Significance
Ewing sarcoma protein (EWS) is essential for various cellular processes but its role in energy metabolism has not been demonstrated. This study identifies a new role of EWS in mitochondria and cellular energy homeostasis. Loss of EWS leads to increased degradation of PGC-1α, a critical protein in maintaining mitochondrial function and energy metabolism. The increased degradation is due to an increased expression of FBXW7, a protein that promotes PGC-1α degradation. In mice lacking EWS, significant decreases in PGC-1α levels and in mitochondria number and function are observed in brown fat, skeletal muscles, and liver. A novel role in mitochondrial and energy homeostasis provides new insights into the expanding role of EWS and suggests an altered energy metabolism in tumors with EWS translocations.
Keywords: EWS, PGC-1alpha, protein stability, mitochondria homeostasis, energy metabolism
Abstract
EWS (Ewing sarcoma) encodes an RNA/ssDNA binding protein that is frequently rearranged in a number of different cancers by chromosomal translocations. Physiologically, EWS has diverse and essential roles in various organ development and cellular processes. In this study, we uncovered a new role of EWS in mitochondrial homeostasis and energy metabolism. Loss of EWS leads to a significant decrease in mitochondria abundance and activity, which is caused by a rapid degradation of Peroxisome proliferator-activated receptor γ Coactivator (PGC-1α), a central regulator of mitochondria biogenesis, function, and cellular energy metabolism. EWS inactivation leads to increased ubiquitination and proteolysis of PGC-1α via proteasome pathway. Complementation of EWS in Ews-deficient cells restores PGC-1α and mitochondrial abundance. We found that expression of E3 ubiquitin ligase, FBXW7 (F-box/WD40 domain protein 7), is increased in the absence of Ews and depletion of Fbxw7 in Ews-null cells restores PGC-1α expression and mitochondrial density. Consistent with these findings, mitochondrial abundance and activity are significantly reduced in brown fat and skeletal muscles of Ews-deficient mice. Furthermore, expression of mitochondrial biogenesis, respiration and fatty acid β-oxidation genes is significantly reduced in the liver of Ews-null mice. These results demonstrate a novel role of EWS in mitochondrial and cellular energy homeostasis by controlling PGC-1α protein stability, and further implicate altered mitochondrial and energy metabolism in cancers harboring the EWS translocation.
Ewing sarcoma breakpoint region 1 (EWSR1, herein termed EWS) was first identified from the Ewing sarcoma chromosomal breakpoint t(11, 22)(q24;q12) region as a translocation-generated fusion gene between EWS and FLI1 (Friend leukemia integration 1) (1), an ETS-family of transcription factor. EWS is a member of the FET (or TET) family of RNA and ssDNA-binding proteins, which includes two other members, FUS/TLS (Fused in Sarcoma/Translocated in Liposarcoma) and TAF15/hTAFII68 (TATA-binding protein-associated factor 15/human TATA-binding protein-associated factor II 68) as well as a Drosophila protein, Cabeza/SARFH (1, 2). A transcriptional role of EWS has been inferred by its association with basic transcription factors (3, 4) and by its modulation of several transcription factor activities (5–8). EWS is also involved in alternative splicing of specific genes in response to DNA damage (9, 10). Animal studies with genetic ablation of Ews have revealed a surprisingly diverse role of EWS in various cellular processes such as pre-B lymphocyte development, meiosis, mitosis and prevention of premature cellular senescence in fibroblasts and hematopoietic stem cells (11–13). Recently, it was shown that EWS is required for determining embryonic brown fat cell fate during development (8), for in vitro white fat differentiation (14), and in the regulation of microRNAs (15).
The transcriptional coactivator PGC-1α is critical for many biological processes that require high oxidative metabolism and is induced by changes in nutrient, environmental and physical status (16, 17). PGC-1α plays essential roles in mitochondrial biogenesis, fatty acid β-oxidation, gluconeogenesis, and heme biosynthesis (18, 19). A close homolog, PGC-1β, has partially redundant roles in many of these processes (18, 19), but is unable to fully compensate for the loss of PGC-1α in maintaining expression of a large number of mitochondrial genes. Therefore, mitochondrial gene expression and functions are reduced in tissues that lack PGC-1α (20, 21).
PGC-1α is regulated at both transcriptional and posttranscriptional levels. PGC-1α is highly expressed in tissues that demand high oxidative metabolism, such as brain, heart and skeletal muscle, whereas its expression is rapidly induced in tissues that are metabolically activated following hormonal or environmental stimuli (22–24). At the protein level, PGC-1α is intrinsically unstable and is degraded by the ubiquitin–proteasome pathway at basal level (25–27). An E3 ligase FBXW7 (F-box/WD40 domain protein7, also termed SCFCdc4) was shown to bind and ubiquitinate PGC-1α in p38 MAPK- and GSK3β-dependent manner (27). These features allow rapid accumulation and activation of PGC-1α by modulating its protein stability and transcription in response to various environmental and dietary conditions. In this study, we present evidence for a new role of EWS in mitochondrial homeostasis through modulation of PGC-1α protein stability.
Results
Loss of Ews in BAT Leads to Reduced Mitochondria Abundance and Structural Anomalies.
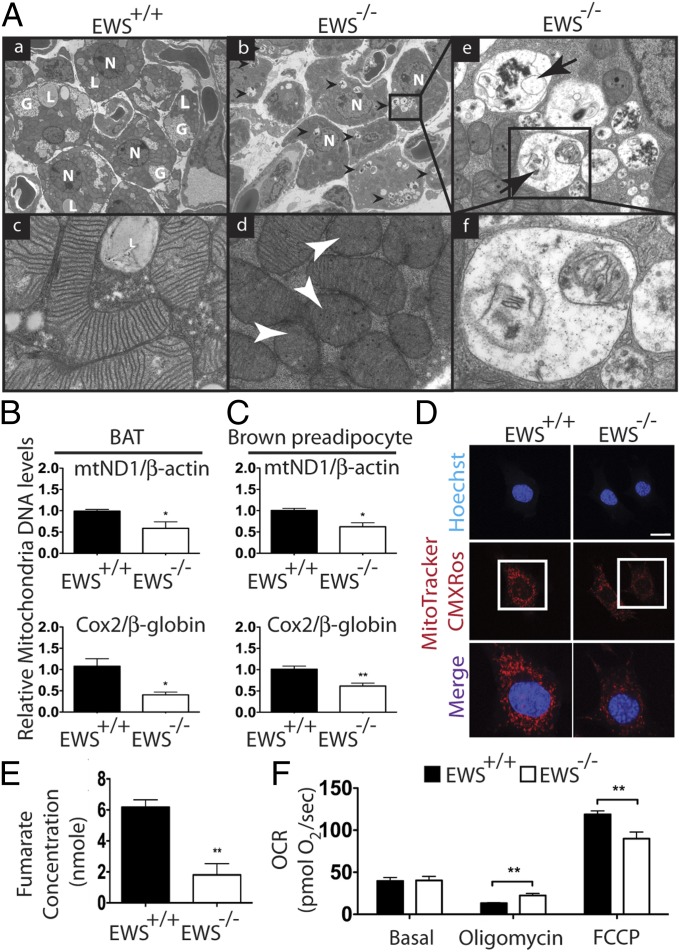
We have recently shown that EWS is required for determining the classical brown fat lineage during development (8). Notably, transmission electron microscopy (TEM) of interscapular brown adipose tissue (iBAT) from newborn Ews−/− (EWS-KO) mice revealed striking morphological anomalies in mitochondria. Although adipocytes from wild-type iBAT contained abundant mitochondria with prominent, densely packed cristae (Fig. 1A a and c), EWS-KO brown fat cells contained fewer mitochondria with less prominent and sparse or absent cristae (Fig. 1A b and d, white arrowheads). The difference in mitochondria abundance was confirmed by quantification of mitochondrial DNA normalized to nuclear DNA (Fig. 1B). Intriguingly, many Ews-null brown fat cells, but not any wild-type cells, contained cytoplasmic vesicles enveloping dark and membraneous structures (Fig. 1A b, black arrowheads). At higher magnifications, structures that resembled remnants of mitochondria with partially degraded cristae were observed inside the vesicles (Fig. 1A e, arrows, and f). A careful inspection revealed that the majority of vesicles were comprised of a single membrane (Fig. 1A f), not the characteristic double membrane of an autophagosome. Furthermore, the ratio of LC3-I and LC3-II and the total LC3-II levels were similar in wild-type or EWS-KO iBAT and brown preadipocytes (Fig. S1 A and B). These results demonstrate that the mitochondria in Ews-deficient brown fat cells are grossly abnormal and are likely undergoing non-autophagy-mediated degradation.
Fig. 1.
Ews inactivation leads to abnormal mitochondria morphology, density and function. (A) TEM of iBATs from Ews+/+ (a and c) and Ews−/− (b and d–f) P0.5 newborn pups (n = 3). (a and b) Cytoplasmic vesicles observed only in Ews−/− cells are indicated by black arrowheads (b). G, glycogen; L, lipid droplets; N, nucleus. Magnification, 4,800×. (c and d) Representative images of mitochondria from Ews+/+ (c) and Ews−/− (d) cells. Mitochondria with sparse cristae are indicated by white arrowheads. Magnification, 60,000×. (e) Higher magnification view (30,000×) of the boxed area in b. Arrows indicate vesicles containing remnants of mitochondria with partial or no cristae. (f) Higher magnification view (120,000×) of the boxed area in e. Cytoplasmic vesicles are enwrapped by single membrane. (B and C) Relative mitochondria DNA quantification of iBAT from P0.5 newborns (n = 3; B) and brown preadipocytes (C). (D) Live brown preadipocytes were stained with MitoTracker Red CMXRos and Hoechst. Representative confocal images are shown. Merged fields contain higher magnification views of the boxed areas. (Scale bar: 20 μm.) (E) Fumarate levels were analyzed in Ews+/+ or Ews−/− brown preadipocytes (n = 2). (F) Ews+/+ or Ews−/− brown preadipocytes (1 × 106) were measured for mitochondria respiration (six replicates). Data in B, C, E, and F are represented as mean ± SEM. Student’s t test, two-tailed, *P < 0.05, **P < 0.01.
Ews Inactivation Leads to Significant Reduction in Mitochondrial Density and Function.
The abnormal mitochondria morphology and abundance in iBAT could be indirectly related to the developmental arrest of classical brown fat in EWS-KO mice (8). Thus, we examined mitochondrial density in immortalized brown preadipocytes before differentiation. In EWS-KO preadipocytes, there was a significant decrease in the number of mitochondria compared with wild-type cells as determined by mitochondria DNA quantification (Fig. 1C). A decline in mitochondrial membrane potential was also evident in the KO cells as shown by MitoTracker Red CMXRos staining (Fig. 1D), a fluorescent dye that specifically accumulates in active mitochondria (28). To determine whether silencing EWS affects mitochondria density in other cell types, we depleted Ews in C2C12 myoblasts using lentivirus expressing Ews shRNA (Fig. S1C). Similar to brown preadipocytes, depletion of Ews in C2C12 myoblasts also resulted in a marked reduction in mitochondria abundance (Fig. S1D).
We next examined whether Ews inactivation also affects mitochondrial function. Expression of essential mitochondrial genes (Cox7a, Cox8b, and Tfam) and the levels of fumarate, a tricarboxylic acid (TCA) cycle intermediate formed by oxidation of succinate, were significantly decreased in EWS-KO brown preadipocytes compared with control cells (Fig. S1E and Fig. 1E). Measurement of oxygen consumption rates revealed that EWS-KO cells have a higher mitochondrial leak respiration (oligomycin-treated) and a lower maximal mitochondrial respiration capacity (FCCP-treated) compared with the control cells (Fig. 1F). Basal mitochondria respiration rate was unaltered. Collectively, these results demonstrate a critical role of EWS in the maintenance of mitochondrial density and function.
EWS Inactivation Leads to Reduced PGC-1α Protein Levels.
PGC-1α is a central protein in mitochondria biogenesis and function (18, 19), and exists in several different isoforms due to alternative promoter use and splice sites as well as posttranslational modifications (29–32). To detect PGC-1α, we used three different antibodies: a mouse monoclonal (4C1.3) antibody against the N terminus of PGC-1α that recognizes a 113-kDa protein (30) or rabbit polyclonal antibodies that recognize smaller 82–90kDa PGC-1α either in the first 300 amino acids (H-300, Santa Cruz) (33) or in the C terminus (amino acids 777–797) (C-term, Millipore). The exact modifications that produce different molecular species of PGC-1α are unknown. Consistent with published reports (29, 30, 33), we detected the 113-kDa and 82- to 90-kDa PGC-1α using the respective PGC-1α antibodies, which were absent in Pgc-1α–deficient cells (Fig. S2 A and B). The specificity of H-300 antibody was also demonstrated by immunoprecipitation (IP)-Western blot and mass spectrometry analyses (Fig. S2 C and D).
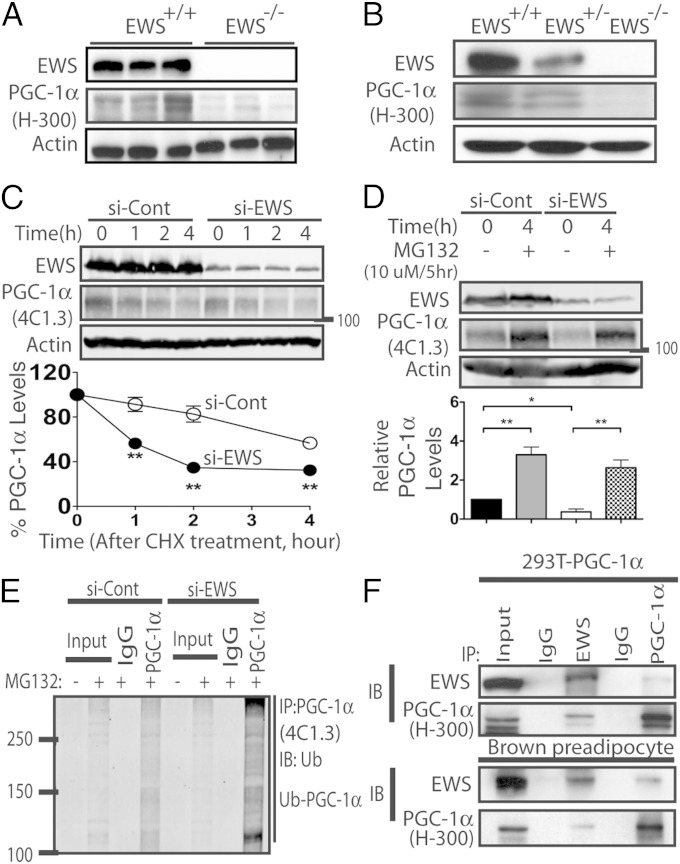
Expression of PGC-1α protein was readily detected in the iBATs from Ews+/+ newborns (P0.5) but was dramatically decreased in EWS-KO iBATs (Fig. 2A). We observed a similar reduction in PGC-1α protein levels in EWS-KO brown adipocytes compared with wild-type (Fig. 2B). In contrast, only a modest decrease in Pgc-1α mRNA levels was observed in EWS-KO cells (Fig. S3A), suggesting that reduction in PGC-1α protein was occurring mainly at the posttranscriptional level. The levels of PGC-1β mRNA and protein were similar or slightly increased in EWS-KO cells (Fig. S3 B and C).
Fig. 2.
Silencing EWS leads to enhanced degradation and ubiquitination of PGC-1α. (A and B) Western blot analysis of EWS and PGC-1α in iBAT of congenic C57BL6 newborn (P0.5) littermates (n = 3; A) and in immortalized brown adipocytes (B). Actin was used as a loading control. (C) HEK293-FLAG-PGC-1α cells were transfected with control (si-Cont) or EWS siRNA (si-EWS) and PGC-1α protein half-life was measured by cycloheximide (CHX)-chase and Western blotting. (Lower) Quantification of PGC-1α from three independent experiments. Data are represented as mean ± SEM. Student’s t test, two-tailed, **P < 0.01. (D) HEK293-FLAG-PGC-1α cells transfected with si-Cont or si-EWS with or without MG132 were harvested and analyzed by Western blotting. A representative blot is shown and quantification data from three independent experiments are represented as mean ± SEM. Student’s t test, two-tailed. (E) HEK293-FLAG-PGC-1α cells transfected with siRNA as in D were immunoprecipitated (IP) with anti-PGC-1α (4C1.3) and immunoblotted with anti-Ubiquitin. (F) HEK293-FLAG-PGC-1α cells (Upper) or wild-type brown preadipocytes (Lower) were immunoprecipitated with either anti-IgG, anti-EWS, or anti-PGC-1α and immunoblotted with the indicated antibodies.
The above results suggest that loss of EWS could be directly affecting the PGC-1α protein level. To test this, we used human embryonic kidney 293 (HEK293) cells stably expressing FLAG-tagged PGC-1α. We silenced EWS using siRNA or a scrambled control and PGC-1α protein half-life was analyzed by cycloheximide-chase experiment. In control siRNA-treated cells, PGC-1α protein levels decreased steadily over time following cycloheximide treatment, whereas silencing EWS resulted in a more rapid loss (t1/2 < 2h) of 113kDa PGC-1α (Fig. 2C) as well as 85–90kDa PGC-1α (Fig. S4A). Addition of proteasome inhibitor MG132 completely blocked PGC-1α proteolysis (Fig. 2D), suggesting that in cells lacking EWS, PGC-1α is degraded more rapidly via the proteasome pathway.
Increased Ubiquitination and Degradation of PGC-1α in the Absence of EWS.
We next examined PGC-1α ubiquitination in EWS depleted cells by IP-Western blot analysis. In HEK293 cells expressing FLAG-PGC-1α, control siRNA transfection showed a modest level of PGC-1α polyubiquitination (Fig. 2E). In contrast, EWS depletion resulted in a marked increase in polyubiquitinated PGC-1α. The use of different IP-Western blot antibodies revealed similar findings (Fig. S4 B and C). These results raised a possibility that EWS might regulate PGC-1α protein stability through protein-protein interaction. Indeed, endogenous EWS interacted, albeit weakly, with FLAG-PGC-1α in HEK293 cells (Fig. 2F, Upper) and with endogenous PGC-1α in wild-type brown preadipocytes (Fig. 2F, Lower).
PGC-1α protein turnover was recently examined in C2C12 myoblasts (34). Thus, we assessed the endogenous PGC-1α protein half-life in C2C12 myoblasts following Ews depletion. Silencing Ews with short-hairpin RNA (shRNA) expressed from lentivirus in C2C12 cells led to a dramatic decrease in PGC-1α protein stability (Fig. S4D). Similarly, depletion of Ews in primary mouse myoblasts also resulted in a severe loss of PGC-1α (Fig. S4E). These results clearly demonstrate that loss of EWS leads to a significant decrease in PGC-1α protein stability.
Complementation of EWS Restores PGC-1α Protein Levels and Mitochondria Abundance.
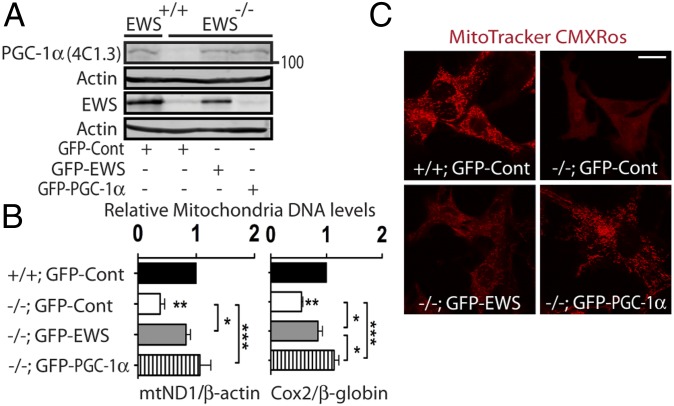
To test whether restoring EWS in EWS-KO cells leads to increased PGC-1α protein levels, we transduced EWS-KO brown preadipocytes with lentivirus expressing GFP alone, EWS and GFP or PGC-1α and GFP (driven by separate promoters). Reintroduction of EWS in EWS-KO cells markedly restored PGC-1α protein levels (Fig. 3A) and mitochondria abundance (Fig. 3B), as did PGC-1α. Ectopic expression of EWS in wild-type brown preadipocytes only modestly increased PGC-1α protein levels (Fig. S5A), but mitochondria abundance remained unaltered (Fig. S5B). Staining of live cells with MitoTracker RedCMXRos also showed restoration of mitochondrial membrane potential in EWS-complemented or PGC-1α–complemented EWS-KO cells (Fig. 3C and Fig. S6). These results demonstrate that EWS is essential in maintaining PGC-1α protein stability and mitochondrial homeostasis.
Fig. 3.
Complementation of EWS or PGC-1α rescues mitochondria abundance and activity in EWS-KO brown preadipocytes. (A) EWS-KO brown preadipocytes were transduced with lentivirus expressing GFP (control), EWS, or PGC-1α, and immunoblotted with antibodies against PGC-1α, EWS and α-Actin. (B) Brown preadipocytes transduced with different lentiviruses as in (A) were analyzed for relative amount of mitochondria DNA. Data from three independent experiments are represented as mean ± SEM. One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001. (C) EWS-KO brown preadipocytes were transduced with indicated lentiviruses and stained with MitoTracker Red CMXRos. Representative images from three independent experiments are shown. (Scale bar: 20 μm.)
FBXW7 E3 Ligase Is Involved in Degradation of PGC-1α in EWS-KO Cells.
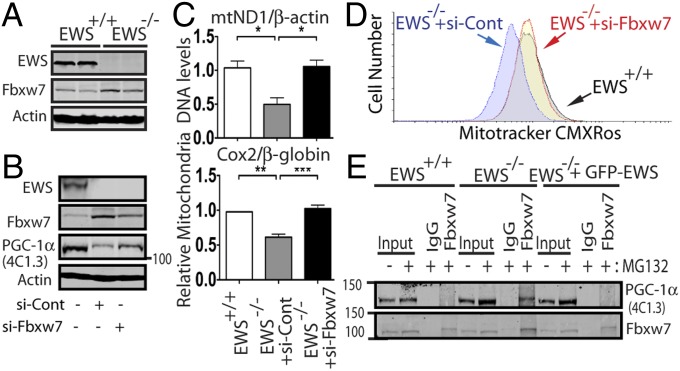
A previous study has identified FBXW7 as the E3 ligase responsible for PGC-1α ubiquitination and degradation (27). Notably, we observed a marked increase in the expression of Fbxw7 in EWS-KO cells compared with controls (Fig. 4A). Depletion of Fbxw7 using siRNA, but not siRNA control, increased PGC-1α protein levels (Fig. 4B) and also led to an increase in mitochondria abundance (Fig. 4C), mitochondrial membrane potential (Fig. 4D), and reduced polyubiquitination of PGC-1α (Fig. S5C). Consistent with these findings, there was an increase in the Fbxw7–PGC-1α interaction in EWS-KO cells compared with control cells, which was mitigated by reintroduction of EWS (Fig. 4E). These results demonstrate that elevated Fbxw7 expression is at least partially responsible for the rapid decrease in PGC-1α and mitochondria abundance in EWS-KO cells.
Fig. 4.
FBXW7 is responsible for the degradation of PGC-1α in the absence of EWS. (A) Western blot analysis of EWS, FBXW7, and Actin in wild-type and EWS-KO brown preadipocytes (n = 2). (B) Western blot analysis of EWS, FBXW7, PGC-1α and Actin in wild-type and EWS-KO brown preadipocytes transfected with si-Cont- or si-Fbxw7. (C) Relative mitochondria DNA quantification in wild-type and EWS-KO brown preadipocytes transfected with si-Cont- or si-Fbxw7. Data from three independent experiments are represented as mean ± SEM. One-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001. (D) Flow cytometry of siRNA-transfected brown preadipocytes (as in C) stained with MitoTracker Red CMXRos. (E) Ews+/+, Ews−/−, and Ews−/− brown preadipocytes complemented with lentivirus expressing EWS were immunoprecipitated with anti-Fbxw7 and immunoblotted with anti-PGC-1α (Upper) or anti-Fbxw7 (Lower).
Inactivation of Ews Leads to Reduced Mitochondrial Activity in Skeletal Muscle.
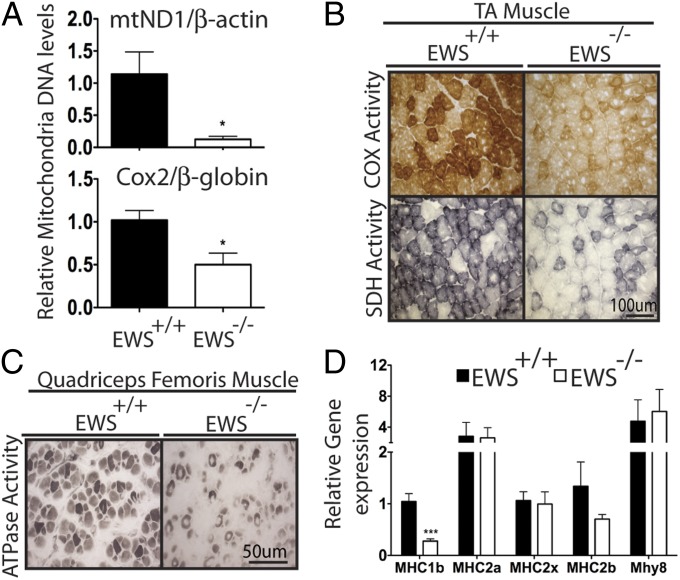
PGC-1α plays an essential role in skeletal muscle composition and physiology (31). Skeletal muscles from Pgc-1α–deficient mice contain fewer slow-twitch muscle fibers and decreased exercise capacity (35). Conversely, transgenic mice expressing PGC-1α in skeletal muscle show an enrichment of slow muscle fibers and greater endurance in exercise training (36). Ews-deficient mice in a congenic C57BL6 strain show complete postnatal lethality within 24 h after birth (8), whereas about 10% of EWS-KO mice survive to adulthood in a 129SvEv/Black Swiss mixed background (11). Therefore, we first examined the abundance of mitochondria in tibialis anterior muscle (TA) of 9- to 12-wk-old wild-type and EWS-KO mice. In the TA muscle of EWS-KO mice, there was a significant reduction in the number of mitochondria, as measured by mitochondria DNA, compared with the TA muscles from widltype littermates (Fig. 5A). TA muscles from EWS-KO mice also contained fewer muscle fibers with high mitochondrial cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) activities compared with controls (Fig. 5B).
Fig. 5.
Decrease in mitochondria abundance, mitochondria activity and slow myosin heavy chain expression in skeletal muscle of EWS-KO mice. (A) Relative mitochondria DNA quantification in tibialis anterior (TA) muscle of Ews+/+ and Ews−/− mice (9-12 wk; n = 4). (B) Representative images of TA muscles cross-sectioned and stained for COX or SDH activity (n = 3). (C) Representative images of quadriceps muscle of P0.5 congenic C57BL6 mouse cross-sectioned and stained for ATPase activity (n = 3). (D) qRT-PCR analysis of slow- and fast-specific myosin heavy chain genes from muscles isolated from whole hind legs of P0.5 mice (n = 3). MHCIb: slow fiber; MHCIIa, MHCIIx, MHCIIb: fast fiber; Mhy8: perinatal MHC. Data in A and D are represented as mean ± SEM. Student’s t test, two-tailed, **P < 0.01, ***P < 0.001.
To confirm these results, we examined quadriceps muscles from congenic C57BL6 Ews+/+ and Ews−/− newborn pups (P0.5). Consistent with our previous findings, quadriceps muscles from Ews−/− newborns contained markedly reduced mitochondrial ATPase, COX, and SDH activities compared with littermate controls (Fig. 5C and Fig. S7A). To examine the relative amount of slow and fast muscle fibers, we analyzed slow- and fast-specific myosin heavy chain (MHC) gene expression by qRT-PCR. The results demonstrated that skeletal muscles from the hind legs of Ews−/− newborns (P0.5) contained significantly reduced (>60%) expression of slow fiber-specific MHC-Ib, whereas fast MHC-IIa, IIx and IIb and perinatal MHC (Mhy8) were unaltered compared with controls (Fig. 5D).
Reduced Hepatic Expression of Essential Mitochondrial Genes and Elevated Serum Lactate Levels in EWS-KO Mice.
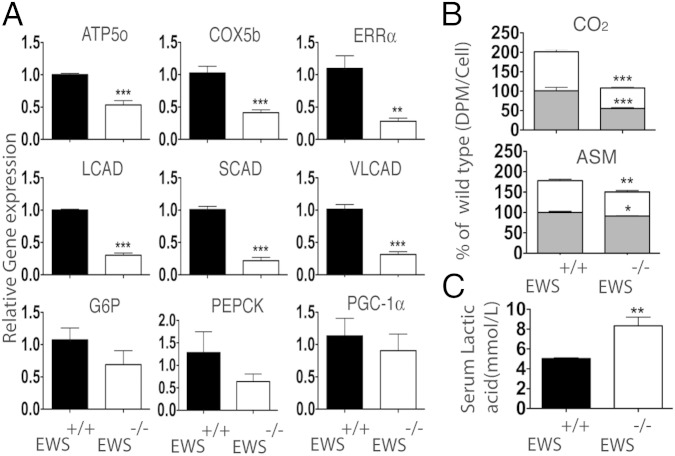
PGC-1α is also important for transcriptional regulation of genes involved in mitochondrial respiration, fatty acid β-oxidation (FAO) and gluconeogenesis (18, 19). To examine their expression levels we isolated total RNA from livers of congenic EWS-KO and wild-type newborns and performed quantitative gene expression analysis. There was a significant decrease in mitochondrial respiration (ATP5o, COX5b), biogenesis (ERRα) and FAO genes (LCAD, SCAD and VLCAD) in the livers of EWS-KO newborns compared with littermate controls (Fig. 6A). In contrast, expression of genes in gluconeogenesis (G6P and PEPCK), although trending lower, was not significantly reduced. Interestingly, depletion of Ews in C2C12 myoblasts also resulted in a significant reduction in the expression of essential mitochondrial genes (Fig. S7B).
Fig. 6.
Reduced expression of essential mitochondrial genes in EWS-KO liver. (A) Total RNA isolated from livers of congenic C57BL6 newborn pups (P0.5; n = 6) was analyzed by qRT-PCR for the indicated genes. (B) Total (gray bar) and mitochondria (white bar) fatty acid oxidation of 14C-oleic acid to CO2 and ASM were significantly decreased in Ews−/− brown preadipocytes. Data from three independent experiments are represented as mean ± SEM. (C) Resting serum lactate levels in Ews+/+ or Ews−/− mice (9–12 wk; n = 6). Data in A–C are represented as mean ± SEM. Student’s t test, two-tailed, *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we measured total and mitochondrial FAO in brown preadipocytes. Ews-deficient cells displayed ∼50% and ∼20% reduction in mitochondria FAO of 14C-oleic acid to CO2 and acid soluble metabolites (ASM), respectively, compared with wild-type cells (Fig. 6B). The reduced mitochondria abundance and activity in the brown fat, skeletal muscle and liver of EWS-KO mice suggest a systemic mitochondrial dysfunction. In accordance, measurement of resting serum lactate levels in 9- to 12-wk-old EWS-KO mice showed significantly elevated serum lactate levels (>8 mmol/L) compared with littermate controls (5 mmol/L) (Fig. 6C).
Discussion
EWS is a multifunctional protein with multiple roles in diverse cellular processes, such as maturation of pre B lymphocytes, meiosis, hypersensitivity to DNA damage, prevention of premature senescence of fibroblasts (11) and hematopoietic stem cells (12), mitosis (13), DNA damage-induced alternative splicing (9, 10), cell-fate determination of classical brown fat (8) and regulation of microRNAs (15). This study ascribes a new function to EWS in mitochondrial homeostasis and cellular energy metabolism. Inactivation of EWS results in concomitant decreases in PGC-1α protein levels and mitochondria abundance and function in brown preadipocytes, myoblasts, skeletal muscles and liver. The role of EWS in mitochondria homeostasis is highly intriguing given that one of the FET members, FUS, is mutated in a subset of amyotrophic lateral sclerosis (ALS) (37, 38) and that expression of mutant FUS in mouse motor neurons causes a significant shortening of mitochondria (39). Moreover, mitochondrial damage and dysfunction is observed in most ALS patients and animal models (40). Thus, FUS mutations in ALS might similarly disrupt mitochondrial homeostasis. In frontotemporal lobar degeneration (FTLD, a neurodegenerative disease similar to ALS), cytoplasmic aggregation of all three FET proteins is observed (41), raising the possibility that reduced PGC-1α and mitochondrial dysfunction could also contribute to FTLD pathogenesis.
Pgc-1α inactivation in mice leads to a decrease in mitochondrial density and function and affects the metabolic activity of several tissues, such as reduced uncoupled respiration and thermogenesis in BAT, decreased oxidative and exercise capacity in skeletal muscles and unregulated gluconeogenesis and lipogenesis in liver (20, 21). EWS is expressed in most tissues, including the highly oxidative BAT, brain, heart, liver and skeletal muscles (8). Consistent with a role in metabolism, Ews inactivation in the mouse leads to metabolic dysfunction in BAT, skeletal muscle and liver. However, because global inactivation of Ews is highly postnatal lethal accompanied by developmental defects in several organ systems including BAT (8, 11, 12, 15), generation of a tissue-specific EWS-KO mouse is necessary to investigate the role of EWS in cellular energy metabolism of other tissues.
How does EWS control PGC-1α protein stability? PGC-1α is an intrinsically unstable protein and its degradation via ubiquitin-mediated proteolysis is mediated by FBXW7 (25–27). Our findings show that FBXW7 expression and its interaction with PGC-1α is increased in the absence of EWS, which further enhances PGC-1α degradation; however, the mechanism by which EWS regulates FBXW7 expression warrants further studies. We found that EWS interacts weakly with PGC-1α; however, given the substoichiometric nature of this interaction, it is highly unlikely that EWS protects PGC-1α through direct interaction. Instead, weak or transient EWS-PGC-1α interaction might alter posttranscriptional modifications of PGC-1α, such as phosphorylation or acetylation, which is critical for PGC-1α stability (42).
PGC-1α is a central regulator of cellular energy metabolism. Our findings demonstrate a novel role of EWS in cellular energy metabolism by controlling PGC-1α protein stability. Notably, at least 15 different chromosomal translocations involving EWS on chromosome 22 have been identified across several types of cancer (43). These translocations invariably result in EWS haploinsufficiency, and this could potentially lead to a reduction in PGC-1α, mitochondrial density, and oxidative phosphorylation in tumor cells harboring the translocation. As cancer cells predominantly use glycolysis for energy production rather than mitochondrial respiration (known as the Warburg effect) (44), it is tantalizing to speculate that EWS haploinsufficiency (and/or EWS fusion oncoproteins working as a dominant inhibitor of wild-type EWS) might contribute to the Warburg effect. Therefore, it will be intriguing to examine the levels of PGC-1α and mitochondrial density and function in cancer cells harboring the EWS translocation.
Materials and Methods
Cell Culture and Transfection.
Mouse brown preadipocytes, C2C12 myoblasts, primary myoblasts and HEK293-FLAG-PGC-1α cell lines were cultured in DMEM containing 10% (vol/vol) FBS, 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen). Brown preadipocytes have been described (8). Primary myoblasts isolation and culture were performed as described (45). Cells were transfected using Lipofectamine 2000 or RNAiMAX (Invitrogen). The siRNA sequences are in SI Materials and Methods.
Animal Care and Serum Lactate Detection.
Ews−/− mice in mixed background (129SvEv/Black Swiss) and in congenic C57BL6 have been described (8, 11). All animal procedures were approved by the Tulane Institutional Animal Care and Use Committee. Resting serum lactate levels were measured using one fresh drop of blood from 9- to 12-wk-old mice with the Accutrend Plus System and lactate detection strip (Roche).
Histological Analysis and Transmission Electron Microscopy.
Muscles from hind limbs or quadriceps were dissected and snap-frozen in liquid nitrogen. Frozen muscles were embedded in OCT and cross-section tissues were analyzed by H&E, COX, SDH, and ATPase staining (n = 3 per genotype) (Histoserv). Interscapular BATs from P0.5 pups (n = 3 per genotype) were isolated and immediately fixed for 4 h in 2.5% glutaraldehyde and processed for TEM analysis as described (8) (JFE Enterprises).
Western Blot Analysis.
The following antibodies were used: EWS (11), PGC-1α (Santa Cruz Biotechnology: H-300 and Millipore: C-Terminal) or monoclonal 4C1.3 antibody (30), FBXW7 (R&D system), FLAG (Sigma), Ubiquitin (Bethyl Laboratories), and Actin (Sigma). For measuring protein half-life, 293T-PGC-1α cells were treated with 10 μg/mL cycloheximide (CHX, Sigma). Quantification of PGC-1α was performed using Odyssey Infrared Imaging System software (Li-COR Biosciences). To detect endogenous PGC-1α, which is very low under a basal condition in brown preadipocytes, C2C12, and primary myoblasts, cells were cultured for 4 d without changing media followed by 1-h incubation at room temperature (cold exposure) and 1 h at 37 °C before harvesting cell lysates or before the addition of 10 μg/mL CHX and chase at 37 °C.
Plasmid DNA.
pSV-PGC-1α plasmid DNA (23) was purchased from Addgene. Full-length mouse Pgc-1α and Ews cDNA was cloned into the SwaI/NotI (Pgc-1α) or EcoRI/NotI (Ews) sites of lentivirus vector pCDH-GFP-Puro (System Biosciences). Lentivirus plasmids containing Ews shRNAs (TRCN0000102385 and TRCN0000102387) were purchased (Sigma).
Mitochondria DNA Quantitation and Functional Analyses.
Total DNA in cells or tissues was extracted using QIAamp DNA mini kit (Qiagen) and quantified using NanoDrop (NanoDrop Technology). Mitochondrial DNA was quantified by measuring the ratio of mitochondria NADH dehydrogenase subunit 1 (mtND1) or cytochrome c oxidase subunit 2 (Cox2) DNA regions to an intron of the nuclear-encoded β-Actin or β-globin using SYBR Green real-time PCR (Bio-Rad). The primer sequences are listed in SI Materials and Methods. Mitochondria imaging was obtained by staining live cells with MitoTracker Red CMXRos (Invitrogen) and DNA was counterstained with Hoechst 33342 (Molecular Probe). Fumarate levels were measured using Fumarate Assay Kit (Sigma). Mitochondrial oxygen consumption rates (OCR) were measured using an Oroboros Oxygraph-2k (Oroboros Instruments). Briefly, one million brown preadipocytes were placed in a magnetically stirred respirometric chamber containing the culture media (2 mL) and maintained at 37 °C during the assay. OCR measurements (six replicates for each cell line) were obtained at baseline and after injection of oligomycin, FCCP and antimycin A. The value of basal mitochondrial respiration, leak respiration, and maximal respiration was determined by subtraction with nonmitochondrial respiration as described in the Oroboros Manual.
Flow Cytometry Analysis.
For flow cytometry analysis, brown preadipocytes were fixed in 4% paraformaldehyde following live staining with MitoTracker Red CMXRos. Flow cytometry was performed using LSRII flow cytometer (BD biosciences).
Fatty Acid β-Oxidation Analysis.
Ews+/+ and Ews−/− brown preadipocytes were cultured overnight and FAO was performed as described (46). Three independent experiments were performed in six replicates per experiment. More detailed information is in SI Materials and Methods.
Gene Expression Analysis by qRT-PCR.
Livers from P0.5 newborns were harvested, immediately frozen and kept at −80 °C. Total RNA was isolated using RNeasy mini kit (Qiagen) and quantified using NanoDrop (NanoDrop Technology). cDNAs were prepared using SuperScript cDNA synthesis kit (Invitrogen) and analyzed by real-time PCR using SYBR Green Master Mix (Applied Biosystems). Primer sequences are listed in SI Materials and Methods.
Statistical Analysis.
Statistical analysis was performed by Student’s t test or analysis of one-way ANOVA using GraphPad Prims 5 program (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Jan Endlich (JFE Enterprises) for transmission electron microscopy, Yun-Ping Wu (National Institute of Diabetes and Digestive and Kidney Diseases, NIDDK) for confocal microscopy, Yihong Ye (NIDDK) for Ubiquitin antibody, Mary Price (Tulane Cancer Center) for flow cytometry, and Thomas Gettys (Pennington Biomedical Research Center) for the 4C1.3 PGC-1α antibody. We thank Richard L. Proia and Elisabetta Mueller (NIDDK) for helpful comments and discussions. This work was supported by the Tulane Startup Fund (S.B.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504391112/-/DCSupplemental.
References
- 1.Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15(18):5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 3.Bertolotti A, et al. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: Interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol. 1998;18(3):1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossow KL, Janknecht R. The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61(6):2690–2695. [PubMed] [Google Scholar]
- 5.Araya N, et al. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J Biol Chem. 2003;278(7):5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 6.Gascoyne DM, Thomas GR, Latchman DS. The effects of Brn-3a on neuronal differentiation and apoptosis are differentially modulated by EWS and its oncogenic derivative EWS/Fli-1. Oncogene. 2004;23(21):3830–3840. doi: 10.1038/sj.onc.1207497. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Rhee BK, Bae GY, Han YM, Kim J. Stimulation of Oct-4 activity by Ewing’s sarcoma protein. Stem Cells. 2005;23(6):738–751. doi: 10.1634/stemcells.2004-0375. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, et al. A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev Cell. 2013;26(4):393–404. doi: 10.1016/j.devcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutertre M, et al. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17(11):1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- 10.Paronetto MP, Miñana B, Valcárcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell. 2011;43(3):353–368. doi: 10.1016/j.molcel.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 11.Li H, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117(5):1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho J, et al. Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence. Blood. 2011;117(4):1156–1166. doi: 10.1182/blood-2010-04-279349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azuma M, Embree LJ, Sabaawy H, Hickstein DD. Ewing sarcoma protein ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS ONE. 2007;2(10):e979. doi: 10.1371/journal.pone.0000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JH, Lee SB. An essential role for Ewing sarcoma gene (EWS) in early white adipogenesis. Obesity. 2015;23(1):138–144. doi: 10.1002/oby.20934. [DOI] [PubMed] [Google Scholar]
- 15.Kim KY, et al. A multifunctional protein EWS regulates the expression of Drosha and microRNAs. Cell Death Differ. 2014;21(1):136–145. doi: 10.1038/cdd.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454(7203):463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppari R, Ramadori G, Elmquist JK. The role of transcriptional regulators in central control of appetite and body weight. Nat Clin Pract Endocrinol Metab. 2009;5(3):160–166. doi: 10.1038/ncpendmet1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Finck BN, Kelly DP. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116(3):615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 23.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 24.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 25.Puigserver P, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8(5):971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 26.Sano M, et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J Biol Chem. 2007;282(35):25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- 27.Olson BL, et al. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22(2):252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Distelmaier F, et al. Life cell quantification of mitochondrial membrane potential at the single organelle level. Cytometry A. 2008;73(2):129–138. doi: 10.1002/cyto.a.20503. [DOI] [PubMed] [Google Scholar]
- 29.Ruas JL, et al. A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151(6):1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J Biol Chem. 2009;284(47):32813–32826. doi: 10.1074/jbc.M109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27(7):728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 32.Chang JS, et al. Regulation of NT-PGC-1alpha subcellular localization and function by protein kinase A-dependent modulation of nuclear export by CRM1. J Biol Chem. 2010;285(23):18039–18050. doi: 10.1074/jbc.M109.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aquilano K, et al. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: Possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285(28):21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adamovich Y, et al. The protein level of PGC-1α, a key metabolic regulator, is controlled by NADH-NQO1. Mol Cell Biol. 2013;33(13):2603–2613. doi: 10.1128/MCB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handschin C, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282(41):30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 36.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 37.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 38.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tradewell ML, et al. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21(1):136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- 40.Cozzolino M, Ferri A, Valle C, Carrì MT. Mitochondria and ALS: Implications from novel genes and pathways. Mol Cell Neurosci. 2013;55:44–49. doi: 10.1016/j.mcn.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Neumann M, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134(Pt 9):2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582(1):46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romeo S, Dei Tos AP. Soft tissue tumors associated with EWSR1 translocation. Virchows Arch. 2010;456(2):219–234. doi: 10.1007/s00428-009-0854-3. [DOI] [PubMed] [Google Scholar]
- 44.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125(6):1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herschey MD, Verdin E. Measuring fatty acid oxidation in tissue homogenates. Protocol Exchange. 2010 doi: 10.1038/nprot.2010.92. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.