Interestingly, Liu and coworkers (29) demonstrated that IL-33 acts on CRC cell lines to enhance carcinoma cell metastatic capacity. Thus, based on this previous study and our expression results, we propose that IL-33 signaling may alter the transcriptional profile of both carcinoma cells and tumor stromal cells to promote CRC metastasis.
In addition to influencing SEMF function, cytokine IL-33 promoted mastocytosis and mast cell activation in ApcMin/+ polyps. In the intestine, mast cells accumulate at sites of parasite infection to induce and participate in Th2-mediated immunity and wound healing (73, 74). Mast cells release a variety of cytokines, proteases, histamine, eicosanoids, and growth factors that influence diverse physiological and pathological processes including epithelial cell proliferation and barrier integrity, granulocyte recruitment, angiogenesis, wound healing, and fibrosis (75). It is through these factors that mast cells establish a microenvironment favorable to polyposis (76). Using Apc mutant and colitis-induced mouse models of CRC, mast cell recruitment and activation have been shown to promote IEC survival and proliferation and tumor-associated angiogenesis (47, 77). It is well recognized that IL-33 induces mast cell progenitor maturation and activates mast cell expression and release of cytokines (e.g., IL-4, IL-13, and IL-6), chemokines, proteases, and lipid mediators (46). Thus, our observation that Il33 deficiency reduced mast cell number and the expression of Il4, Il13, Il6, and several proteases in polyps is consistent with the established role for IL-33 in mast cell activation and suggests that IL-33 is an important mediator of polyp-associated mastocytosis.
In conclusion, we have demonstrated that the IL-33 signaling network is induced in human CRC and plays an important role in intestinal polyposis in the ApcMin/+ mouse model. Our observations imply a model in which the tumor epithelium is a source of cytokine IL-33 during polyposis and that IL-33 activates at least two cell types, SEMFs and mast cells, to form a tissue microenvironment favorable to polyposis. Whether these two cell types independently influence different aspects (e.g., proliferation versus angiogenesis) or stages (e.g., initiation versus growth) of polyposis or cooperatively induce several protumorigenic phenotypes remains to be resolved. However, because SEMF- and mast cell-derived factors (e.g., matrix metalloproteinases from SEMFs and chymases from mast cells) interact to regulate ECM remodeling and IEC homeostasis (78), cooperative functions during polyposis are possible. Importantly, results from our assessment of IL-33 expression in CRC patients agree with previous analyses (29, 30) and suggest that IL-33 plays a similar role in progression of the human disease. Significantly, the ability of an IL1RL1 antagonist antibody to reduce tumor initiation and growth in the ApcMin/+ mouse model suggests that IL-33 signaling may be a useful target for CRC prevention or therapy.
Materials and Methods
Human Tissues and Cell Lines.
For serum samples, patients with metastatic colon cancer treated at the University of Southern California/Norris Comprehensive Cancer Center (USC/NCCC) or the Los Angeles County + USC Medical Center (LAC+USCMC) between 1992 and 2003 were eligible for this study. This population included only metastatic or recurrent colon cancer patients. All patients in this study signed informed consents and enrolled in protocols designed to study the molecular determinants of colon cancer. These protocols permitted blood collection and/or tissue collection. All enrolled patients were followed with an institutional database. Patient information was collected through database review and retrospective chart review when additional patient information was necessary. A large number of the patients (220/318 = 69%) were initially treated at an outside institution until, because of failure to respond to previous treatment, they were referred to USC/NCCC or LAC+USCMC for future treatments. All 318 patients were enrolled in at least one chemotherapy clinical trial while being treated at this institution (USC/NCCC or LAC+USCMC). All patients were treated with 5-fluorouracil–based chemotherapy regimens. Response to chemotherapy was not investigated as an end point for this study. This is a heavily pretreated cohort, with 20 patients (6%) treated with one line of chemotherapy, 19 patients (6%) treated with two different chemotherapy regimens, 183 patents (58%) treated with three different chemotherapy regimens, and 96 patients (30%) treated with four chemotherapy regimens. Although the treatment regimens varied among patients, the majority of patients were exposed to similar chemotherapies. All 318 patients (100%) received treatment with 5-fluorouracil, 298 patients (94%) received treatment with 5-fluorouracil/irinotecan (CPT-11), and 279 patients (88%) received treatment with 5-fluorouracil/oxaliplatin. For RNA samples, the patient population from which colon adenocarcinoma and adjacent normal tissue biopsies were collected and cell lines were derived was previously described (79). Representative examples of colonic adenocarcinomas and corresponding normal colonic mucosa were obtained as discarded tissue samples from the Tissue Procurement Service of the Case Comprehensive Cancer Center under an Internal Review Board-approved protocol at University Hospitals Case Medical Center.
Quantification of IL-33 Serum Protein Levels.
Whole blood was drawn from patients with stage 4 metastatic, colorectal adenocarcinomas and healthy individuals using a 22-gauge syringe into red-topped vaccutainers and centrifuged for 10 min at 500 × g. Serum was collected from each sample, immediately aliquoted, and stored at –80 °C until further assayed. All studies were approved by the Internal Review Board of the Keck School of Medicine of USC. Human serum IL-33 concentrations were measured using the IL-33 (human) ELISA kit (Enzo Life Sciences).
Microarray Reanalysis of IL33 Expression in Human CRC.
Primary sources for the tumor expression data obtained from the Oncomine Database (www.oncomine.org) were as follows: Skrzypczak colorectal 1 and 2 (31) and Sabates-Bellver Colon (32). Data are available in the Gene Expression Omnibus (GEO) database [accession nos. GSE2091 (Skrzypczak colorectal 1 and 2) and GSE871 (Sabates-Bellver Colon) at www.ncbi.nlm.nih.gov/geo].
Animals.
C57BL/6J (B6, JR000664), C57BL/6J-ApcMin/+/J (B6.ApcMin/+, JR002020), and B6.C-Tg(CMV-cre)1Cgn/J (B6.CMV-Cre, JR006054) mice were purchased from The Jackson Laboratory. C57BL/6NCrl mice harboring an engineered Il33 knockout–LacZ knock-in allele (Il33tm1(KOMP)Vlcg) (Fig. S2A) were obtained from Regeneron through the Knockout Mouse Project (KOMP) and backcrossed for 10 generations to C57BL/6J. Before use in studies, breeding heterozygous B6-Il33tm1(KOMP)Vlcg/+ mice to B6.CMV-Cre mice removed the loxP flanked neomycin selectable marker in the knockout allele. PCR genotyping with primers Flox-1 5′-GCAATAGCATCACAAATTTCACA-3′, Flox-2 5′-CCCAAGTCCCGTCCTAAAAT-3′, and Flox-3 5′-TCGGTTGTTTTCTTGTTTTGC-3′ verified excision of the neomycin selection cassette (Fig. S2B). A PCR assay using primers WT-F 5′-TCCTGCCTCCCTGAGTACAT-3′, WT-R 5′-TTGCTCTTGGTCTTTTCCAGA-3′, KO-F 5′-AACAGTACTGAGATTTCAACAC-3′, and KO-R 5′-GTCTGTCCTAGCTTCCTCACTG-3′ was used to identify WT, heterozygous, and homozygous Il33 knockout mice (Fig. S2C).
Animals were housed in the Transgenic Mouse Facility at Baylor College of Medicine (BCM) with the exception of those used for antibody injection studies, which were housed at the Wolstein Research Facility at Case Western Reserve University (CWRU). Animals were maintained on a 12-h light/dark cycle and fed the Purina 5010 Lab Diet (CWRU) or the Harlan 2920x diet (BCM). For IL1RL1 antagonism studies, B6-ApcMin/+ were injected intraperitoneally with 150 μg of monoclonal anti-IL1RL1 (mu-IgG1-FC–anti-muST2, Amgen) or control IgG (4D2-mu-IgG1–anti-huAGP3-Pb, Amgen) twice a week from 21 to 90 d of age. For all studies, 90-d-old mice were euthanized by carbon dioxide, and the small and large intestines were removed, flushed with cold PBS, and cut longitudinally. Polyps were counted and their diameter was measured in the small intestine and colon using a Leica MZ10F Modular Stereomicroscope. All procedures were approved and conducted in compliance with Institutional Animal Care and Use Committee standards at BCM and CWRU.
qRT-PCR.
RNA from frozen colon adenocarcinoma tissues and adjacent control tissues was extracted using a GIT/cesium gradient. Size-matched (1 mm) polyps from B6.ApcMin/+ mice and normal mucosa from B6.Apc+/+ age-matched littermates were scraped from the ileum or jejunum of the small intestine, and RNA was extracted using the Qiagen RNeasy Mini Kit. RNA was reverse transcribed with the SuperScript First-Strand Synthesis System (Life Technologies), and qRT-PCR was performed using the Roche FastStart Universal SYBR Green Master Mix (04913850001). Relative expression was calculated using Rpl7 and 18s as a housekeeping gene for mouse and human tissues, respectively, and the ΔΔCt analysis method. Primer sequences are available upon request.
Immunohistochemistry.
Intestinal tissues (ileum and jejunum) from 90-d-old mice and human CRC biopsies (colon) were fixed in 4% (wt/vol) paraformaldehyde, embedded into paraffin blocks, and cut into 5-μm sections. Tissue sections were deparaffinized, processed for antigen retrieval in sodium citrate buffer (pH 6.0), and blocked in 5% (vol/vol) donkey serum, 3% (wt/vol) BSA, and 0.3% Triton X-100 in 1× PBS. Sections were incubated with primary antibodies (Table S1) overnight at 4 °C. For angiogenesis assays, antigen retrieval was performed in Target Retrieval Solution (S170084, Dako), and sections were incubated with primary antibody for 1.5 h at room temperature. For detection by fluorescence microscopy, sections were incubated with 1:400 dilutions of donkey anti-goat AlexaFluor 488 (A-11055, Life Technologies), donkey anti-mouse 594 (A-21203, Life Technologies), donkey anti-rabbit 594 (A21207, Life Technologies), donkey anti-rat 594 (A21209, Life Technologies), or donkey anti-chicken 594 (703-585-155, Jackson ImmunoResearch) secondary antibodies. For confocal microscopy, sections were incubated with 1:400 dilutions of donkey anti-goat AlexaFluor 488, donkey anti-mouse 555 (A31570, Life Technologies), or donkey anti-chicken 647 (703-606-155, Jackson ImmunoResearch) secondary antibodies. Nuclei were counterstained with DAPI. For secondary detection by immunohistochemistry, sections were incubated with 1:300 dilutions of biotin-labeled anti-rabbit (ab6801) or anti-goat (ab6884) antibodies and stained using streptavidin–horseradish peroxidase (HRP) (20774, Millipore) and 3,3′-diaminobenzidine (DAB) substrate (SK-4100, Vector Labs) according to the manufacturers’ instructions.
Proliferation and Apoptosis Assays.
For BrdU incorporation assays, mice were injected at 90 d of age with 400 μL of 10 mg/mL BrdU (BD Pharmingen). Four hours after injection, animals were euthanized and the small and large intestines were immediately removed, flushed with cold PBS, fixed overnight at 4 °C in 4% paraformaldehyde, and processed for paraffin embedding. Deparaffinized sections of polyps (5 μm) were immunolabeled for BrdU incorporation using the Millipore BrdU Cell Proliferation Kit (2750) according to the manufacturer’s instructions and counterstained with hematoxylin. For TUNEL assays, deparaffinized sections of polyps (5 μm) were labeled with the ApopTag Direct In Situ Apoptosis Detection Kit (Millipore, S7160) according to the manufacturer’s instructions. Digoxigenin-labeled NTPs and a fluorescein-conjugated antidigoxigenin antibody were used to visualize products of the terminal deoxynucleotidyl transferase (Tdt) reactions. Nuclei were counterstained with DAPI.
Human SEMF Cell Line Culture and Whole Transcriptome Sequencing (RNA-seq).
Cell line CCD18Co (ATCC, CRL-1459) was cultured in T-75 flasks in Eagle's minimum essential medium (EMEM) (112-018-101, Quality Biologicals) supplemented with 10% (vol/vol) FBS (10437, Life Technologies), 1× nonessential amino acids (11140, Life Technologies), and 100 U/mL penicillin–streptomycin (15140, Life Technologies) at 37 °C and 5% CO2. At 80% confluency, new medium supplemented with 10 ng/mL recombinant human IL-33 (3625-IL/CF, R&D Systems) or vehicle control (1× PBS) was added, and cells were cultured for an additional 6 h. RNA was processed in triplicate for each condition using the Qiagen RNeasy Mini Kit. Sequencing libraries were prepared and 6-plex barcoded with the Illumina TruSeq RNA Sample Prep Kit V2 and subjected to 100 cycles of paired-end sequencing on two flow cell lanes using an Illumina HiSEq. 2000 platform. Sequencing resulted in an average of 65 million paired-end reads per sample. RNA-seq data were assessed for quality using FastQC. Adapter sequences were removed and low-quality bases were trimmed from reads, reads were quality filtered, and sample reads were concatenated. The TopHat and Cufflinks V2.1.1 software packages were used to align and assemble reads using the UCSC hg19 genome and transcript annotation files (support.illumina.com/sequencing/sequencing_software/igenome.html) as references. Differential expression between control and IL-33–treated conditions was assessed using the Cuffdiff module of Cufflinks with a false discovery rate of 0.05. IL-33–regulated mRNAs were defined as those with more than a 1.5-fold higher or lower expression (P < 0.05). All software was accessed through the Galaxy Server (galaxyproject.org). Functional annotation of up- and down-regulated mRNAs (1.5-fold, P < 0.05) was performed using the KEGG Pathways and GO Biological Processes annotations accessed through GeneCodis (genecodis.cnb.csic.es) (80). Data are available in the GEO database (accession no. GSE62518).
CAE Staining.
Intestinal tissues were fixed in 4% paraformaldehyde, embedded in paraffin blocks, and cut into 5-μm sections. Deparaffinized sections were stained for CAE with the Naphthol-AS-D chloroacetate (specific esterase) kit (91C-1KT, Sigma) and counterstained with hematoxylin according to the manufacturer’s instructions.
Image Acquisition and Analysis.
Bright field and fluorescence microscopy images of tissue sections were collected with a Zeiss Axioplan2 microscope. Confocal images of sections were taken with a Nikon A1-Rs inverted Laser Scanning Microscope. For BrdU and TUNEL assays, nuclei were counted in images (magnification, 400×) with the ImageJ Software program and the plugin “ITCN” to determine total cell number. BrdU- and TUNEL-positive cells in sections were enumerated with the ImageJ plugin “Cell Counter,” and the percentage of positive cells per section was calculated in four polyp sections from four mice for each genotype (a total of 16 polyp sections analyzed per genotype). Vessel area density was calculated using methods previously described (47). Briefly, the ImageJ Software command “color threshold” was used to identify regions of vWF-positive cell staining and tissue background staining in 400× images of polyp sections. The total area occupied by vWF-positive vessels (≥60 μm2), including hollow structures, and background tissue staining was determined using the command “analyze particles.” Vessel area density was calculated as the percentage of the vWF-positive area divided by the total tumor area in four polyp sections from four mice for each genotype (a total of 16 polyp sections analyzed per genotype). CAE-positive cells in mucosal and polyp sections were counted (magnification, 400×) with the ImageJ plugin Cell Counter.
Statistical Analysis.
Analyses were performed with the Prism6 software. One-way ANOVA, 95% confidence intervals, with Bonferroni’s multiple comparisons posttests was used for analyses of mouse and human gene expression data. ELISA results were analyzed by unpaired t test with Welch’s correction. Polyp number, polyp size, CAE staining, BrdU incorporation, and TUNEL assay results were analyzed by unpaired t tests.
Supplementary Material
Acknowledgments
We thank Dr. Graham Casey (USC) for his assistance with identifying CRC patient serum samples and his helpful discussions. We also thank Kay Washington (Vanderbilt University) for assistance with tumor pathology. Finally, we thank Dirk Smith, PhD, and Amgen for providing the IL1RL1 antagonist and control antibodies. We thank the Baylor College of Medicine Genetically Engineered Mouse, Human Tissue Acquisition and Pathology, Genomic and RNA Profiling, and Integrated Microscopy Advanced Technologies Cores for assistance with mouse rederivation, tissue processing, next generation whole transcriptome sequencing, and confocal microscopy, respectively. Resources accessed through all cores were supported by National Institutes of Health (NIH)–National Cancer Institute Grant CA125123 to the Dan L. Duncan Cancer Center. Additionally, NIH Grants HD007495 and DK56338 and a grant from the John S. Dunn Gulf Coast Consortium for Chemical Genomics supported resources accessed through the Integrated Microscopy Core. This work was supported by the Caroline Wiess Law Fund for Research in Molecular Medicine (to J.D.H.), the De Gregorio Family Foundation (to T.T.P.), and NIH Grants CA150964 (to S.D.M.), RR12305 (to J.H.N.), and DK056762 (to T.T.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE62518).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422445112/-/DCSupplemental.
References
- 1.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Powell SM, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359(6392):235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 3.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1(1):55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 4.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138(6):2101–2114, e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 5.Powell DW, Adegboyega PA, Di Mari JF, Mifflin RC. Epithelial cells and their neighbors I. Role of intestinal myofibroblasts in development, repair, and cancer. Am J Physiol Gastrointest Liver Physiol. 2005;289(1):G2–G7. doi: 10.1152/ajpgi.00075.2005. [DOI] [PubMed] [Google Scholar]
- 6.Gause WC, Wynn TA, Allen JE. Type 2 immunity and wound healing: Evolutionary refinement of adaptive immunity by helminths. Nat Rev Immunol. 2013;13(8):607–614. doi: 10.1038/nri3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owens BM, Simmons A. Intestinal stromal cells in mucosal immunity and homeostasis. Mucosal Immunol. 2013;6(2):224–234. doi: 10.1038/mi.2012.125. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432(7015):332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264(1):169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 11.Calon A, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsushima H, et al. Circulating transforming growth factor beta 1 as a predictor of liver metastasis after resection in colorectal cancer. Clin Cancer Res. 2001;7(5):1258–1262. [PubMed] [Google Scholar]
- 13.Lopetuso LR, Scaldaferri F, Pizarro TT. Emerging role of the interleukin (IL)-33/ST2 axis in gut mucosal wound healing and fibrosis. Fibrogenesis Tissue Repair. 2012;5(1):18. doi: 10.1186/1755-1536-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood IS, Wang B, Trayhurn P. IL-33, a recently identified interleukin-1 gene family member, is expressed in human adipocytes. Biochem Biophys Res Commun. 2009;384(1):105–109. doi: 10.1016/j.bbrc.2009.04.081. [DOI] [PubMed] [Google Scholar]
- 15.Moussion C, Ortega N, Girard JP. The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: A novel ‘alarmin’? PLoS ONE. 2008;3(10):e3331. doi: 10.1371/journal.pone.0003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz J, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Carriere V, et al. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci USA. 2007;104(1):282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: Epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lüthi AU, et al. Suppression of interleukin-33 bioactivity through proteolysis by apoptotic caspases. Immunity. 2009;31(1):84–98. doi: 10.1016/j.immuni.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Chackerian AA, et al. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179(4):2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- 21.Hayakawa H, Hayakawa M, Kume A, Tominaga S. Soluble ST2 blocks interleukin-33 signaling in allergic airway inflammation. J Biol Chem. 2007;282(36):26369–26380. doi: 10.1074/jbc.M704916200. [DOI] [PubMed] [Google Scholar]
- 22.Lopetuso LR, Chowdhry S, Pizarro TT. Opposing functions of classic and novel IL-1 family members in gut health and disease. Front Immunol. 2013;4:181. doi: 10.3389/fimmu.2013.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphreys NE, Xu D, Hepworth MR, Liew FY, Grencis RK. IL-33, a potent inducer of adaptive immunity to intestinal nematodes. J Immunol. 2008;180(4):2443–2449. doi: 10.4049/jimmunol.180.4.2443. [DOI] [PubMed] [Google Scholar]
- 24.Beltrán CJ, et al. Characterization of the novel ST2/IL-33 system in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2010;16(7):1097–1107. doi: 10.1002/ibd.21175. [DOI] [PubMed] [Google Scholar]
- 25.Kobori A, et al. Interleukin-33 expression is specifically enhanced in inflamed mucosa of ulcerative colitis. J Gastroenterol. 2010;45(10):999–1007. doi: 10.1007/s00535-010-0245-1. [DOI] [PubMed] [Google Scholar]
- 26.Pastorelli L, et al. Epithelial-derived IL-33 and its receptor ST2 are dysregulated in ulcerative colitis and in experimental Th1/Th2 driven enteritis. Proc Natl Acad Sci USA. 2010;107(17):8017–8022. doi: 10.1073/pnas.0912678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seidelin JB, et al. IL-33 is upregulated in colonocytes of ulcerative colitis. Immunol Lett. 2010;128(1):80–85. doi: 10.1016/j.imlet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Sponheim J, et al. Inflammatory bowel disease-associated interleukin-33 is preferentially expressed in ulceration-associated myofibroblasts. Am J Pathol. 2010;177(6):2804–2815. doi: 10.2353/ajpath.2010.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui G, et al. Dynamics of the IL-33/ST2 network in the progression of human colorectal adenoma to sporadic colorectal cancer. Cancer Immunol Immunother. 2015;64(2):181–190. doi: 10.1007/s00262-014-1624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, et al. IL-33/ST2 pathway contributes to metastasis of human colorectal cancer. Biochem Biophys Res Commun. 2014;453(3):486–492. doi: 10.1016/j.bbrc.2014.09.106. [DOI] [PubMed] [Google Scholar]
- 31.Skrzypczak M, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabates-Bellver J, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5(12):1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 33.Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136(3):780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 34.Palmer G, et al. Inhibition of interleukin-33 signaling attenuates the severity of experimental arthritis. Arthritis Rheum. 2009;60(3):738–749. doi: 10.1002/art.24305. [DOI] [PubMed] [Google Scholar]
- 35.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1(5):a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi YS, et al. Interleukin-33 induces angiogenesis and vascular permeability through ST2/TRAF6-mediated endothelial nitric oxide production. Blood. 2009;114(14):3117–3126. doi: 10.1182/blood-2009-02-203372. [DOI] [PubMed] [Google Scholar]
- 37.Powell DW, et al. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol. 1999;277(2 Pt 1):C183–C201. doi: 10.1152/ajpcell.1999.277.2.C183. [DOI] [PubMed] [Google Scholar]
- 38.Nishida A, et al. Expression of interleukin 1-like cytokine interleukin 33 and its receptor complex (ST2L and IL1RAcP) in human pancreatic myofibroblasts. Gut. 2010;59(4):531–541. doi: 10.1136/gut.2009.193599. [DOI] [PubMed] [Google Scholar]
- 39.Andoh A, Bamba S, Brittan M, Fujiyama Y, Wright NA. Role of intestinal subepithelial myofibroblasts in inflammation and regenerative response in the gut. Pharmacol Ther. 2007;114(1):94–106. doi: 10.1016/j.pharmthera.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Andoh A, Bamba S, Fujiyama Y, Brittan M, Wright NA. Colonic subepithelial myofibroblasts in mucosal inflammation and repair: Contribution of bone marrow-derived stem cells to the gut regenerative response. J Gastroenterol. 2005;40(12):1089–1099. doi: 10.1007/s00535-005-1727-4. [DOI] [PubMed] [Google Scholar]
- 41.Andoh A, Fujino S, Okuno T, Fujiyama Y, Bamba T. Intestinal subepithelial myofibroblasts in inflammatory bowel diseases. J Gastroenterol. 2002;37(Suppl 14):33–37. doi: 10.1007/BF03326410. [DOI] [PubMed] [Google Scholar]
- 42.Hinterleitner TA, Saada JI, Berschneider HM, Powell DW, Valentich JD. IL-1 stimulates intestinal myofibroblast COX gene expression and augments activation of Cl- secretion in T84 cells. Am J Physiol. 1996;271(4 Pt 1):C1262–C1268. doi: 10.1152/ajpcell.1996.271.4.C1262. [DOI] [PubMed] [Google Scholar]
- 43.Zucker S, Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 2004;23(1-2):101–117. doi: 10.1023/a:1025867130437. [DOI] [PubMed] [Google Scholar]
- 44.Mukaratirwa S, de Witte E, van Ederen AM, Nederbragt H. Tenascin expression in relation to stromal tumour cells in canine gastrointestinal epithelial tumours. J Comp Pathol. 2003;129(2-3):137–146. doi: 10.1016/s0021-9975(03)00021-5. [DOI] [PubMed] [Google Scholar]
- 45.Lunderius-Andersson C, Enoksson M, Nilsson G. Mast cells respond to cell injury through the recognition of IL-33. Front Immunol. 2012;3:82. doi: 10.3389/fimmu.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enoksson M, et al. Mast cells as sensors of cell injury through IL-33 recognition. J Immunol. 2011;186(4):2523–2528. doi: 10.4049/jimmunol.1003383. [DOI] [PubMed] [Google Scholar]
- 47.Gounaris E, et al. Mast cells are an essential hematopoietic component for polyp development. Proc Natl Acad Sci USA. 2007;104(50):19977–19982. doi: 10.1073/pnas.0704620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blatner NR, et al. In colorectal cancer mast cells contribute to systemic regulatory T-cell dysfunction. Proc Natl Acad Sci USA. 2010;107(14):6430–6435. doi: 10.1073/pnas.0913683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheon EC, et al. Mast cell 5-lipoxygenase activity promotes intestinal polyposis in APCDelta468 mice. Cancer Res. 2011;71(5):1627–1636. doi: 10.1158/0008-5472.CAN-10-1923. [DOI] [PubMed] [Google Scholar]
- 50.Gurish MF, Boyce JA. Mast cells: Ontogeny, homing, and recruitment of a unique innate effector cell. J Allergy Clin Immunol. 2006;117(6):1285–1291. doi: 10.1016/j.jaci.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 51.Friend DS, et al. Mast cells that reside at different locations in the jejunum of mice infected with Trichinella spiralis exhibit sequential changes in their granule ultrastructure and chymase phenotype. J Cell Biol. 1996;135(1):279–290. doi: 10.1083/jcb.135.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalgleish AG, O’Byrne KJ. Chronic immune activation and inflammation in the pathogenesis of AIDS and cancer. Adv Cancer Res. 2002;84:231–276. doi: 10.1016/s0065-230x(02)84008-8. [DOI] [PubMed] [Google Scholar]
- 53.Kidd P. Th1/Th2 balance: The hypothesis, its limitations, and implications for health and disease. Altern Med Rev. 2003;8(3):223–246. [PubMed] [Google Scholar]
- 54.Terabe M, Park JM, Berzofsky JA. Role of IL-13 in regulation of anti-tumor immunity and tumor growth. Cancer Immunol Immunother. 2004;53(2):79–85. doi: 10.1007/s00262-003-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romagnani S. The Th1/Th2 paradigm. Immunol Today. 1997;18(6):263–266. doi: 10.1016/s0167-5699(97)80019-9. [DOI] [PubMed] [Google Scholar]
- 56.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12(3):170–180. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 57.O’Hara RJ, et al. Advanced colorectal cancer is associated with impaired interleukin 12 and enhanced interleukin 10 production. Clin Cancer Res. 1998;4(8):1943–1948. [PubMed] [Google Scholar]
- 58.Pellegrini P, et al. Disregulation in TH1 and TH2 subsets of CD4+ T cells in peripheral blood of colorectal cancer patients and involvement in cancer establishment and progression. Cancer Immunol Immunother. 1996;42(1):1–8. doi: 10.1007/s002620050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibata M, et al. Decreased production of interleukin-12 and type 2 immune responses are marked in cachectic patients with colorectal and gastric cancer. J Clin Gastroenterol. 2002;34(4):416–420. doi: 10.1097/00004836-200204000-00006. [DOI] [PubMed] [Google Scholar]
- 60.Kurzrock R. Cytokine deregulation in cancer. Biomed Pharmacother. 2001;55(9-10):543–547. doi: 10.1016/s0753-3322(01)00140-8. [DOI] [PubMed] [Google Scholar]
- 61.Nakanishi Y, et al. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in ApcMin/+ mouse polyps. Carcinogenesis. 2011;32(9):1333–1339. doi: 10.1093/carcin/bgr128. [DOI] [PubMed] [Google Scholar]
- 62.Kojima M, et al. Association of enhanced cyclooxygenase-2 expression with possible local immunosuppression in human colorectal carcinomas. Ann Surg Oncol. 2001;8(5):458–465. doi: 10.1007/s10434-001-0458-x. [DOI] [PubMed] [Google Scholar]
- 63.Kalinski P. Regulation of immune responses by prostaglandin E2. J Immunol. 2012;188(1):21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagawa Y, Suzuki M, Matsumoto M, Togashi H. Prostaglandin E(2) enhances IL-33 production by dendritic cells. Immunol Lett. 2011;141(1):55–60. doi: 10.1016/j.imlet.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Jovanovic I, et al. ST2 deletion enhances innate and acquired immunity to murine mammary carcinoma. Eur J Immunol. 2011;41(7):1902–1912. doi: 10.1002/eji.201141417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jovanovic IP, et al. Interleukin-33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int J Cancer. 2014;134(7):1669–1682. doi: 10.1002/ijc.28481. [DOI] [PubMed] [Google Scholar]
- 67.Chen SF, et al. The paracrine effect of cancer-associated fibroblast-induced interleukin-33 regulates the invasiveness of head and neck squamous cell carcinoma. J Pathol. 2013;231(2):180–189. doi: 10.1002/path.4226. [DOI] [PubMed] [Google Scholar]
- 68.Schmieder A, Multhoff G, Radons J. Interleukin-33 acts as a pro-inflammatory cytokine and modulates its receptor gene expression in highly metastatic human pancreatic carcinoma cells. Cytokine. 2012;60(2):514–521. doi: 10.1016/j.cyto.2012.06.286. [DOI] [PubMed] [Google Scholar]
- 69.Li J, et al. Biliary repair and carcinogenesis are mediated by IL-33-dependent cholangiocyte proliferation. J Clin Invest. 2014;124(7):3241–3251. doi: 10.1172/JCI73742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbst A, et al. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics. 2014;15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 72.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14(21):6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 73.Voehringer D. Protective and pathological roles of mast cells and basophils. Nat Rev Immunol. 2013;13(5):362–375. doi: 10.1038/nri3427. [DOI] [PubMed] [Google Scholar]
- 74.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10(6):440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bischoff SC. Physiological and pathophysiological functions of intestinal mast cells. Semin Immunopathol. 2009;31(2):185–205. doi: 10.1007/s00281-009-0165-4. [DOI] [PubMed] [Google Scholar]
- 76.Maltby S, Khazaie K, McNagny KM. Mast cells in tumor growth: Angiogenesis, tissue remodelling and immune-modulation. Biochim Biophys Acta. 2009;1796(1):19–26. doi: 10.1016/j.bbcan.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wedemeyer J, Galli SJ. Decreased susceptibility of mast cell-deficient Kit(W)/Kit(W-v) mice to the development of 1, 2-dimethylhydrazine-induced intestinal tumors. Lab Invest. 2005;85(3):388–396. doi: 10.1038/labinvest.3700232. [DOI] [PubMed] [Google Scholar]
- 78.Groschwitz KR, Wu D, Osterfeld H, Ahrens R, Hogan SP. Chymase-mediated intestinal epithelial permeability is regulated by a protease-activating receptor/matrix metalloproteinase-2-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2013;304(5):G479–G489. doi: 10.1152/ajpgi.00186.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 80.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8(1):R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.