The maintenance of the human genome is dependent upon several cellular processes including DNA replication. Ordinarily, DNA replication is an exceptionally faithful process, with approximately one error occurring for every 109–1010 nucleotides (1, 2). High-fidelity replicative DNA polymerases with exonucleolytic proofreading activity, along with DNA mismatch repair machinery, are responsible for accurate DNA synthesis. DNA polymerase δ (pol δ) and polymerase ε (pol ε) are two essential replicative lagging and leading strand polymerases, respectively, that ensure efficient and high-fidelity genome replication (3–5). Replicative polymerase variants have recently been identified in human tumors that harbor enormous numbers of mutations. In recent studies reported in PNAS, Mertz et al. and Williams et al. provide evidence that this hypermutator phenotype results from expansion of deoxyribonucleoside triphosphate (dNTP) pools (6, 7).
The concept of a mutator phenotype leading to cancer was introduced by Lawrence Loeb in 1974 to account for the disparity between the low mutation rates in normal cells and the large numbers of mutations present in many human tumors. This hypothesis has evolved over time, with better understanding of the molecular basis for tumorigenesis, and it is now known that a mutator phenotype results from mutations in genes that maintain genome stability (8–13). The recently identified germ-line and somatic mutations of POLE and POLD, which encode DNA polymerases ε and δ, respectively, and which are associated with hypermutated tumors, provide additional strong evidence for the mutator phenotype hypothesis.
Mutator variants of pol ε and pol δ have recently been characterized in Saccharomyces cerevisiae by Williams et al. and Mertz et al. (6, 7). Williams et al. characterized the pol2-4 proofreading defective and pol2-M644G base selectivity-deficient alleles of pol2, which encode pol ε (5, 14). Mertz and colleagues studied the pol3-R696W S. cerevisiae mimic of a POLD variant that is deficient in base selectivity that was identified in two colorectal cancer cell lines, DLD1 and HCT15, which were derived from the same tumor (15–17).
Interestingly, Williams et al. and Mertz et al. find the mutator phenotypes of these POLE and POLD variants depend upon the Dun1 effector kinase, which plays a role in the regulation of dNTP pools and the transcriptional response to DNA damage (Fig. 1) (6, 7, 18). In fact, cells with the pol2-M644G allele are dependent upon Dun1 for survival. It has been well established that dNTP pools are precisely regulated during the cell cycle by ribonucleotide reductase (RNR) (19, 20). In addition, Chabes et al. demonstrated that dNTP levels rise six- to eightfold as a result of DNA damage (21). These increased pools were a consequence of activation of a DNA damage cascade involving Mec1, Rad53, and Dun1, which in turn stimulates higher RNR activity. Importantly, this pool expansion following DNA damage is accompanied by an increased error rate, resulting in the dNTP mutator phenotype (19). Imbalanced ratios of dNTPs are thought to have the most mutagenic potential (22), but proportional increases in dNTP levels can also result in mutagenesis (23). In contrast, significant dNTP pool increases following DNA damage have not been observed in mammalian cells (20).
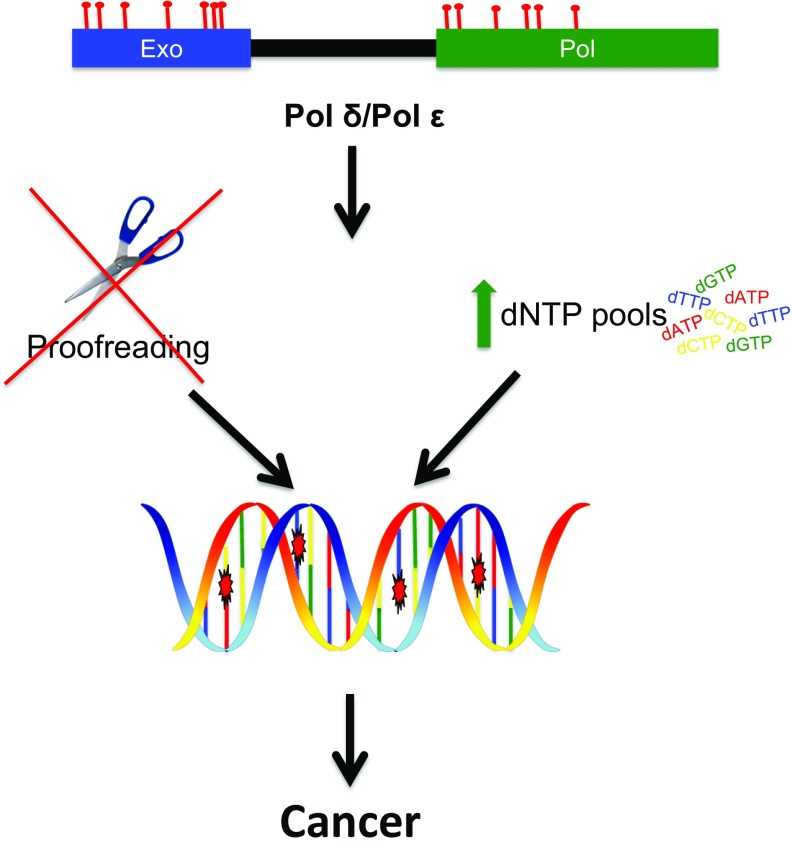
Fig. 1.
Replicative polymerase mutations and dNTP pools can contribute to cancer. Somatic and germ-line mutations in pol ε or pol δ, commonly located in the exonuclease domain, can lead to a loss of proofreading ability. Mutations in the catalytic domain result in deficient substrate discrimination. These mutations initiate signaling pathways that result in increased dNTP pools, further increasing the rate of mutation. This mutator phenotype leads to cancer.
Critically absent from previous analyses of pol ε and pol δ mutants is an exploration of their contribution to dNTP pool levels. For the first time, Williams et al. and Mertz et al. have identified a link between the mutations in the yeast pol ε and pol δ homologs (pol2 and pol3, respectively) and the alteration of dNTP pools as an underlying mechanism of the mutator phenotype (6, 7). Williams et al. (7) show that Dun1 directly influences the mutagenic potential of the error-prone pol2-M644G mutation by altering dNTP pool levels. The increase in mutation rate resulting from the pol2-M644G allele activates Dun1, which in turn induces RNR1 and RNR3 expression through the Mec1/Rad53 pathway, resulting in increased dNTP pools. Similarly, Mertz et al. (6) show that the mutator phenotype in a yeast strain expressing pol3-R696W is mediated by Dun1-dependent alteration of dNTP pools. Interestingly, the pol3-R696W yeast strain exhibited a significant increase in dNTP levels and changes in relative ratios, whereas wild-type dNTP pool levels and mutagenic rates were restored in a dun1Δ pol3-R696W strain. One important caveat of these studies is that yeast and mammalian cells show differences in dNTP pool modulation following DNA damage, and that although several members of the yeast Mec1/Rad53/Dun1 signaling cascade have human homologs, Dun1 and Sml1 in particular are not conserved. This finding suggests that additional pathways may be at work in human pol ε and pol δ mutations implicated in cancer.
Classic studies on proofreading by DNA polymerase I (pol I) of Escherichia coli by Brutlag and Kornberg showed that fraying of the DNA at a mismatched nascent base pair was critical for excision of the misinserted base by pol I (24). In order for incorrect bases to be removed from the primer strand, it must be transferred to the proofreading exonuclease active site of the polymerase, which is distant from the polymerase catalytic site. Recent work shows that pol ε M644G is less efficient than wild-type pol ε in transferring the 3′-terminus containing the incorrect base to the exonuclease active site (25), despite intact proofreading. Therefore, cells expressing a polymerase variant with reduced dNTP discrimination are likely to slow replication, leading to replication stress and checkpoint activation; this could lead to up-regulation of dNTP pools, facilitating the extension of nascent mispairs.
Although the error rate of the pol2–4 proofreading-deficient mutant is significantly increased compared to the wild-type cells, it is lower than what is observed in the pol2-M644G mutant with reduced base discrimination. In addition, there appears to be no checkpoint activation in the pol2–4 strain, even though its observed mutator phenotype is dependent upon the Dun1 effector kinase. The increase in dNTP pools
Overall, the work by Williams et al. and Mertz et al. establishes for the first time a relationship between altered dNTP pools and specific pol ε or pol δ mutations.
in pol2–4 cells is only observed in the absence of Sml1 and Crt1, which are repressors of RNR. Importantly, this increase in dNTP pool levels is equivalent in the presence or absence of the pol2–4 allele. Thus, up-regulated dNTP pools may not be directly responsible for the mutator phenotype of proofreading exonuclease-deficient pol ε, even though the mutator phenotype is dependent on Dun1. Alternatively, up-regulation of dNTP pools may be necessary, but because base selectivity is largely intact in the pol2–4 strain, misincorporation may not occur often enough within the population of cells being analyzed, making detection of checkpoint activation difficult. Single-cell analysis using more sensitive methods to detect activation of the checkpoint would be informative. Alternatively, the conformational dynamics of a proofreading-deficient polymerase may be inherently different from those of a polymerase compromised in dNTP discrimination base selectivity, leading to entirely different downstream events.
Overall, the work by Williams et al. and Mertz et al. (6, 7) establishes for the first time a relationship between altered dNTP pools and specific pol ε or pol δ mutations. Understanding the downstream effects of exonuclease mutations in these replicative polymerases is critical, as many of these mutations have been implicated in a host of human cancers. It is likely that other mechanisms resulting in a mutator phenotype may also be at play because of differences in dNTP pool modulation in yeast versus mammalian cells. Identifying the mechanisms of tumorigenesis resulting from pol ε or pol δ mutations will have significant impact on understanding and treating cancer.
Acknowledgments
This work was supported by Grant R01 CA080830 from the National Cancer Institute (to J.B.S.).
Footnotes
References
- 1.Bielas JH, Loeb LA. Mutator phenotype in cancer: Timing and perspectives. Environ Mol Mutagen. 2005;45(2-3):206–213. doi: 10.1002/em.20111. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279(17):16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 3.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7(12):e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pursell ZF, Kunkel TA. DNA polymerase epsilon: A polymerase of unusual size (and complexity) Prog Nucleic Acid Res Mol Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mertz TM, Sharma S, Chabes A, Shcherbakova PV. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc Natl Acad Sci USA. 2015;112:E2467–E2476. doi: 10.1073/pnas.1422934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams LN, et al. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc Natl Acad Sci USA. 2015;112:E2457–E2466. doi: 10.1073/pnas.1422948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51(12):3075–3079. [PubMed] [Google Scholar]
- 9.Loeb LA. Microsatellite instability: Marker of a mutator phenotype in cancer. Cancer Res. 1994;54(19):5059–5063. [PubMed] [Google Scholar]
- 10.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10(7):705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 11.Takama F, et al. Mutation analysis of the hMTH1 gene in sporadic human ovarian cancer. Int J Oncol. 2000;17(3):467–471. doi: 10.3892/ijo.17.3.467. [DOI] [PubMed] [Google Scholar]
- 12.Yamtich J, Nemec AA, Keh A, Sweasy JB. A germline polymorphism of DNA polymerase beta induces genomic instability and cellular transformation. PLoS Genet. 2012;8(11):e1003052. doi: 10.1371/journal.pgen.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eshleman JR, Markowitz SD. Mismatch repair defects in human carcinogenesis. Hum Mol Genet. 1996;5(Spec No):1489–1494. doi: 10.1093/hmg/5.supplement_1.1489. [DOI] [PubMed] [Google Scholar]
- 14.Williams LN, Herr AJ, Preston BD. Emergence of DNA polymerase ε antimutators that escape error-induced extinction in yeast. Genetics. 2013;193(3):751–770. doi: 10.1534/genetics.112.146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flohr T, et al. Detection of mutations in the DNA polymerase delta gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer. 1999;80(6):919–929. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 16.Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase delta variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci USA. 2010;107(1):157–162. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abaan OD, et al. The exomes of the NCI-60 panel: A genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73(14):4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaehnig EJ, Kuo D, Hombauer H, Ideker TG, Kolodner RD. Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Reports. 2013;4(1):174–188. doi: 10.1016/j.celrep.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20(9):1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 20.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281(12):7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 21.Chabes A, et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112(3):391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 22.Kumar D, et al. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39(4):1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(48):19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972;247(1):241–248. [PubMed] [Google Scholar]
- 25.Ganai RA, Bylund GO, Johansson E. Switching between polymerase and exonuclease sites in DNA polymerase ε. Nucleic Acids Res. 2015;43(2):932–942. doi: 10.1093/nar/gku1353. [DOI] [PMC free article] [PubMed] [Google Scholar]