Significance
Exposure to a high-altitude (HA) hypobaric hypoxia environment produces physiological changes. Among these, the changes in the apelin signaling system are significant because this system regulates vascular and oxygen homeostasis. This study demonstrates that the HA environment stimulates the apelin system to distinguish genetic variants and the methylation profile of CpG islands that may impair or improve pulmonary vascular function, thereby resulting in HA pulmonary edema (HAPE) in patients or adaptation in healthy controls. Of the several variants of this system, apelin rs3761581G and rs2235312T, individually and in combination, and a greater methylation of a CpG island in the 5′ UTR, associated with low levels of apelin-13 and nitrite in HAPE, whereas a reverse trend was observed in the control groups.
Keywords: high-altitude pulmonary edema, haplotype, DNA methylation, correlation, healthy Ladakhi highland natives
Abstract
Hypoxia-inducible factor stimulates the expression of apelin, a potent vasodilator, in response to reduced blood arterial oxygen saturation. However, aberrations in the apelin system impair pulmonary vascular function, potentially resulting in the development of high-altitude (HA)-related disorders. This study aimed to elucidate the genetic and epigenetic regulation of apelin, apelin receptor (APLNR), and endothelial nitric oxide synthase (NOS3) in HA adaptation and HA pulmonary edema (HAPE). A genome-wide association study and sequencing identified variants of apelin, APLNR, and NOS3 that were validated in a larger sample size of HAPE-patients (HAPE-p), HAPE-free controls (HAPE-f), and healthy highland natives (HLs). Apelin-13 and nitrite levels and apelin and NOS3 expression were down-regulated in HAPE-p (P < 0.001). Among the several studied polymorphisms, apelin rs3761581, rs2235312, and rs3115757; APLNR rs11544374 and rs2282623; and NOS3 4b/4a, rs1799983, and rs7830 were associated with HAPE (P < 0.03). The risk allele rs3761581G was associated with a 58.6% reduction in gene expression (P = 0.017), and the risk alleles rs3761581G and rs2235312T were associated with low levels of apelin-13 and nitrite (P < 0.05). The latter two levels decreased further when both of these risk alleles were present in the patients (P < 0.05). Methylation of the apelin CpG island was significantly higher in HAPE-p at 11.92% than in HAPE-f and HLs at ≤7.1% (P < 0.05). Moreover, the methylation effect was 9% stronger in the 5′ UTR and was associated with decreased apelin expression and apelin-13 levels. The rs3761581 and rs2235312 polymorphisms and methylation of the CpG island influence the expression of apelin in HAPE.
Apelin plays a crucial role in the adaptive and nonadaptive physiological responses of the vascular endothelium and smooth muscles (1). It is a potent regulator of vascular and oxygen homeostasis, which are pertinent to the high-altitude (HA) physiology. Therefore, elucidating apelin function under hypobaric hypoxia is of significant importance. Hypobaric hypoxia of HA lowers the blood arterial oxygen saturation (SaO2) in the body. To restore this cellular O2 content, an array of adaptive responses occurs that are mediated through hypoxia-inducible factor (HIF) (2–4). Stimulation of the apelin signaling system is one of these responses, and any aberration in this response impairs pulmonary vascular function, which may result in the development of HA-related disorders (5, 6).
Apelin induces endothelium-dependent vasodilation by activating endothelial nitric oxide synthase (NOS3) via the AKT pathway (7). The release of nitric oxide (NO) by NOS3 activates soluble guanylate cyclase in vascular smooth muscle cells (VSMCs), resulting in an increased level of cyclic guanosine monophosphate (8). These events enhance vasodilation, an important phenomenon required for increasing blood circulation, which in turn improves tissue oxygenation in the body (9). However, in an impaired endothelium, apelin binds to its receptor, apelin receptor (APLNR), which is present in VSMCs, to induce VSMC-dependent vasoconstriction (1). Thus, apelin, APLNR and NOS3 may contribute to pathophysiological manifestations associated with various HA disorders (10, 11). HA pulmonary edema (HAPE) is one such HA disorder, characterized by pulmonary vasoconstriction, endothelial dysfunction, and intravascular fluid retention, which develops in otherwise healthy individuals upon rapid ascent to altitudes above 2,500 m (12).
In addition to the local environment, physiological regulatory mechanisms are also governed by genetics (12), and the loci of several genes that are associated with HA adaptation and disease susceptibility have been identified (13–18). However, the genetic and epigenetic regulations of apelin signaling have not been investigated in HAPE and HA adaptation, even though variants of the genes involved in this signaling have been extensively investigated in several disease conditions (17–24). Our previous reports on NOS3 revealed a selection of polymorphisms in both HA adaptation and HAPE (17, 18). Epigenetic regulatory mechanisms help elucidate the complex interactions between the genome and the hypobaric hypoxia environment (25). Among these epigenetic mechanisms, DNA methylation plays a crucial role in regulating the genes and therefore the physiology (26). Alterations in the DNA methylation of the CpG islands of several genes have been found in various cancers and other diseases (26, 27). A CpG island is a short stretch of DNA that is enriched with CpG sites in the 5′ end of a gene; although it remains unmethylated, it tends to undergo aberrant methylation upon exposure to certain environmental conditions (28). Thus, it may explain the physiological consequences of the complex interactions among aberrant DNA methylation, the genome, and the hypobaric hypoxia environment.
To elucidate the apelin signaling system at HA, we used a genome-wide association study (GWAS) and sequencing to identify novel and known variants of apelin, APLNR, and NOS3 in HAPE-patients (HAPE-p), HAPE-free controls (HAPE-f), and healthy highland natives (HLs). The identified variants were further validated in a larger sample size and through luciferase activity assays. Additionally, the plasma levels of apelin-13 and nitrite, the expression levels of the three genes and the CpG island methylation status were also assessed in these subjects. The functional consequences of associated SNPs and methylated CpG islands were determined by performing several association, correlation, and regression analyses.
Results
Clinical Assessment, Biochemical Characterization, and Gene Expression.
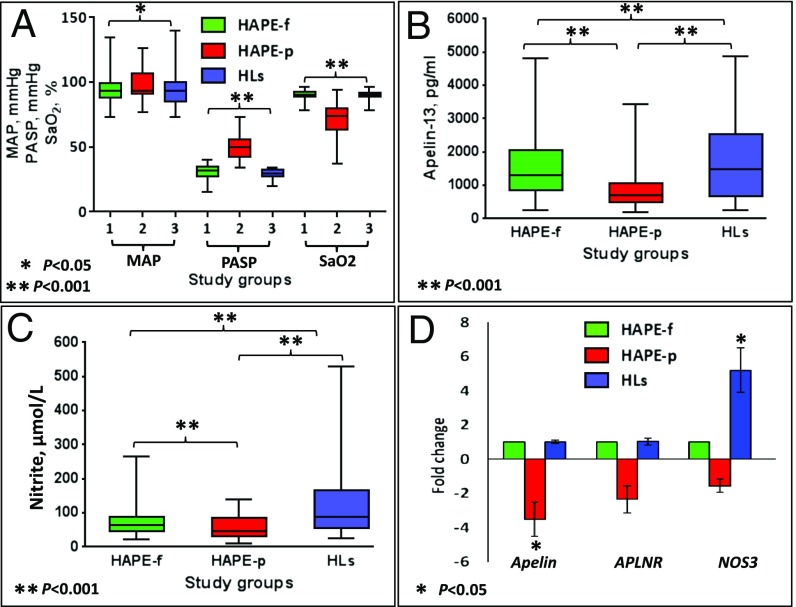
There was no difference in age between the HAPE-p and HAPE-f (Table S1). The mean arterial pressure (MAP) was significantly higher in the HAPE-p compared with the HAPE-f and HLs (P ≤ 0.021) (Fig. 1A). The pulmonary arterial systolic pressure (PASP) was 50.1 ± 9.0 mmHg in the HAPE-p, which was significantly higher than 29.20 ± 3.77 mmHg in the HAPE-f and 30.33 ± 6.35 mmHg in the HLs (P < 0.001) (Fig. 1A). The SaO2 level was 71.7 ± 12.5% in the HAPE-p, which was significantly lower compared with the levels 91.2 ± 3.7% and 89.6 ± 3.2% in the two healthy HAPE-f and HLs groups, respectively (P < 0.001) (Fig. 1A). The Apelin-13 level was 945.7 ± 664.7 pg/mL in the HAPE-p, which was significantly lower compared with the levels 1515.0 ± 1017.6 pg/mL in the HAPE-f and 1734.0 ± 1211.0 pg/mL in the HLs (P < 0.0001) (Fig. 1B). The nitrite level was 57.67 ± 35.50 μmol/L in the HAPE-p, which was significantly lower compared with the levels 73.75 ± 49.90 μmol/L in the HAPE-f and 122.45 ± 103.30 μmol/L in the HLs (P < 0.0001) (Fig. 1C). The expression of apelin and APLNR was down-regulated 3.52- and 2.35-fold, respectively, in the HAPE-p compared with the HAPE-f (P = 0.003 and 0.068, respectively). However, the HLs and HAPE-f exhibited similar expression levels of these two genes. Interestingly, NOS3 expression was up-regulated 5.2-fold in the HLs (P = 1.5E-03) and down-regulated 1.55-fold in the HAPE-p (P < 0.05) compared with the HAPE-f (Fig. 1D). All levels are reported as the means ± SD.
Fig. 1.
Clinical levels, gene expression and biochemical parameters of the study. (A) Clinical levels of the MAP, mmHg; PASP, mmHg and SaO2, percent in HAPE-p, HAPE-f, and HLs. HAPE-p had higher MAP and PASP values compared with the HAPE-f (P = 0.012 and < 0.0001, respectively) and HLs (P = 0.021 and < 0.0001, respectively). SaO2 levels were lower in the HAPE-p compared with the HAPE-f and HLs (P < 0.0001). (B) Apelin-13, pg/mL, and (C) nitrite, μmol/L. Apelin-13 and nitrite levels were lower in the HAPE-p compared with HAPE-f and HLs (P < 0.0001). (D) The relative expression levels of apelin, APLNR, and NOS3 were evaluated by real-time PCR and are expressed as foldchange in the HAPE-p and HLs compared with the HAPE-f. Apelin and APLNR were down-regulated 3.52- and 2.35-fold in the HAPE-p (P = 0.003 and 0.068, respectively), and NOS3 was up-regulated 5.2-fold in the HLs (P = 0.0015). Bars show the mean ± SD. Each box plot in A, B, and C shows the minimum, 25th percentile, median, 75th percentile, and maximum values.
GWAS-Pathway Based Findings.
A total of 138 SNPs that belonged to the following 10 genes involved in the apelin pathway were identified: apelin, APLNR, NOS3, AMP-activated, α 1 catalytic subunit, Kruppel-like factor 2 and 4 (KLF2/4), angiotensinogen, angiotensin receptor 1, angiotensin converting enzyme 2, and vascular endothelial growth factor (Table S2). Subsequently, the associated loci were analyzed to determine their roles in the adaptive and nonadaptive physiological responses of the vascular endothelial and smooth muscle cells.
Genotype and Allele Distribution.
The three groups were in Hardy–Weinberg equilibrium for the studied SNPs. Of the 10 genotyped apelin polymorphisms, rs3761581T/G, rs2235312T/C, and rs3115757G/C differed significantly between the HAPE-p and the HAPE-f (P ≤ 3.2E-03) (SI Results and Table S3). Of the seven genotyped APLNR polymorphisms, the SNPs rs11544374G/A and rs2282623G/A differed significantly between the HAPE-p and the HAPE-f (P ≤ 0.011) (SI Results and Table S4). Of the 11 genotyped NOS3 polymorphisms, 4b/4a, rs1799983G/T, and rs7830A/C differed significantly between the HAPE-p and the HAPE-f (P ≤ 0.016) (SI Results and Table S5).
Within-Gene Interactions.
Maximum-likelihood analysis by Phase identified four haplotypes for each gene, apelin, APLNR, and NOS3, which differed significantly between the groups (P < 0.05) (SI Results and Table S6). These haplotypes exceeded the cut-off frequency of >2%.
Gene–Gene Interactions.
The MDR model evaluated the gene–gene interactions. A four-locus model comprising apelin rs3761581T/G and rs2235312T/C and APLNR rs11544374G/A and rs2282623G/A emerged as the strongest interaction model (SI Results and Fig. S1 A, i). Similarly, a two-locus model comprising apelin rs2235312T/C and NOS3 rs7830A/C emerged as the strongest interaction model (SI Results and Fig. S1 A, ii).
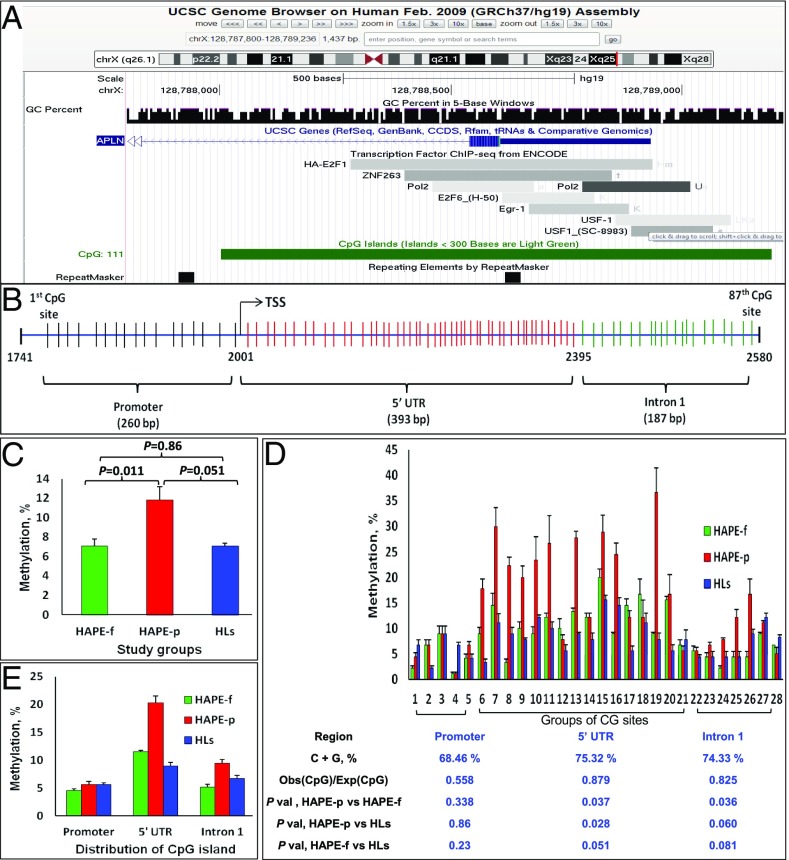
Methylation Profile of a CpG Island in apelin.
Fig. 2A displays the identified CpG island as confirmed by the University of California, Santa Cruz (UCSC) genome browser. The Ion Torrent platform profiled the identified 840-bp island as having 87 CpG sites (Fig. 2B). The CpG island covered three regions of the gene: the promoter, the 5′ UTR and intron 1, with 16 CpG sites in 260 bp of the promoter, 49 CpG sites in 393 bp of the 5′ UTR, and 22 CpG sites in 187 bp of intron 1 (Fig. 2B). Moreover, the methylation distribution was 11.92% in the HAPE-p, which was significantly higher compared with the 6.9% and 7.1% distributions in the HAPE-f and the HLs, respectively (P = 0.011 and 0.051, respectively) (Fig. 2C). In Fig. 2D, three CpG sites were pooled for each single bar for 84 CpG sites for each group. Here, the percentage of methylation distribution of the CpG sites in the 5′ UTR was significantly higher in the HAPE-p than in the two healthy control groups (P ≤ 0.05). Furthermore, the region-wise methylation distribution was 20% for the HAPE-p, 11.53% for the HAPE-f, and 8.96% for the HLs (P ≤ 0.037) (Fig. 2E). A CpG island was designated methylated and unmethylated at a cut-off percentage of 10%.
Fig. 2.
Methylation status of the apelin CpG island. (A) The CpG island as identified by the UCSC genome browser. (B) The graphical representation of the CpG island spanning the promoter, 5′ UTR, and intron 1 of the gene. (C) Methylation distribution in the CpG island in the three groups. The HAPE-p had a higher amount of CpG island methylation compared with the HAPE-f and HLs (P = 0.011 and 0.051, respectively). (D) An exhaustive resolution of the methylation distribution of all of the CpG sites in this island. Each bar represents three CpG sites, forming 28 groups of bars that represent a total of 84 CpG sites. Each group of sites in the 5′ UTR of the island exhibited higher methylation in the HAPE-p. (E) Methylation distribution in the CpG island of the promoter, 5′ UTR, and intron 1 in the three groups. The CpG sites in the 5′ UTR of the island exhibited significantly higher methylation in the HAPE-p compared with the two control groups (P < 0.05). Bars represent the mean ± SE. TSS, transcription start site.
Correlation Findings.
Correlation between apelin-13 and nitrite levels.
A positive correlation existed between the apelin-13 and nitrite levels in the HAPE-p, HAPE-f, and HLs (P ≤ 0.014) (Fig. S2).
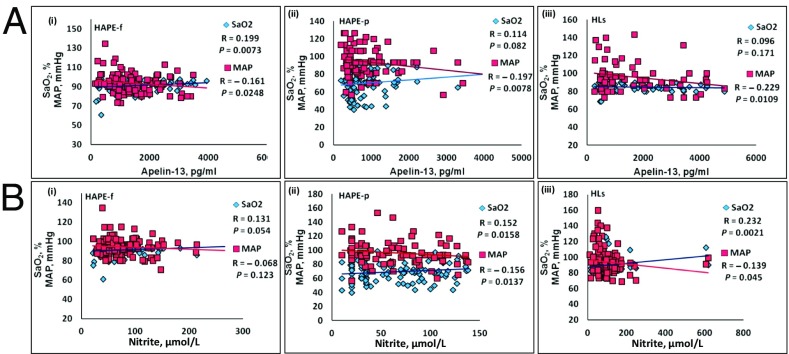
Correlation between clinical parameters and bio-levels.
The apelin-13 level correlated positively with the SaO2 level in only the HAPE-f (P = 0.007) (Fig. 3 A, i) and was inversely correlated with the MAP in the HAPE-f, HAPE-p, and HLs (P ≤ 0.025) (Fig. 3A). Furthermore, in the HAPE-p and HLs, the nitrite level was positively correlated with the SaO2 level (P = 0.015 and 0.002, respectively) (Fig. 3 B, ii and iii) but was inversely correlated with the MAP (P = 0.013 and 0.045, respectively) (Fig. 3 B, ii and iii).
Fig. 3.
Scatter plot between the clinical parameters and bio-levels. The clinical parameters SaO2, percent, and MAP, mmHg, in relation to (A) the apelin-13 and (B) nitrite levels in (i) the HAPE-f, (ii) HAPE-p, and (iii) HLs. The SaO2 level is positively correlated with the apelin-13 level in the HAPE-f (P = 0.0073) and with the nitrite levels in the HAPE-p and HLs (P = 0.016 and 0.002, respectively). The MAP is negatively correlated with the apelin-13 levels in the HAPE-f, HAPE-p, and HLs (P = 0.0248, 0.0078, and 0.011, respectively) and negatively correlated with the nitrite levels in the HAPE-p and HLs (P = 0.0137 and 0.045, respectively).
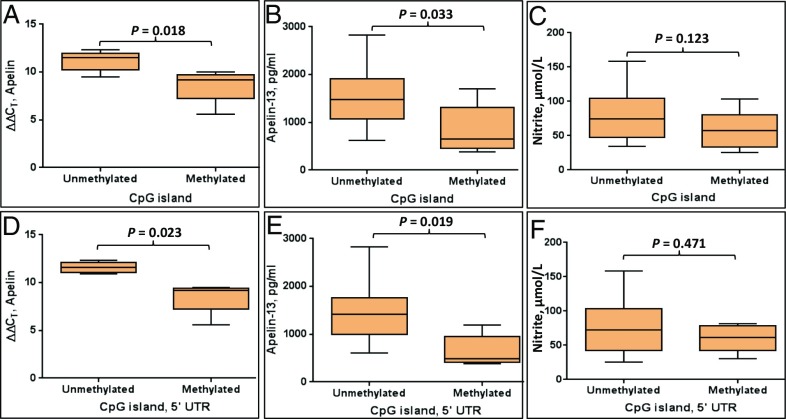
Correlation of methylated CpG island with apelin expression and bio-levels.
CpG islands with greater than 10% methylation in each group demonstrated a 5.9-fold reduction in apelin expression and a 43.6% reduction in the apelin-13 level compared with the unmethylated islands (P = 0.018 and 0.033, respectively) (Fig. 4 A and B). In particular, the methylation of the CpG sites exclusively in the 5′ UTR was associated with a 8.5-fold reduction in apelin expression and a 55.9% reduction in the apelin-13 level (P = 0.023 and 0.019, respectively) (Fig. 4 D and E). This down-regulation of apelin expression and the apelin-13 level associated with the CpG sites in the 5′ UTR was a 2.6-fold further reduction in apelin expression and a 12.3% further decrease in the apelin-13 level compared with the values for the entire methylated island. Methylated islands also correlated with decreased nitrite levels, but this correlation was not significant (Fig. 4 C and F).
Fig. 4.
The influence of apelin CpG island methylation on gene expression and the levels of apelin-13 and nitrite. Methylated and unmethylated CpG island with respect to (A) apelin gene expression, (B) the apelin-13 level, and (C) the nitrite level. The unmethylated CpG island was associated with higher apelin expression and higher apelin-13 and nitrite levels (P = 0.018, 0.033 and 0.123, respectively). Methylated and unmethylated CpG island spanning only the 5′ UTR with respect to (D) apelin gene expression, (E) the apelin-13 level, and (F) the nitrite level. The unmethylated CpG island in the 5′ UTR was associated with higher apelin expression and a higher apelin-13 level (P = 0.023 and 0.0193, respectively) Each box plot shows the minimum, 25th percentile, median, 75th percentile, and maximum values.
Causal Role of the Associated SNPs.
Contribution of the risk alleles and haplotypes to the apelin-13 level.
The regression coefficient showed that there was an association between the apelin risk allele rs3761581G and a decreased apelin-13 level in the three groups (P ≤ 0.036) (Table 1, SI Results, and Fig. S3A). The apelin risk allele rs2235312T was associated with a decreased apelin-13 level in only the HAPE-p and HLs (P = 0.004 and 0.003, respectively) (Table 1, SI Results, and Fig. S3B). The APLNR risk allele rs11544374A was associated with a decreased apelin-13 level in the HAPE-p and HLs (P = 0.030 and 0.048, respectively). The apelin risk haplotypes G-T-G and T-T-G demonstrated significant associations with a decreased apelin-13 level in the HAPE-p (P = 0.015 and 0.021, respectively). Understandably, the protective haplotype T-C-C was positively and significantly associated with the apelin-13 level in the HAPE-p (P = 0.001). There was another positive association with the protective haplotype T-C-G in the HAPE-f and HLs (P = 0.039 and 0.002, respectively). The APLNR risk haplotype A-G was inversely correlated with the apelin-13 level in the HAPE-p and HLs (P = 0.002 and 0.036, respectively) and the protective haplotype G-A was positively associated with the apelin-13 level in the HAPE-p (P = 0.008) (Table 1). The NOS3 risk alleles and haplotypes did not demonstrate associations with the apelin-13 level.
Table 1.
Risk alleles and major haplotypes of apelin and APLNR in relation to apelin-13 levels in the three studied groups
| Gene polymorphisms | Alleles/haplotypes | HAPE-p | HAPE-f | HLs | |||
| β | P | β | P | β | P | ||
| Apelin | Risk alleles | ||||||
| rs3761581 | G | −0.189 | 0.0087 | −0.171 | 0.0176 | −0.157 | 0.036 |
| rs2235312 | T | −0.208 | 0.0039 | −0.123 | 0.087 | −0.219 | 0.003 |
| Major haplotypes | |||||||
| rs3761581-rs2235312- | G-T-G | −0.145 | 0.015 | −0.069 | 0.178 | −0.027 | 0.584 |
| -rs3115757 | T-T-G | −0.137 | 0.021 | −0.095 | 0.061 | −0.148 | 0.009 |
| T-C-C | 0.184 | 0.001 | 0.057 | 0.263 | 0.047 | 0.349 | |
| T-C-G | 0.107 | 0.108 | 0.126 | 0.039 | 0.17 | 0.002 | |
| APLNR | Risk alleles | ||||||
| rs11544374 | A | −0.111 | 0.030 | −0.062 | 0.222 | −0.105 | 0.048 |
| Major haplotypes | |||||||
| rs11544374- | A-G | −0.168 | 0.002 | −0.048 | 0.343 | −0.125 | 0.036 |
| -rs2282623 | G-A | 0.146 | 0.008 | 0.067 | 0.191 | 0.039 | 0.464 |
P values were obtained by multinomial logistic regression analysis using SPSS and Bonferroni’s multiple correction test. η = frequency of subjects, β = regression coefficient.
Contribution of the risk alleles and major haplotypes to the nitrite level.
The regression coefficient showed that there was an association between the apelin risk allele rs3761581G and a decreased nitrite level in the three groups (P ≤ 0.044) (Table 2, SI Results, and Fig. S3C). Another risk allele, rs2235312T, was also associated with a decreased nitrite level in the HAPE-p and HLs (P = 6.0E-04) (Table 2, SI Results, and Fig. S3D). Similarly, the NOS3 risk alleles 4a, rs1799983T, and rs7830C, were also associated with a decreased nitrite level in the HAPE-p and HLs (P ≤ 0.042). The associations of the apelin and NOS3 haplotypes with the nitrite level were equally noteworthy (Table 2). The apelin susceptible haplotypes G-T-G and T-T-G were associated with a decreased nitrite level in the HAPE-p (P = 0.001). The apelin protective haplotype T-C-C was associated with an increased nitrite level in the three groups (P ≤ 0.05). The other apelin protective haplotype, T-C-G, was associated with an increased nitrite level in the HAPE-p and HAPE-f (P = 8.2E-05 and 3.4E-06, respectively). In the case of the NOS3 haplotypes, the risk haplotype a-G-C was associated with a decreased nitrite level (P ≤ 0.002) and the protective haplotype b-G-A was associated with an increased nitrite level in the three groups (P ≤ 0.027). The APLNR risk alleles and haplotypes did not demonstrate any type of association with the nitrite level.
Table 2.
Risk alleles and major haplotypes of apelin and NOS3 in relation to nitrite levels in the three studied groups
| Gene polymorphisms | Alleles/haplotypes | HAPE-p | HAPE-f | HLs | |||
| β | P | β | P | β | P | ||
| Apelin | Risk alleles | ||||||
| rs3761581 | G | −0.152 | 0.036 | −0.167 | 0.020 | −0.151 | 0.044 |
| rs2235312 | T | −0.245 | 0.0006 | −0.130 | 0.072 | −0.252 | 0.0006 |
| Major haplotypes | |||||||
| rs3761581-rs2235312- | G-T-G | −0.178 | 0.001 | −0.098 | 0.054 | −0.100 | 0.132 |
| -rs3115757 | T-T-G | −0.181 | 0.001 | −0.019 | 0.710 | −0.219 | 2.7E-05 |
| T-C-C | 0.098 | 0.050 | 0.145 | 0.012 | 0.129 | 0.027 | |
| T-C-G | 0.213 | 8.2E-05 | 0.245 | 3.45E-06 | 0.048 | 0.339 | |
| NOS3 | Risk alleles | ||||||
| 4b/4a | 4a | −0.131 | 0.010 | −0.085 | 0.094 | −0.115 | 0.030 |
| rs1799983 | T | −0.104 | 0.042 | −0.100 | 0.050 | −0.115 | 0.031 |
| rs7830 | C | −0.176 | 0.0005 | −0.089 | 0.080 | −0.107 | 0.042 |
| Major haplotypes | |||||||
| 4b/4a-rs1799983- | a-T-C | −0.063 | 0.219 | — | — | −0.113 | 0.018 |
| -rs7830 | a-G-C | −0.169 | 0.002 | −0.161 | 0.005 | −0.154 | 0.0006 |
| b-G-A | 0.192 | 0.0003 | 0.132 | 0.027 | 0.197 | 5.85E-06 | |
| b-T-A | 0.024 | 0.636 | 0.093 | 0.069 | 0.229 | 8.85E-08 | |
P values were obtained by multinomial logistic regression analysis using SPSS and Bonferroni’s multiple correction test. η = frequency of subjects, β = regression coefficient.
Apelin rs3761581T/G affects transcriptional activity.
The rs3761581T or rs3761581G allele of the apelin promoter polymorphism rs3761581 was inserted into the promoterless pGL3 basic Firefly luciferase vector and expressed in HEK-293 cells. As shown in Fig. S4, the vector with the risk allele rs3761581G decreased the relative luciferase activity 58.6% compared with the protective allele rs3761581T (P = 0.016).
Discussion
The present study revealed the contribution of apelin signaling to the adaptive physiological and pathophysiological responses to HA by demonstrating the associations of genetic markers. Additionally, the decreased circulating levels of apelin-13 and nitrite in HAPE suggested that there was impaired vasodilation. Consistent with the two levels, the gene expression of apelin and NOS3 was also significantly down-regulated in HAPE. Conversely, HLs and healthy lowlanders had elevated levels of apelin-13 and nitrite. Equally pertinent was the assessment of clinical parameters in the three groups. The significantly decreased SaO2 level and elevated MAP and PASP depicted the pathophysiology of HAPE, whereas opposite trends were observed in the two types of healthy controls, HAPE-f and HLs.
Multivariate-logistic regression analysis revealed that there was an overrepresentation of the apelin alleles rs3761581G, rs2235312T, and rs3115757G; the APLNR alleles rs11544374A and rs2282623G; and the NOS3 alleles 4a, rs1799983T, and rs7830C in HAPE. The effect of any individual polymorphism on the body physiology is modest (29), which was amply demonstrated by the greater influence of both within-gene interactions (haplotype) and between-gene interactions. Overrepresentation of the risk allele-bearing haplotypes of the three genes was observed in the HAPE-p, which conferred a four-times greater risk than the individual alleles. The protective haplotypes conferred three-times more protection in the HAPE-f and HLs. Equally pertinent was the between-gene interactions that influenced HA physiology. The interaction between apelin and APLNR was demonstrated by the best disease predicting four-locus MDR model rs3761581TT-rs2235312GG-rs2282623GG-rs11544374AG. The interaction between apelin and NOS3 was demonstrated by the model rs2235312TT-rs7830CC. These interactions clearly emphasized the contribution of the apelin alleles rs3761581G and rs2235312T in conferring the risk of HAPE and in the counter alleles maintaining health. Multivariate regression analyses of genetic variants with the apelin-13 and nitrite levels also emphasized the functionality of these two apelin SNPs.
Because rs3761581G is a promoter risk allele, we could validate its functional relevance using a luciferase activity assay. The assay revealed decreased expression of the gene, which was consistent with the association between rs3761581G and the decreased levels of apelin-13 and nitrite in the three groups. Similarly, the association between the risk allele rs2235312T and the decreased levels of apelin-13 and nitrite also demonstrated its functional relevance. Although rs2235312 is an intronic allele, the HapMap data for this SNP predicted it to be an informative SNP. It is in complete linkage disequilibrium with the SNPs rs2281068, rs3115759, rs909657, rs2235308, and rs3115758 of the same gene in CEU (European), CHB (Chinese), and JPT (Japanese) populations in HapMap. The findings from a previous study by Chandra et al. support our findings (6). Although this group did not study the genetics but the physiological outcome, their in vivo and in vitro analyses support our finding. This study reported that apelin-null mice exposed to chronic hypoxia developed severe pulmonary hypertension (PH) compared with wild-type mice. Furthermore, the apelin-null mice demonstrated a significant down-regulation of NOS3 and KLF2, resulting in a decreased production of NO and the development of PH (6). Furthermore, NOS3 and KLF2 expression have also been shown to be decreased in apelin-null human pulmonary artery endothelial cells. Moreover, the apelin level was reduced in patients with PH compared with healthy controls. These results suggested that the damaged intima prevented apelin from performing its physiological function of NO-dependent vasodilation (6, 7). Correlations between the apelin-13 and nitrite levels with clinical parameters, such as the MAP and the SaO2 level, were another highlight of this study. A lower SaO2 level and higher MAP are predictive of HAPE pathophysiology (30, 31). Here, the positive correlation between the levels of apelin-13 and NO with the SaO2 level and their inverse correlation with the MAP signified their contribution to the functioning of the body at HA.
Regulation of the transcription and translation of a gene in a given environment depends on several factors, including epigenetics (32). Identification of a CpG island in the 5′ end of the apelin gene was pivotal to this study. Interestingly, the presence of a hypoxic responsive element immediately downstream of this CpG island further implicated it in the regulation of this gene. A significantly higher methylation distribution that was associated with reductions in apelin expression and the apelin-13 level was observed in HAPE patients. Apelin is HIF-inducible; therefore, methylation of this region may hinder the binding of HIF to the gene, resulting in the down-regulation of its transcription (Fig. 5) (5). The reduction in apelin expression and the apelin-13 level that subsequently resulted in a reduced NO level in the HAPE patients in our study supported our presumption and emphasized the relevance of the higher methylation of the CpG island in impairing vasodilation and subsequently inducing the progression of HAPE. Furthermore, differentially methylated sites within the crucial regulatory areas of the genome, such as in the promoter or within the immediate vicinity of the transcription start site, have a more pronounced effect on the functioning of genes (32). For example, methylation in the 5′ end of the gene is known to block the initiation of transcription (32). This knowledge prompted us to look at the relevance of methylation of CpG sites in regions such as the promoter, 5′ UTR, and intron 1. The methylation effect was stronger in the 5′ UTR than in the other regions of the CpG island as it lowered apelin expression 2.6-fold and the apelin-13 level by 12% compared with the methylated CpG sites of the entire CpG island.
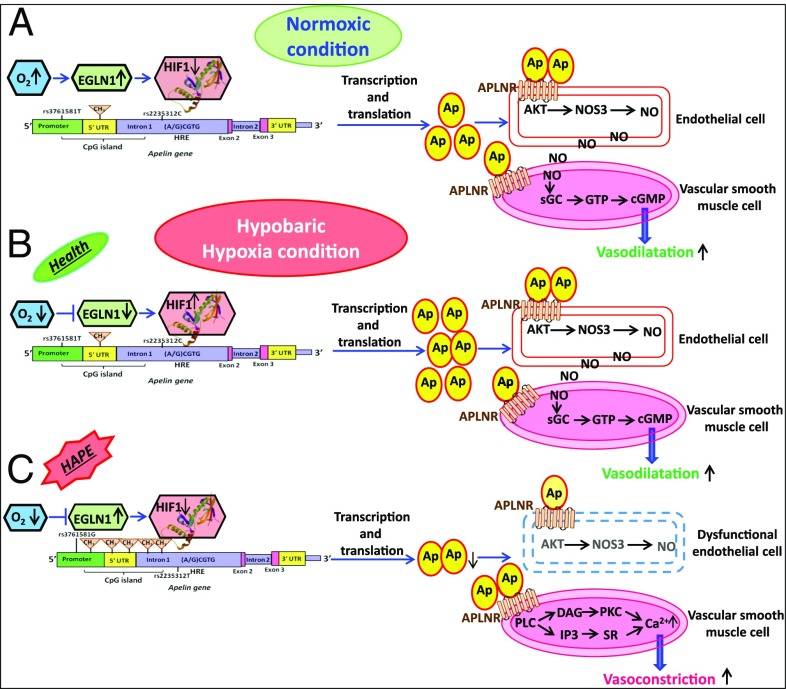
Fig. 5.
Apelin signaling under hypobaric hypoxia. (A) Under normoxic conditions, apelin is normally transcribed and translated to induce vasodilation through NOS3 activation. (B) Under hypobaric hypoxic conditions, apelin is induced by HIF and the resulting augmented apelin induces vasodilation, improving blood oxygenation and maintaining health at HA. (C) However, a disease condition, such as HAPE, exhibits significantly reduced level of apelin. These low levels may be attributed to the influence of the risk alleles together with the increased methylation of the apelin CpG island. AKT, protein kinase B; Ap, apelin; cGMP, cyclic guanosine monophosphate; CH3, methyl group; DAG, diacylglycerol; EGLN1, HIF-prolyl hydroxylase 2; HIF-1, hypoxia inducible factor-1; HRE, hypoxia response element; IP3, inositol triphosphate; NO, nitric oxide; NOS3, endothelial nitric oxide synthase; O2, oxygen; PKC, protein kinase C; sGC, soluble guanylate cyclase; SR, sarcoplasmic reticulum.
In conclusion, apelin under normoxic conditions is normally transcribed and translated to induce vasodilation (Fig. 5A). Apelin activates the AKT-NOS3 pathway to increase the production of NO, which is transferred to muscle cells in which the guanylate system is activated to induce vasodilation. However, under hypobaric hypoxia, apelin is induced by the HIF transcription factor (Fig. 5B) (5). Thus, more apelin molecules are activated to maintain normal vasodilation (33, 34). It is likely that the protective apelin alleles rs3761581T and rs2235312C, individually and together, contribute to a normal physiological process. In contrast, the alleles rs3761581G and rs2235312T, which are correlated with low levels of apelin-13 and nitrite and a greater methylation of the CpG island of apelin, contribute to the HAPE pathophysiology. Here, the aberrant methylation may hinder the binding of HIF to the apelin hypoxic responsive element, thereby repressing transcription and reducing the apelin-13 level. The latter contributes to repressing NOS3, which reduces NO production (Fig. 5C). Reduced vasodilation caused by the impairment of apelin and NO also hinders the oxygenation of hemoglobin under hypobaric hypoxia, and the significantly low SaO2 level in HAPE patients supports this phenomenon. The hypothesis that the apelin signaling system is operative under hypobaric hypoxia, however, needs to be validated. The prevalence of risk alleles in HAPE-p and protective alleles in HLs provided us with allelic variants at the same locus that were involved in disease and adaptation. Furthermore, the associations of these polymorphisms with the apelin-13 and nitrite levels contributed to uncovering their functional relevance in the hypobaric hypoxic environment.
Materials and Methods
Detailed materials and methods are reported in SI Materials and Methods.
Study Subjects.
In a cross-sectional study, 200 subjects were categorized into each of the following three well-defined groups: (i) HAPE-patients (HAPE-p), (ii) HAPE-free sojourners (HAPE-f), and (iii) healthy highland natives (HLs). The details of these subjects are described in SI Materials and Methods.
The human ethical committee of the Council of Scientific and Industrial Research-Institute of Genomics and Integrative Biology, Delhi, India, and the human ethical committee of Sonam Norboo Memorial Hospital, Leh, India, approved the investigation. The HAPE selection criterion is described in SI Materials and Methods. Ten milliliters of blood sample was collected from each subject (SI Materials and Methods).
Quantification of Apelin-13 and Nitrite Levels.
Of the several apelin peptides that activate APLNR, apelin-13 is resistant to enzymatic breakdown and is capable of retaining its biological activity for longer periods. Therefore, it is the preferred marker (34). The plasma apelin-13 level was measured using an immunoassay kit (USCN Life Science) and the plasma nitrite level was estimated using an enzymatic assay kit (Cayman Chemical), as described in SI Materials and Methods.
Quantitative RT-PCR Analysis of Apelin, APLNR, and NOS3 Genes.
Gene expression analysis was performed on 10 samples from each group, HAPE-p, HAPE-f, and HLs, as described in SI Materials and Methods (Table S7).
Identification of the Apelin Pathway in a GWAS.
A genome scan was conducted on 288 subjects (96 subjects from each group, HAPE-p, HAPE-f, and HLs) using Illumina HumanOmni1-Quad BeadChips (Illumina) as described in SI Materials and Methods.
Apelin and APLNR Gene Sequencing.
Of the three genes, apelin and APLNR have not been studied in relation to HA. Therefore, the identification of novel variants by direct sequencing was performed along the total length of apelin, 11,694 bp, and APLNR, 5,659 bp, as described in SI Materials and Methods.
Selection and Genotyping of Polymorphisms.
Ten polymorphisms in apelin, 7 polymorphisms in APLNR, and 11 polymorphisms in NOS3 were selected for validation in a larger sample size, as described in SI Materials and Methods (Tables S8, S9, and S10).
Evaluation of the Functional Role of the Associated SNPs.
The functional roles of the promoter SNPs apelin rs3761581 and APLNR rs11544374 were validated by constructing reporter plasmids that were transiently transfected into cells for use in a luciferase activity assay as described in SI Materials and Methods (Table S11). Genetic associations between the associated SNPs and haplotypes with the apelin-13 and serum nitrite levels were evaluated (SI Materials and Methods).
CpG Island of Apelin Gene.
While sequencing, one CpG island was identified in apelin and was later confirmed by the UCSC genome browser (genome.ucsc.edu/). The methylation status was assessed in the three groups as described in SI Materials and Methods (Table S12).
Statistical Analyses.
All of the statistical analyses performed are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Director of the Council of Scientific and Industrial Research-Institute of Genomics and Integrative Biology for support and encouragement; Satish Sati, who helped us evaluate the CpG island methylation status; the staff at Sonam Norboo Memorial Hospital, Leh for technical support; and all the participants. This work was supported by the Council of Scientific and Industrial Research, India through Projects MLP1401, SIP0006, and BSC0123.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422759112/-/DCSupplemental.
References
- 1.Japp AG, Newby DE. The apelin-APJ system in heart failure: Pathophysiologic relevance and therapeutic potential. Biochem Pharmacol. 2008;75(10):1882–1892. doi: 10.1016/j.bcp.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Hohenhaus E, Paul A, McCullough RE, Kücherer H, Bärtsch P. Ventilatory and pulmonary vascular response to hypoxia and susceptibility to high altitude pulmonary oedema. Eur Respir J. 1995;8(11):1825–1833. doi: 10.1183/09031936.95.08111825. [DOI] [PubMed] [Google Scholar]
- 3.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92(3):967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyries M, et al. Hypoxia-induced apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ Res. 2008;103(4):432–440. doi: 10.1161/CIRCRESAHA.108.179333. [DOI] [PubMed] [Google Scholar]
- 6.Chandra SM, et al. Disruption of the apelin-APJ system worsens hypoxia-induced pulmonary hypertension. Arterioscler Thromb Vasc Biol. 2011;31(4):814–820. doi: 10.1161/ATVBAHA.110.219980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Japp AG, et al. Vascular effects of apelin in vivo in man. J Am Coll Cardiol. 2008;52(11):908–913. doi: 10.1016/j.jacc.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Arnold WP, Mittal CK, Katsuki S, Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc Natl Acad Sci USA. 1977;74(8):3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatemoto K, et al. The novel peptide apelin lowers blood pressure via a nitric oxide-dependent mechanism. Regul Pept. 2001;99(2-3):87–92. doi: 10.1016/s0167-0115(01)00236-1. [DOI] [PubMed] [Google Scholar]
- 10.Kleinz MJ, Davenport AP. Emerging roles of apelin in biology and medicine. Pharmacol Ther. 2005;107(2):198–211. doi: 10.1016/j.pharmthera.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Penaloza D, Arias-Stella J. The heart and pulmonary circulation at high altitudes: Healthy highlanders and chronic mountain sickness. Circulation. 2007;115(9):1132–1146. doi: 10.1161/CIRCULATIONAHA.106.624544. [DOI] [PubMed] [Google Scholar]
- 12.Bärtsch P, Mairbäurl H, Maggiorini M, Swenson ER. Physiological aspects of high-altitude pulmonary edema. J Appl Physiol (1985) 2005;98(3):1101–1110. doi: 10.1152/japplphysiol.01167.2004. [DOI] [PubMed] [Google Scholar]
- 13.Bigham A, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6(9):e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329(5987):72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 16.Qadar Pasha MA, Newman JH. High-altitude disorders: Pulmonary hypertension: Pulmonary vascular disease: The global perspective. Chest. 2010;137(6 suppl):13S–19S. doi: 10.1378/chest.09-2445. [DOI] [PubMed] [Google Scholar]
- 17.Ahsan A, Norboo T, Baig MA, Qadar Pasha MA. Simultaneous selection of the wild-type genotypes of the G894T and 4B/ 4A polymorphisms of NOS3 associate with high-altitude adaptation. Ann Hum Genet. 2005;69(Pt 3):260–267. doi: 10.1046/j.1529-8817.2005.00158.x. [DOI] [PubMed] [Google Scholar]
- 18.Ahsan A, et al. eNOS allelic variants at the same locus associate with HAPE and adaptation. Thorax. 2004;59(11):1000–1002. doi: 10.1136/thx.2004.029124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castan-Laurell I, Dray C, Knauf C, Kunduzova O, Valet P. Apelin, a promising target for type 2 diabetes treatment? Trends Endocrinol Metab. 2012;23(5):234–241. doi: 10.1016/j.tem.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Zhang R, et al. Associations of common variants at APLN and hypertension in Chinese subjects with and without diabetes. Exp Diabetes Res. 2012;2012:917496. doi: 10.1155/2012/917496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao YC, et al. Apelin gene polymorphism influences apelin expression and obesity phenotypes in Chinese women. Am J Clin Nutr. 2011;94(3):921–928. doi: 10.3945/ajcn.110.008813. [DOI] [PubMed] [Google Scholar]
- 22.Li WW, et al. Family-based analysis of apelin and AGTRL1 gene polymorphisms with hypertension in Han Chinese. J Hypertens. 2009;27(6):1194–1201. doi: 10.1097/HJH.0b013e32832a3eb1. [DOI] [PubMed] [Google Scholar]
- 23.Jin W, et al. Interactive association of five candidate polymorphisms in Apelin/APJ pathway with coronary artery disease among Chinese hypertensive patients. PLoS ONE. 2012;7(12):e51123. doi: 10.1371/journal.pone.0051123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Q, et al. Association of genetic variants in the apelin-APJ system and ACE2 with blood pressure responses to potassium supplementation: The GenSalt study. Am J Hypertens. 2010;23(6):606–613. doi: 10.1038/ajh.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 27.Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647(1-2):30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han L, Su B, Li WH, Zhao Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008;9(5):R79. doi: 10.1186/gb-2008-9-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bärtsch P, Gibbs JS. Effect of altitude on the heart and the lungs. Circulation. 2007;116(19):2191–2202. doi: 10.1161/CIRCULATIONAHA.106.650796. [DOI] [PubMed] [Google Scholar]
- 31.Gallagher SA, Hackett PH. High-altitude illness. Emerg Med Clin North Am. 2004;22(2):329–355, viii. doi: 10.1016/j.emc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 33.Weir EK, López-Barneo J, Buckler KJ, Archer SL. Acute oxygen-sensing mechanisms. N Engl J Med. 2005;353(19):2042–2055. doi: 10.1056/NEJMra050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashley EA, et al. The endogenous peptide apelin potently improves cardiac contractility and reduces cardiac loading in vivo. Cardiovasc Res. 2005;65(1):73–82. doi: 10.1016/j.cardiores.2004.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.