Significance
The human mitochondrial chaperonin is vital for proper cell function because it assists in folding of mitochondrial proteins. Additionally, it participates in extramitochondrial processes, such as apoptosis, inflammation, and carcinogenesis. In this study, we report the crystal structure of mitochondrial chaperonins. The model shows an “American football”-shaped intermediate, composed of two 7-membered chaperonin rings capped at each end by a cochaperonin ring. This complex was captured in the early stages of dissociation. The extensive interface contacts between its rings, the asymmetry that exists within each ring, and symmetric binding of nucleotide cofactors that exists between the rings suggest that the mitochondrial system operates via a mechanism that is distinct from the mechanism of the canonical Escherichia coli (GroEL/GroES) system.
Keywords: mitochondrial chaperonin, symmetrical complex, Hsp60, chaperone, Hsp10
Abstract
Human mitochondria harbor a single type I chaperonin system that is generally thought to function via a unique single-ring intermediate. To date, no crystal structure has been published for any mammalian type I chaperonin complex. In this study, we describe the crystal structure of a football-shaped, double-ring human mitochondrial chaperonin complex at 3.15 Å, which is a novel intermediate, likely representing the complex in an early stage of dissociation. Interestingly, the mitochondrial chaperonin was captured in a state that exhibits subunit asymmetry within the rings and nucleotide symmetry between the rings. Moreover, the chaperonin tetradecamers show a different interring subunit arrangement when compared to GroEL. Our findings suggest that the mitochondrial chaperonins use a mechanism that is distinct from the mechanism of the well-studied Escherichia coli system.
The type I chaperonin family is essential for the life of all eukaryotes, because the main function of its members is to mediate actively the folding of cellular proteins (1–4). The bacterial chaperonin GroEL/GroES system is the most intensively studied chaperonin system, owing to its profound stability, and it serves as a prototype for the less stable 60-kDa chaperonins from mitochondria and chloroplasts. The GroEL molecule is a tetradecamer whose subunits are organized in two rings, producing a barrel-like structure. The central cavity of the barrel provides an isolated environment for protein folding, a process that requires the assistance of the cochaperonin GroES as well as ATP hydrolysis (5, 6).
Several intermediate complexes were suggested to be key players in the GroEL/GroES reaction cycle. The transition between the various intermediates is governed by the state of the nucleotide molecules bound to GroEL. In vitro, the acceptor state for the unfolded protein substrate is either the apo or the asymmetric form of the GroEL–GroES complex (a bullet-shaped complex containing one molecule of GroEL and one molecule of GroES). The protein-folding cycle begins when GroES binds to the ring occupied by substrate protein and ATP (the cis ring). ATP hydrolysis in the cis ring is followed by binding of ATP to the trans ring, which results in dissociation of the cis complex, thereby releasing the folded substrate protein and resetting the cycle (7–9). However, numerous studies have demonstrated that an additional complex is formed between GroEL and GroES during the protein-folding reaction cycle. This symmetric complex, called the “football” (American football-shaped), was shown to be a functional intermediate of the chaperonin folding reaction cycle (10–13). Of all the GroEL/GroES chaperonin intermediates, the symmetric complex is the only form whose structure has not been determined until recently (published during the review of this paper), apparently because of its transient nature (14, 15).
Human mitochondria harbor one chaperonin system whose proteins, human mitochondrial heat shock protein 60 (mHsp60) and its cochaperonin, human mitochondrial heat shock protein 10 (mHsp10), exhibit 51% and 33% identity to GroEL and GroES, respectively. The accepted model of the protein-folding reaction cycle by mHsp60 suggests that this folding nanomachine functions via a mechanism distinct from the mechanism of GroEL/GroES and acts as a single heptameric ring (16–18). This chaperonin system is crucial for mitochondrial function and cellular viability, as demonstrated by the embryonic lethality in mice that results from inactivation of the mHsp60 gene, as well as by the identification of three human neurodegenerative genetic disorders associated with mutations in this protein (19–23). In addition to their essential protein-folding activity in mitochondria, studies have implicated the mammalian mitochondrial chaperonins in a wide range of other extramitochondrial and extracellular activities, including modulation of apoptosis (24, 25), inflammation (26), and carcinogenesis (reviewed in refs. 27–30).
Here we report, the first crystal structure of mHsp60 in complex with its cochaperonin. The model that we obtained provides a snapshot of a unique intermediate, likely in a step preceding the dissociation of the complex into its components, that exhibits three unique properties: (i) the mHsp60–mHsp10 complex forms a symmetric double-ring, football-like structure [mHsp6014–(mHsp107)2 complex] that displays extensive interring contacts; (ii) the symmetric nature of the chaperonin subunits within each ring, which is observed in GroEL, is not preserved; and (iii) the interring nucleotide asymmetry that defines the bacterial folding cycle is absent, because both mHsp60 rings are in the ADP-bound state.
Results
Structure Determination and Refinement.
The mHsp60–mHsp10 complex is known for its intrinsic instability (16, 17). To facilitate crystallization of this complex, we eliminated the salt bridge between E321 and K176 (mHsp60E321K) that is known to break upon transition from the closed conformation to the open conformation during the reaction cycle, leading to the formation of a high-affinity complex. We previously isolated this mutant and showed that it has acquired the ability to function with GroES both in vivo and in vitro, although losing the ability to function with its endogenous partner, mHsp10 (31) (Fig. S1). This lack of functionality was shown to be due to its inability to release mHsp10 (detailed in ref. 31). Further experiments with the mHsp60E321K mutant using the substrate EGFP revealed that the mutant is able to functionally encapsulate and refold HCl-denatured EGFP and to release it within the chaperonin cavity. The mutant has only lost the ability to open the cavity and to discharge the folded protein (Fig. S2). This method was used previously to characterize the SR1 mutant of GroEL, which is arrested in the same stage as our mutant, as described by Weissman et al. (32). These results indicate that the stable mHsp60–mHsp10 complex represents a true intermediate of the mHsp60 folding cycle. Crystallization trials were carried out with purified mHsp60E321K lacking the C-terminal GGM motif. Removing the GGM tail, which is predicted to be unstructured, improved data resolution.
Crystallization of the mHsp60E321K mutant in the presence of mHsp10 turned out to be advantageous on two counts: (i) The high stability of the mHsp60E321K–mHsp10 complex helped to overcome the labile nature of the mHsp60 molecule, and (ii) the E321K mutation stabilizes the “open” conformation of the molecule, making it possible to obtain crystals of mHsp60 that are fully complexed with mHsp10. mHsp60E321K and His-tagged mHsp10 were cocrystallized in the presence of ATP, as we reported previously for the complex of human mHsp60E321K with mouse mitochondrial Hsp10 (33). Dehydration of the crystals extended the diffraction resolution from 7 Å to 3.15 Å (Table S1). The structure of the complex was determined by molecular replacement, using the mHsp60E321K-mouse–Hsp10 complex as a search model.
Architecture of the mHsp60–mHsp10 Complex.
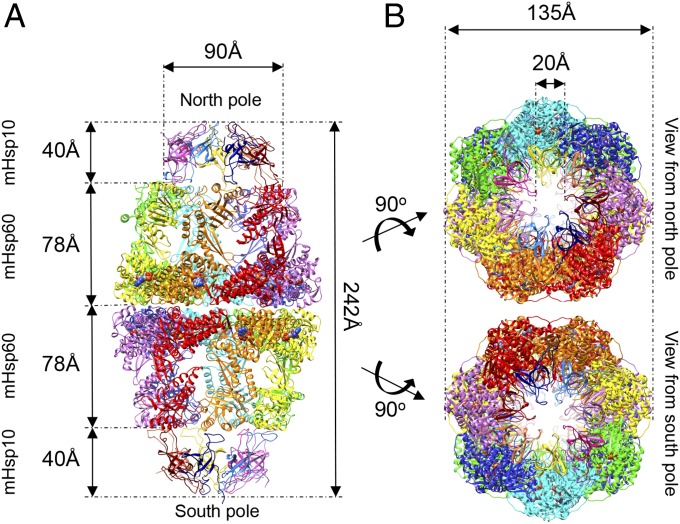
The overall architecture of the complex preserves the well-known domain assembly of chaperonin subunits (Fig. 1). Within this framework, the subunit of the mHsp60 contains an apical, an intermediate, and an equatorial domain. Also conserved is the basic domain structure of the mHsp10 subunit, comprising a seven-strand, β-barrel structure and the prominent mobile loop (Fig. 1A and Fig. S3). Finally, the oligomeric states of the individual mHsp60 (tetradecamers) and mHsp10 molecules (heptamers) are preserved (Fig. 1). Notably, mHsp60 assembles into a double-ring structure that binds two mHsp10 molecules, one at each end of the tetradecamer, forming the symmetric chaperonin complex [mHsp6014–(mHsp107)2 complex], previously termed the “football complex.” This complex was found to play an important role in the chaperonin protein-folding reaction cycle (34–41). To distinguish between the two symmetric halves of the football, we called one of the halves the “north pole” and the other the “south pole” (the full nomenclature of subunits is provided in Fig. S4).
Fig. 1.
Overview of the mHsp60–mHsp10 football complex. (A) View perpendicular to the long axis of the complex. The seven subunits in each mHsp60 ring are colored red, orange, yellow, green, cyan, blue, and purple. The mHsp10 subunits in each ring are colored blue, light blue, purple, dark purple, yellow, pink, and dark red. Structure dimensions are marked by arrows (distances between Cα). Atoms of the 14 ADP molecules are rendered as spheres. (B) Views from the north and south poles of the mHsp6014–(mHsp107)2 complex. The coloring scheme is as in A.
Our efforts to crystallize mHsp60 in the presence of mHsp10 lacking the C-terminal His-tag were unsuccessful. The 3.15-Å resolution structure explains why: The C-terminal His-tag of subunit Z from the south poles of each football complex protrudes toward the neighboring symmetry-related complexes and makes contact with subunits F and G of the north pole, apparently stabilizing the crystal lattice (Fig. S5 A and B; a detailed description of the packing arrangement of the complex is provided in SI Materials and Methods). To rule out the possibility that the observed double-ring structure is induced by the His-tag, we examined the oligomeric state of mHsp60–mHsp10 complexes in solution, using SEC-MALS (size-exclusion chromatography multi-angle static light scattering) (Fig. S6). In these experiments, we used two mHsp10 constructs, only one of which contained the His-tag tail. Our data show that both constructs lead to the formation of similar oligomeric complexes (Fig. S6 C and D). The results of our experiments also confirm our prediction that WT mHsp60 is less stable than the mHsp60E321K mutant, because we detected large amounts of monomers during examination of the WT protein (Fig. S6 A and B).
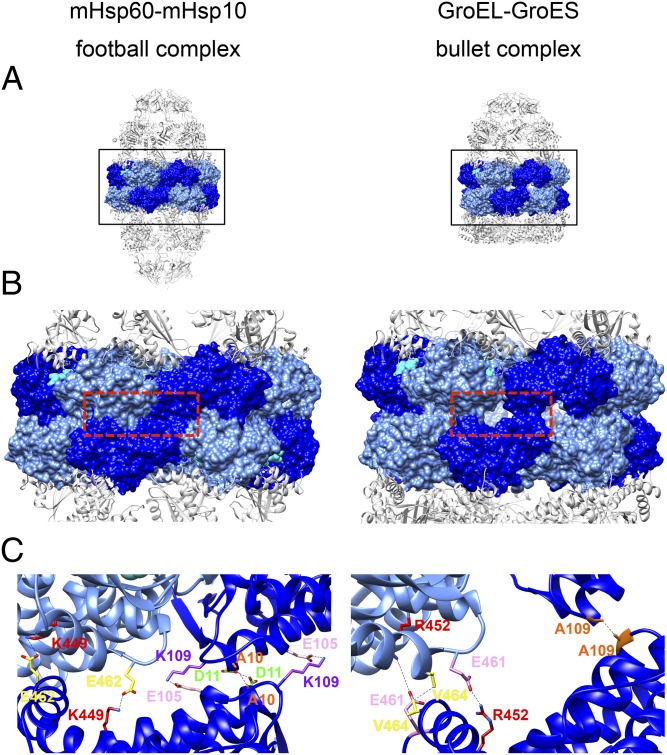
To gain more insight on the assembly of mHsp60 into double rings, we examined the contacts between the mHsp60 rings more closely. Using the PISA program (42), we found that contacts between two heptameric rings of mHsp60 in the football structure are more than twice as extensive as those contacts observed for the two heptameric GroEL rings in the bullet GroEL/GroES structure [Protein Data Bank (PDB) ID code 1AON], with contact surface areas of 5,820 Å2 and 2,466 Å2, respectively. Magnification of a selected area in the interring region shows that different contacts are indeed formed in the football structure than in the bullet structure of GroEL (Fig. 2). Inspection of the football structure in a direction perpendicular to the long axis of the complex provides an explanation for this observation: The mHsp60 rings are rotated around the long axis in opposite directions by ∼5° relative to their orientation in GroEL. This movement enables the equatorial domains of each ring to be positioned deeper into each other in a zipper-like formation, making it possible for new contacts to be formed in the football structure. In this newly formed interface, the subunits maintain their two symmetric contact points that exist in GroEL, but the residues participating in the contacts have changed (Fig. 2C). For example, at one of the two existing contact points, the symmetric key interring contact of A109 in GroEL is switched with a salt bridge between K109 and E105 (in mHsp60, the amino acid is K in position 109 instead of A). In addition, a new symmetric hydrophobic interaction is formed between two A10, whereas a new symmetric hydrogen bond is formed between two D11. At the other contact point, the salt bridge between E461 and R452 that is formed in GroEL is replaced by a salt bridge between E462 and K449 (in mHsp60, there is an M in the equivalent position of R452). To evaluate the importance of the above residues for formation of the double-ring structure, we mutated three residues at the ring interface (on a background of the WT mHsp60). The mutations in this triple mutant (mHsp60TM) were E105A, K109Q, and E462A. Interestingly, mHsp60TM was extremely unstable in the apo state and eluted as ∼2,000-kDa aggregates when examined using SEC-MALS. When ATP and mHsp10 were added, they induced formation of mainly single-ring molecules (∼420 kDa; Fig. S7A). When we tested the ability of mHsp60TM to refold malate dehydrogenase, it was found to be as active as the WT mHsp60 (Fig. S7B). In summary, these findings reinforce the observation from the crystal structure that the interring arrangement of mHsp60 is different from the interring arrangement for GroEL, and that although mHsp60 is isolated as a single ring, it is found in equilibrium between single- and double-ring structures in the presence of mHsp10 and ATP, supporting a previous report (43) and challenging the common view that this protein acts exclusively as a single-ring protein (18).
Fig. 2.
Hsp60 interring contacts in the symmetric human football complex (Left) and in the asymmetric bacterial bullet complex (Right). (A) Full side views of the mitochondrial complex and the bacterial (PDB ID code 1AON) complex. The subunit equatorial domains are rendered as a molecular surface in alternating blue and light blue. The other domains in the Hsp60 subunits and Hsp10 subunits are presented as gray cartoons. (B) Zoom-in view of the boxed area in A, showing the Hsp60 interring contact area. ADP atoms are presented as cyan spheres. (C) Three subunits from each complex were taken from the red boxed area in B and presented as cartoons with stripped surfaces, colored as in A and B. Residues forming the main interring contacts are presented as sticks: in the mitochondrial complex (Left) [A10 (orange), D11 (green), E105 (pink), K109 (purple), K449 (red), and E462 (yellow)] and in the bacterial complex (Right) [A109, (orange), R452 (red), E461 (pink), and V464, (yellow)]. The bonds between the interacting residues are shown as black dashed lines. Subunits of mHsp60 assemble in a zipper-like conformation in which the contact surface is larger and contacts are tighter than in GroEL (B and C), allowing new contacts to be formed.
Intraring Asymmetry.
One notable feature of the canonical type I chaperonin reaction cycle is that conversion of one reaction intermediate to another (e.g., from apo GroEL to an asymmetric bullet complex) requires the concerted movement of all seven apical domains in one ring toward the open conformation (44, 45).
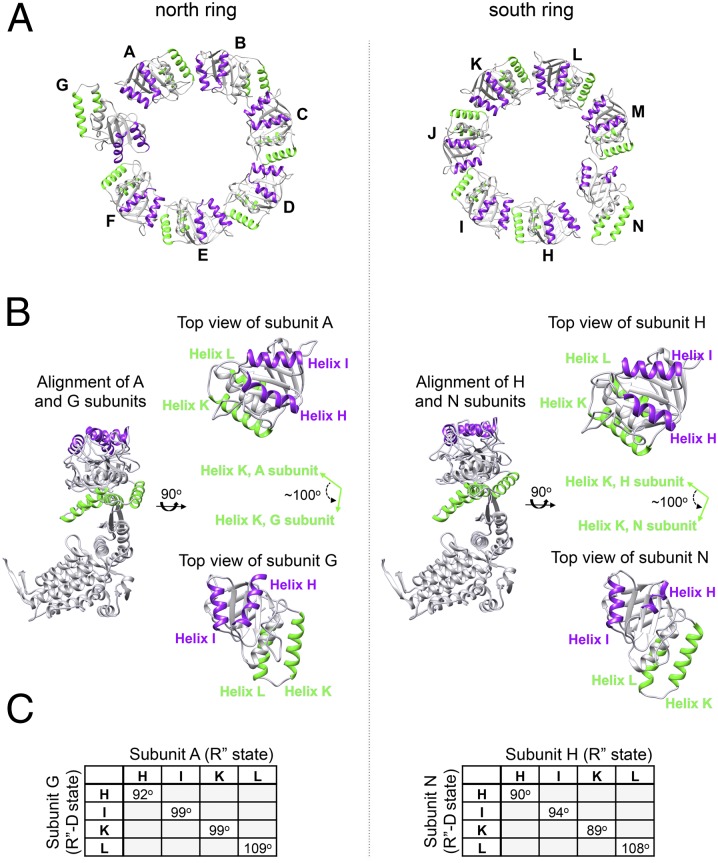
Examination of the football complex looking down from each of its poles yielded a surprising observation: Whereas six of the mHsp60 subunits are almost identical in their tertiary conformation, the seventh subunit is completely different, exhibiting an ∼100° counterclockwise rigid body movement of the apical domain, thus breaking the symmetry within the individual rings (Fig. 3, Fig. S8, and Table S2). Previous studies have shown that following ATP and cochaperonin binding, the apical domain undergoes two conformational changes: The first movement is a 60° elevation, and the second movement is a ∼90° clockwise twisting movement that exposes the GroES-binding site to mobile loop binding and allows the release of substrate into the central chamber (46). We suggest that subunits G and N have begun the reverse movement, namely, a counterclockwise movement of 100° toward dissociation of the complex, but the process is arrested at this stage and further “down” movements do not occur. The unique intermediate that was captured in these two subunits was termed by us the R′′-D state (relaxed state that started the dissociation phase).
Fig. 3.
Asymmetry of subunits within mHsp60 rings. (A) Top view of the apical domains in a ring (residues 192–375) is presented: north pole (Left) and south pole (Right). Helices H and I are colored purple, and helices K and L are colored green. Subunits of mHsp60 are identified by letters. In each ring, there is one subunit that breaks the symmetry of the ring (subunit G in the north pole and subunit N in the south pole). (B) Alignment of subunits A and G (north ring) and subunits H and N (south ring). Coloring is as in A. (Right) In each column, separate top views of the aligned subunits are shown. The apical domains of subunits G and N have rotated counterclockwise nearly 100° from the original position of the other six subunits in each ring, as demonstrated here by comparing helices K of subunits A and G or of subunits H and N (green arrows). (C) Tables showing the crossing angles of helices H, I, K, and L between the R′′ state (relaxed state after ATP hydrolysis) and the newly formed R′′-D state (R′′ state that started the dissociation process) in subunits G and N. Angles were calculated using the UCSF Chimera package (64).
Two observations support our assumption that the structure determined here likely represents an event in the cycle just before mHsp10 dissociates from mHsp60: (i) Surface plasmon resonance measurements showed that the association step of mHsp60E321K with mHsp10 is similar to the association step of the WT Hsp60, whereas in the case of the mutant, the dissociation step is markedly prolonged (31), and (ii) despite the fact that the complex was crystallized in the presence of ATP, the nucleotide occupying the binding site in the football structure is ADP in all of the subunits (as will be discussed below).
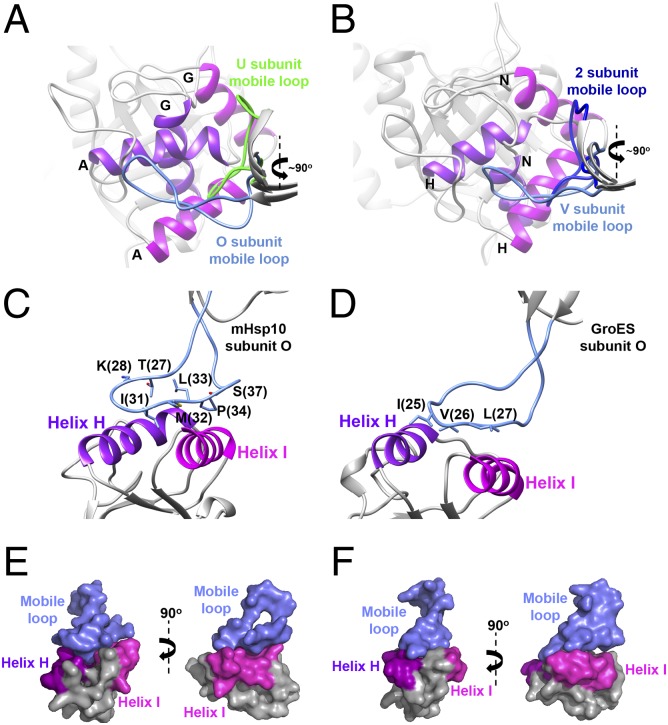
We can see that the mHsp10 heptamers deviate from symmetry as well (Fig. 4 and Table S3). Despite the occurrence of a significant back movement of 100° in the apical domain, the mobile loop follows the apical domain of the G and N subunits and remains associated with it (Fig. 4 A and B). Thus, the mobile loops of mHsp10 subunits U and 2 (the nomenclature is provided in Fig. S4) evidently initiate the dissociation move but do not proceed further, owing to stabilization of the open state of mHsp60E321K (31). We can speculate that complete dissociation of the mobile loop from mHsp60 is gradual, and occurs once the apical domain begins the downward movement toward its apo conformation.
Fig. 4.
Conformations of the mobile loop in the mHsp6014–(mHsp107)2 complex. (A) View from the north pole on the alignment of mHsp60 subunits A and G, showing the binding sites with mHsp10 mobile loops of subunits O and U, respectively. Mobile loops are colored light blue (subunit O) and green (subunit U). Helix H is colored purple, and helix I is colored magenta. Subunit identities of helices H and I are shown. (B) Same as A, but for the view from the south pole on the alignment of subunits H and N while bound to mobile loops of subunits V and 2, respectively. Mobile loops are colored light blue (subunit V) and blue (subunit 2). Views show the mHsp10 mobile loop/mHsp60-binding site (C) compared with the GroES mobile loop/GroEL-binding site (D). In both images, the mobile loop is colored blue and helices H and I are colored as in A. The residues in the mobile loop that mediate the binding to mHsp60/GroEL apical domains are displayed as sticks. (E and F) Surface presentation of C and D, respectively (Left), and 90° image rotation of the same views (Right). The color scheme is as in C and D.
Interestingly, the number of residues in the mobile loop of mHsp10 that participate in mHsp60 binding to helices H and I is greater than in GroEL/GroES (Fig. 4 C and D). In the mobile loop of GroES, the well-known IVL amino acid triad is responsible for interaction with the GroEL apical domain. In mHsp10, there are four residues (T27, K28, P34, and S37), in addition to the IML amino acid triad (equivalent to GroES IVL) that participate in the interaction between the mobile loop and the neighboring apical domain. Comparison of the interface between the mHsp10 mobile loop and mHsp60 with the interface of GroEL/GroES shows that the mobile loop adopts a flattened conformation that is more buried and more tightly bound in the interacting helices (Fig. 4 C–F). Furthermore, the contact interface area between the mHsp10 mobile loop and mHsp60, as examined by the PISA program, is ∼54 Å2 larger than the contact area of GroES with GroEL (average of 427 Å2 and 373.5 Å2, respectively). These observations support a previous suggestion that mHsp10 has a higher affinity for chaperonins than other cochaperonin homologs (47).
The lack of conformational identity of subunits in each heptamer of mHsp60 and mHsp10 contradicts the accepted view of perfect sevenfold symmetry and concerted release of cochaperonin that exists in the GroEL/GroES system. The breakage of perfect symmetry was reported in the past, but to a milder extent, because it was shown that the refined apo GroEL structure and the (GroEL-KMgATP)14 structure (PDB ID codes 1OEL and 1KP8, respectively) present a flexible apical domain that can be attributed to the ability of the chaperonin to bind different substrates or to the involvement of the apical domains in substrate unfolding processes (48, 49). Similarly, a lack of symmetry exists in the cis ring of the Thermus thermophilus chaperonin (50). This deviation was suggested to result from encapsulation of peptide substrates, thus representing the functional conformation of the complex. In more recent studies, it was also shown that a deviation from symmetry exists between subunits of one ring in the crystal structure of the GroEL–R-ADP complex and in the structures of the GroEL–GroES football complexes, published during the reviewing process of this paper [PDB ID codes 4KI8 (R-ADP), 4PKO, and 4PKN (football)] (14, 51). The observed intraring asymmetry in our structure is more intense than most of the flexible movements in the above studies, and it is accompanied by similarly asymmetric movements of mHsp10 mobile loops. Still, the fact that this rotation is observed in both rings, which present different lattice contacts and slightly different rotation angles (as can be seen in Fig. 3C and Fig. S5 C and D), implies that these conformations are not solely lattice-induced. Our results suggest that the movement of subunits within the mitochondrial chaperonin ring is not concerted, although we cannot exclude the possibility that the movement of the subunits is concerted during the binding event but not during the dissociation.
Interring Symmetry.
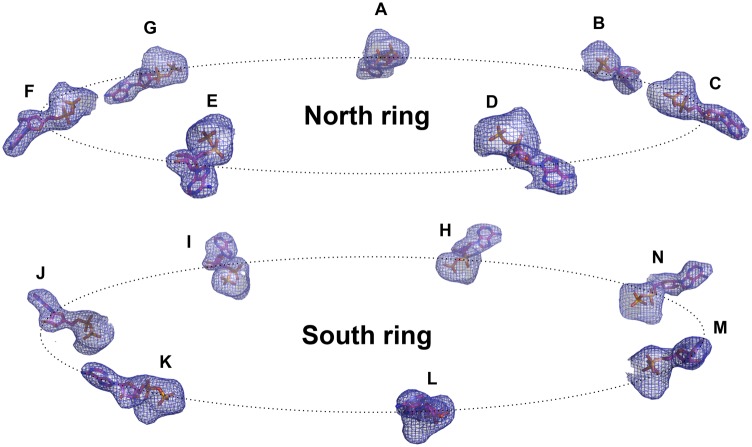
Another feature of the GroEL/GroES mechanism is that negative cooperativity of ATP hydrolysis exists between the two chaperonin rings (44). According to this model, both rings of the chaperonin will not be occupied concomitantly by ADP. It is expected that both rings will be occupied by ATP or that one will be occupied by ADP and the other by ATP, where the former is the ring that is about to release the cochaperonin. During the protein crystallization process, ATP nucleotide was included in the mixture of mHsp60E321K and mHsp10. Notably, examination of the football structure shows that the nucleotide asymmetry that characterizes GroEL rings was not observed: The nucleotide-binding sites in all mHsp60 subunits are occupied by ADP and magnesium (Fig. 5). The latter finding implies that the mHsp60–mHsp10 complex reaction cycle proceeds without significant negative cooperativity between the two rings, which would block ATP hydrolysis in the opposite (trans) ring. However, the observation that the same rotation is seen in the equivalent subunits (G and N) within the complex may imply that there is some coordination between the two rings.
Fig. 5.
Both mHsp60 rings are in the post ATP-hydrolysis state. A side view of the 14 ADP molecules as they are positioned in the structure is shown. Nucleotides are shown as sticks and are labeled according to their subunits. In addition, a Fo-Fc omit electron density map of the 14 nucleotides is shown. The map was calculated by omitting the nucleotides and magnesium atoms from the final model and performing five cycles of refinement using REFMAC [version 5.7.32 (58)]. The map was contoured at a 3σ cutoff.
Discussion
The crystal structure of the football mHsp6014–(mHsp107)2 complex reported here suggests that this chaperonin system is mechanistically unique in two aspects. First, movement of the apical domains within the heptameric rings is not concerted. Second, the fact that all subunits in both rings are occupied by ADP suggests that no significant negative cooperativity exists between the two rings in this complex. The fact that the mHsp60–mHsp10 complex crystallized as a double-ring football assembly is intriguing, given that the apo form of the complex is found mainly as a single ring. Our results suggest the following as the main steps in the mitochondrial chaperonin reaction cycle: (i) The apo form of the molecule exists as a single ring; it can bind ATP and unfolded substrate protein, leading to a slight shift in the equilibrium toward formation of double rings; (ii) upon binding of cochaperonin, the mHsp10-bound rings associate to form the double-ring football structures; and (iii) following ATP hydrolysis, which occurs independently in each ring, the cochaperonin, ADP, and the folded substrate are released (18) and the apo form of mHsp60 is restored. Thus, it is tempting to speculate that for the majority of the mHsp60 molecules, the productive cycle starts with a single ring, which assembles to form a double-ring football structure, with no need for an intermediate asymmetric bullet structure. Alternatively, it is possible that the mHsp60 double-ring oligomer keeps one ring occupied with mHsp10, resulting in a catalytic cycle that alternates between symmetric and asymmetric complexes. Recent studies have suggested that the GroEL/GroES chaperonin system undergoes two cycles: the asymmetric “bullet” cycle and the symmetric football cycle (52, 53). It was shown that the symmetric cycle is preferred in the presence of substrate protein. The results of the present study suggest that the symmetric cycle of the mHsp60–mHsp10 complex chaperonin system is preferred even in the absence of substrate protein.
Materials and Methods
Crystallization.
Crystallization trials with the mHsp60E321K–mHsp10 complex (His-tag mHsp10) were carried out as described by Nisemblat et al. (33). Crystallization of the selenomethionine (SeM)–mHsp60E321K–mHsp10 complex was carried out under the same conditions. Before diffraction measurements, the crystals were dehydrated as described (33).
Data Collection and Processing.
The dehydrated crystals were harvested from hanging drops using cryoloops, oriented along the longest axis, plunged into liquid nitrogen, and placed in pucks for transportation to the synchrotron at cryogenic temperature. Crystals were mounted in the diffraction position using the standard sample changer maintained at 100 K at the European Synchrotron Radiation Facility (Grenoble, France). Diffraction data were collected on ID23-1 and ID29 beam lines set to wavelengths 0.95370 and 0.97908, respectively, using ADSC Q315 and Pilatus 6M area detectors. Data were integrated and scaled using DENZO and SCALEPACK as implemented in HKL2000 (54) and XDS (55). Statistics of the data processing are shown in Table S1.
Crystal Structure Determination and Refinement.
The structure of the human mHsp60–mouse mHsp10 complex (more information is found in ref. 33) was initially determined at 3.34 Å by the molecular replacement program MOLREP [version 11.1.03 (56)] as implemented in the CCP4 suite [version 6.3.0 (57)], using the coordinates of the GroEL/GroES R′′-state cis ring [GroEL7-GroES7-ADP; PDB ID code 1AON (46)] as a model [having 48.6% and 32.8% identity with the human chaperonin and mouse cochaperonin constructs, respectively (construct sequences are provided in Fig. S3)]. Solute molecules, nucleotide cofactors, and magnesium atoms were deleted from the model coordinate file. The model coordinates were not trimmed to the unequivocal structure. The molecular replacement solution (score = 0.233, translation function peak/noise = 17.92, and contrast = 12.38) revealed 14 mHsp60 and 14 mHsp10 molecules arranged in an elliptical football-like assembly. In addition to the sevenfold noncrystallographic symmetry (NCS) of chaperonin and cochaperonin rings, the complex possessed a twofold NCS symmetry axis located as annotated in Fig. S4A. Refinement of the structure started at Rmodel = 0.5169 and Rfree = 0.5164, and reached Rmodel = 0.3595 and Rfree = 0.4176 after 100 cycles of refinement using REFMAC [version 5.7.32 (58)] with the activated jelly-body option, the use of secondary structure restraints as generated by PROSMART [version 0.816 (59)], and the application of local NCS restraints, unequivocally supporting the correctness of the molecular replacement solution.
When 3.15-Å resolution diffraction data of the human Hsp60–human Hsp10 complex became available, the model obtained for the human Hsp60–mouse Hsp10 complex was mutated to a human Hsp60–human Hsp10 complex (with differences in four amino acid residues in Hsp10). The model was further refined and rebuilt by cycling through REFMAC5 [version 5.8.0069 (58)] and Coot (60). Close to the final stages of refinement, proper weighting of the experimental information and the applied restraints were determined using the PDB_REDO Web Server (61). Refinement was implemented with the activated jelly-body option, the use of secondary structure restraints as generated by PROSMART, and the application of local NCS restraints. The apical domains of subunits G and N showed a significant deviation from the other 12 subunits, and they had to be further adjusted manually in Coot (60). General correctness of sequence and position was confirmed by the anomalous map of the SeM derivative of mHsp60E321K, calculated using PHENIX (62) at 3.8 Å resolution (Table S1). This map exhibited correlation of the Se peak positions with the peak positions of the methionine sulfur atoms, verifying their position in the model. Final cycles of refinement were performed using PHENIX (version 1.9_1692) and converged to Rwork = 0.241 and Rfree = 0.270. The final model has 93% of residues in allowed regions on a Ramachandran plot (63). In addition, 2.3% of the residues in the most disordered parts of the structure are in disallowed regions of the Ramachandran plot.
DEDICATION.
On August 28, 2014 we lost one of the top Israeli crystallographers, Professor Felix Frolow. Felix was a devoted teacher, a friend and a leader in the field of crystallography. This paper is dedicated to his memory, and to the tremendous effort, skill, and knowledge he put into solving the structure of the human mitochondrial chaperonin complex. We are grateful for having had the opportunity to work side-by-side with him and we will cherish his memory.
Supplementary Material
Acknowledgments
We thank Drs. Celeste Weiss and Anna Vitlin Gruber for their useful comments and discussions, Profs. Oded Livnah and Joel Hirsch for their valuable suggestions relating to crystallographic aspects of the manuscript, Dr. Yehezkel Sasson for his assistance with the SEC-MALS experiments, and Olga Levin for the help with the EGFP cloning. We are grateful to the European Synchrotron Radiation Facility (Grenoble, France) for the use of its macromolecular crystallographic data collection facilities, and, in particular, to the ID29 and ID23-1 staff for their assistance. This work was supported by an Eshkol Fellowship from the Israel Ministry of Science (to S.N.), the Legacy Heritage Biomedical Program of the Israel Science Foundation (Grant 1902/08 to A.A.), and a grant from the Israel Science Foundation (Grant 1507/13 to A.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4PJ1).
1Deceased August 28, 2014.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411718112/-/DCSupplemental.
References
- 1.Cheng MY, et al. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337(6208):620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 2.Ostermann J, Horwich AL, Neupert W, Hartl FU. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341(6238):125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 3.Goloubinoff P, Gatenby AA, Lorimer GH. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337(6202):44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 4.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989;342(6252):884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 5.Saibil HR, Ranson NA. The chaperonin folding machine. Trends Biochem Sci. 2002;27(12):627–632. doi: 10.1016/s0968-0004(02)02211-9. [DOI] [PubMed] [Google Scholar]
- 6.Horwich AL, Farr GW, Fenton WA. GroEL-GroES-mediated protein folding. Chem Rev. 2006;106(5):1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 7.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42(2):83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 8.Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and allostery of the chaperonin GroEL. J Mol Biol. 2013;425(9):1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 9.Horovitz A. Putting handcuffs on the chaperonin GroEL. Proc Natl Acad Sci USA. 2013;110(27):10884–10885. doi: 10.1073/pnas.1309581110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt M, et al. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science. 1994;265(5172):656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
- 11.Azem A, Kessel M, Goloubinoff P. Characterization of a functional GroEL14(GroES7)2 chaperonin hetero-oligomer. Science. 1994;265(5172):653–656. doi: 10.1126/science.7913553. [DOI] [PubMed] [Google Scholar]
- 12.Todd MJ, Viitanen PV, Lorimer GH. Dynamics of the chaperonin ATPase cycle: Implications for facilitated protein folding. Science. 1994;265(5172):659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 13.Llorca O, Marco S, Carrascosa JL, Valpuesta JM. The formation of symmetrical GroEL-GroES complexes in the presence of ATP. FEBS Lett. 1994;345(2-3):181–186. doi: 10.1016/0014-5793(94)00432-3. [DOI] [PubMed] [Google Scholar]
- 14.Fei X, Ye X, LaRonde NA, Lorimer GH. Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc Natl Acad Sci USA. 2014;111(35):12775–12780. doi: 10.1073/pnas.1412922111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike-Takeshita A, Arakawa T, Taguchi H, Shimamura T. Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8Å reveals rearrangement between two GroEL rings. J Mol Biol. 2014;426(21):3634–3641. doi: 10.1016/j.jmb.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Viitanen PV, et al. Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J Biol Chem. 1992;267(2):695–698. [PubMed] [Google Scholar]
- 17.Viitanen PV, et al. Purification of mammalian mitochondrial chaperonin 60 through in vitro reconstitution of active oligomers. Methods Enzymol. 1998;290:203–217. doi: 10.1016/s0076-6879(98)90020-9. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen KL, Cowan NJ. A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol Cell. 1998;2(1):93–99. doi: 10.1016/s1097-2765(00)80117-3. [DOI] [PubMed] [Google Scholar]
- 19.Christensen JH, et al. Inactivation of the hereditary spastic paraplegia-associated Hspd1 gene encoding the Hsp60 chaperone results in early embryonic lethality in mice. Cell Stress Chaperones. 2010;15(6):851–863. doi: 10.1007/s12192-010-0194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen JJ, et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70(5):1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen J, et al. A novel mutation in the HSPD1 gene in a patient with hereditary spastic paraplegia. J Neurol. 2007;254(7):897–900. doi: 10.1007/s00415-006-0470-y. [DOI] [PubMed] [Google Scholar]
- 22.Magen D, et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am J Hum Genet. 2008;83(1):30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bross P, Magnoni R, Bie AS. Molecular chaperone disorders: Defective Hsp60 in neurodegeneration. Curr Top Med Chem. 2012;12(22):2491–2503. doi: 10.2174/1568026611212220005. [DOI] [PubMed] [Google Scholar]
- 24.Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J. 1999;18(8):2040–2048. doi: 10.1093/emboj/18.8.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta S, Knowlton AA. HSP60, Bax, apoptosis and the heart. J Cell Mol Med. 2005;9(1):51–58. doi: 10.1111/j.1582-4934.2005.tb00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osterloh A, et al. Lipopolysaccharide-free heat shock protein 60 activates T cells. J Biol Chem. 2004;279(46):47906–47911. doi: 10.1074/jbc.M408440200. [DOI] [PubMed] [Google Scholar]
- 27.Deocaris CC, Kaul SC, Wadhwa R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones. 2006;11(2):116–128. doi: 10.1379/CSC-144R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cappello F, Conway de Macario E, Marasà L, Zummo G, Macario AJ. Hsp60 expression, new locations, functions and perspectives for cancer diagnosis and therapy. Cancer Biol Ther. 2008;7(6):801–809. doi: 10.4161/cbt.7.6.6281. [DOI] [PubMed] [Google Scholar]
- 29.Czarnecka AM, Campanella C, Zummo G, Cappello F. Mitochondrial chaperones in cancer: From molecular biology to clinical diagnostics. Cancer Biol Ther. 2006;5(7):714–720. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- 30.David S, et al. Hsp10: Anatomic distribution, functions, and involvement in human disease. Front Biosci (Elite Ed) 2013;5:768–778. doi: 10.2741/e657. [DOI] [PubMed] [Google Scholar]
- 31.Parnas A, et al. Identification of elements that dictate the specificity of mitochondrial Hsp60 for its co-chaperonin. PLoS ONE. 2012;7(12):e50318. doi: 10.1371/journal.pone.0050318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84(3):481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 33.Nisemblat S, Parnas A, Yaniv O, Azem A, Frolow F. Crystallization and structure determination of a symmetrical ‘football’ complex of the mammalian mitochondrial Hsp60-Hsp10 chaperonins. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 1):116–119. doi: 10.1107/S2053230X1303389X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corrales FJ, Fersht AR. Kinetic significance of GroEL14.(GroES7)2 complexes in molecular chaperone activity. Fold Des. 1996;1(4):265–273. doi: 10.1016/s1359-0278(96)00040-5. [DOI] [PubMed] [Google Scholar]
- 35.Grallert H, Buchner J. Review: A structural view of the GroE chaperone cycle. J Struct Biol. 2001;135(2):95–103. doi: 10.1006/jsbi.2001.4387. [DOI] [PubMed] [Google Scholar]
- 36.Inobe T, et al. Asymmetry of the GroEL-GroES complex under physiological conditions as revealed by small-angle x-ray scattering. Biophys J. 2008;94(4):1392–1402. doi: 10.1529/biophysj.107.114710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koike-Takeshita A, Yoshida M, Taguchi H. Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant. J Biol Chem. 2008;283(35):23774–23781. doi: 10.1074/jbc.M802542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sameshima T, et al. Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J Biol Chem. 2008;283(35):23765–23773. doi: 10.1074/jbc.M802541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nojima T, Yoshida M. Probing open conformation of GroEL rings by cross-linking reveals single and double open ring structures of GroEL in ADP and ATP. J Biol Chem. 2009;284(34):22834–22839. doi: 10.1074/jbc.M109.020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sameshima T, Iizuka R, Ueno T, Funatsu T. Denatured proteins facilitate the formation of the football-shaped GroEL-(GroES)2 complex. Biochem J. 2010;427(2):247–254. doi: 10.1042/BJ20091845. [DOI] [PubMed] [Google Scholar]
- 41.Sameshima T, et al. Single-molecule study on the decay process of the football-shaped GroEL-GroES complex using zero-mode waveguides. J Biol Chem. 2010;285(30):23159–23164. doi: 10.1074/jbc.M110.122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Levy-Rimler G, et al. The effect of nucleotides and mitochondrial chaperonin 10 on the structure and chaperone activity of mitochondrial chaperonin 60. Eur J Biochem. 2001;268(12):3465–3472. doi: 10.1046/j.1432-1327.2001.02243.x. [DOI] [PubMed] [Google Scholar]
- 44.Yifrach O, Horovitz A. Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry. 1995;34(16):5303–5308. doi: 10.1021/bi00016a001. [DOI] [PubMed] [Google Scholar]
- 45.Yifrach O, Horovitz A. Allosteric control by ATP of non-folded protein binding to GroEL. J Mol Biol. 1996;255(3):356–361. doi: 10.1006/jmbi.1996.0028. [DOI] [PubMed] [Google Scholar]
- 46.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388(6644):741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 47.Richardson A, Schwager F, Landry SJ, Georgopoulos C. The importance of a mobile loop in regulating chaperonin/ co-chaperonin interaction: Humans versus Escherichia coli. J Biol Chem. 2001;276(7):4981–4987. doi: 10.1074/jbc.M008628200. [DOI] [PubMed] [Google Scholar]
- 48.Braig K, Adams PD, Brünger AT. Conformational variability in the refined structure of the chaperonin GroEL at 2.8 A resolution. Nat Struct Biol. 1995;2(12):1083–1094. doi: 10.1038/nsb1295-1083. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Boisvert DC. Structural basis for GroEL-assisted protein folding from the crystal structure of (GroEL-KMgATP)14 at 2.0A resolution. J Mol Biol. 2003;327(4):843–855. doi: 10.1016/s0022-2836(03)00184-0. [DOI] [PubMed] [Google Scholar]
- 50.Shimamura T, et al. Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity. Structure. 2004;12(8):1471–1480. doi: 10.1016/j.str.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 51.Fei X, Yang D, LaRonde-LeBlanc N, Lorimer GH. Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 Å resolution. Proc Natl Acad Sci USA. 2013;110(32):E2958–E2966. doi: 10.1073/pnas.1311996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye X, Lorimer GH. Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc Natl Acad Sci USA. 2013;110(46):E4289–E4297. doi: 10.1073/pnas.1317702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang D, Ye X, Lorimer GH. Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc Natl Acad Sci USA. 2013;110(46):E4298–E4305. doi: 10.1073/pnas.1318862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276(Macromolecular Crystallography, part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 57.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joosten RP, Joosten K, Murshudov GN, Perrakis A. PDB_REDO: Constructive validation, more than just looking for errors. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):484–496. doi: 10.1107/S0907444911054515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.