Significance
Cell sheets often organize in tubular structures, for example, in the kidney. Also, cells from epithelial tumors are known to wrap around vessels or muscle fibers as they migrate collectively. By plating cells on thin glass wires, we mimic these physiological conditions in vitro and show that high curvature favors cell detachment at the front edge. This switch from collective to individual migration may reproduce features often observed in cancer invasion. High curvature also induces a circumferential organization of the actin cytoskeleton reminiscent of in vivo embryonic morphogenesis situations where tissues develop on a cylindrical template. Finally, monolayer migration is halted at submicron radii, and the tissue reconfigures into hollow cysts at its leading tip.
Keywords: collective cell behaviors, out-of-plane curvature, epithelial tubes
Abstract
In a wide range of epithelial tissues such as kidney tubules or breast acini, cells organize into bidimensional monolayers experiencing an out-of-plane curvature. Cancer cells can also migrate collectively from epithelial tumors by wrapping around vessels or muscle fibers. However, in vitro experiments dealing with epithelia are mostly performed on flat substrates, neglecting this out-of-plane component. In this paper, we study the development and migration of epithelial tissues on glass wires of well-defined radii varying from less than 1 µm up to 85 µm. To uncouple the effect of out-of-plane curvature from the lateral confinement experienced by the cells in these geometries, we compare our results to experiments performed on narrow adhesive tracks. Because of lateral confinement, the velocity of collective migration increases for radii smaller than typically 20 µm. The monolayer dynamics is then controlled by front-edge protrusions. Conversely, high curvature is identified as the inducer of frequent cell detachments at the front edge, a phenotype reminiscent of the Epithelial−Mesenchymal Transition. High curvature also induces a circumferential alignment of the actin cytoskeleton, stabilized by multiple focal adhesions. This organization of the cytoskeleton is reminiscent of in vivo situations such as the development of the trachea of the Drosophila embryo. Finally, submicron radii halt the monolayer, which then reconfigures into hollow cysts.
In the last years, the physical microenvironment has been repeatedly demonstrated to be crucial in defining cellular behaviors and phenotypes. For example, the rigidity of a substrate on which a cell adheres can dictate its morphology, phenotype, and even fate (1, 2). Also, the substrate’s microtopography or nanotopography can change cell adhesion and orientation (3, 4). Few studies, however, have focused on quantifying the role of out-of-plane curvature on cells or cell assemblies. This is surprising because epithelial sheets are often naturally curved, organizing into tubes, cysts, crypts, or vili whose radius of curvature is typically on the order of a few cells or tens of microns (5). Tubules can even be formed out of a cohesive chain of single cells, each enclosing a central lumen (6), as has been observed for the Drosophila trachea tip cells (7) and in some parts of the Caenorhabditis elegans digestive tract (8).
In vitro, curved substrates provide a controlled way to study the role of an out-of-plane curvature on a system otherwise identical to the classic 2D culture assay on flat substrates. Because multicellular assemblies are largely controlled by the interactions between cells, the influence of curvature on epithelial tissues must be explored on monolayers. In that case, most of the reported experiments have been performed in negative curvature conditions (i.e., tubes or channels) under perfusion (9, 10). In contrast, experiments on positively curved substrates (i.e., wires) have been mostly conducted on single cells. In particular, Schwann cells (11) or fibroblasts (12–15) seeded on glass wires of radii of the order of 10 µm have been shown to align with the wire long axis. More recently, it has been observed that the response of cells to positive curvature depends strongly on the cell type: Epithelial single cells spread in all directions and favor a circumferential alignment of their actin cytoskeleton (16), while fibroblasts align their cell body and their actin fibers longitudinally (16, 17). Some of these differences are captured by a theoretical model in which cell adhesion and active contractility balance the anisotropic bending stiffness of the stress fibers (18). Very recently, monolayers formed from different endothelial cell types cultured on wires were shown to exhibit cell type-dependent orientations (19).
Since growing monolayers on cylindrical wires intrinsically induces a lateral, although borderless, confinement, it is possible that confinement rather than curvature dictates the behavior of these epithelia. To uncouple these two contributions, we grew cells on smooth cylindrical rods of varying radii and systematically compared our results with experiments conducted on confining tracks of equivalent confinement. We note that, here, the basal side of the cell faces the wire and is inside the tube. The cells therefore have an inverted polarity compared with tubule formation during development. Conversely, the in vivo situation of collective migration on blood vessels, nerves, or myofibers (20–22), in particular originating from tumors, is directly comparable to our experiments.
We show that below a radius of the order of 40 µm, the curvature imposes a transverse actin alignment. This cell architecture is coupled with an increase in the number of focal adhesions (FAs) and with a curvature-induced breaking off of the cells at the front edge. In contrast, confinement rather than curvature controls the dynamics of collective migration. Moreover, we show that there exists a critical submicrometric radius under which cells are unable to migrate collectively, resulting in the formation of hollow cysts.
Results
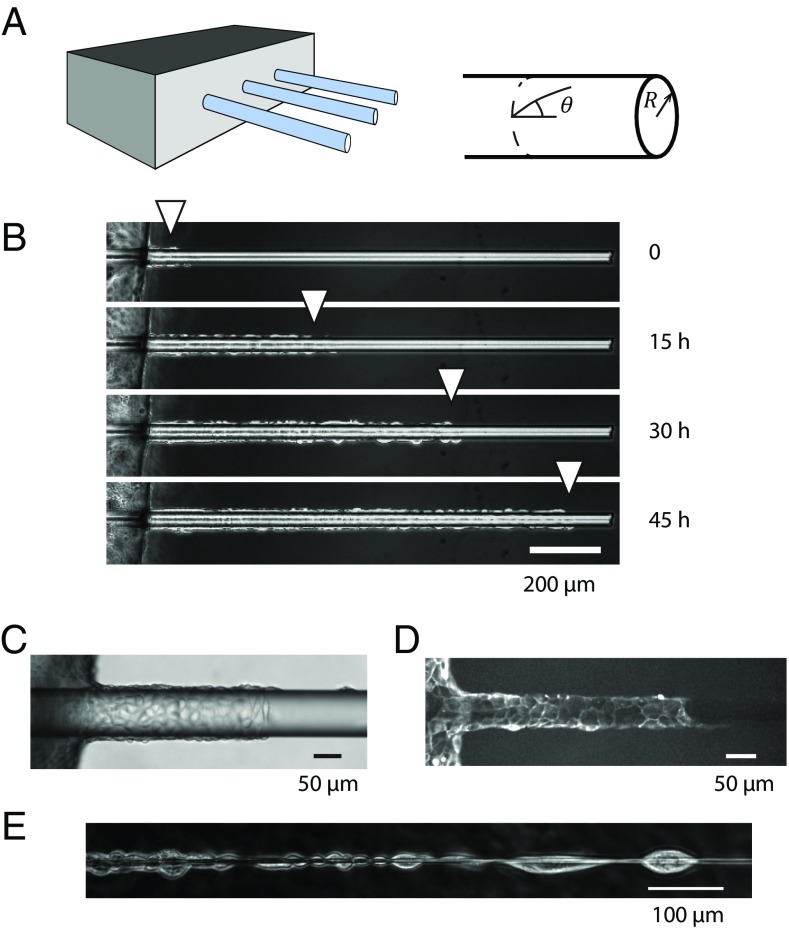
Identical smooth glass wires of radius R varying from 2 µm to 85 µm were aligned on the edge of a piece of silicone gel, fixed in place with cross-linked polydimethylsiloxane (PDMS) (Fig. 1A and SI Appendix, Fig. S1). Unless otherwise specified, the wires were coated with fibronectin. Madin Darby Canine Kidney (MDCK) epithelial cells were then seeded on the fibronectin-coated PDMS supports. We note that these kidney-derived cells assemble into tubes in 3D environments when stimulated by growth factors (23).
Fig. 1.
(A) Schematic of the experimental device with the glass wires aligned on the edge of a PDMS support. (B) The cells grow to confluence on the support at the left of the pictures before invading the wire collectively (R = 20 µm) (C and D) Phase contrast (C) and fluorescence image (D) of cells migrating on wires (distinct experiments, R ≈ 50 µm). (E) For small radii (R < 5 µm), there is room for one cell only around the wire. Cells then arrange in the form of chains of cells. Note that they still adhere together at their front end and back end. (B and E) MDCK wild-type cells; (C and D) MDCK LifeAct cells.
A monolayer grew on the supports to confluence and then progressed onto the wires, which were suspended in the medium. The cells wrapped fully around the wires and moved collectively along them while maintaining cell−cell adhesions (Fig. 1 B−D). Cell migration was monitored for typically 2 days. To uncouple curvature and lateral confinement, cells were also cultured in 2D micropatterned adhesive tracks of varying widths (24, 25). In the following, confinement is defined as the tracks’ width (w) or the wires’ perimeter (2πR).
Fiber Radius Controls the Migratory Phenotype.
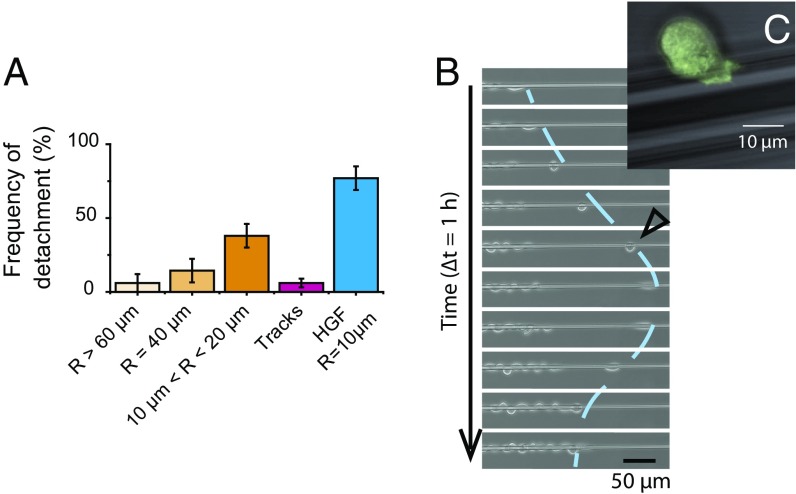
Cells on relatively thick wires (R > 40 µm) remained cohesive and did not escape the monolayer (Movie S1 and Fig. 2A). In contrast, when the wire radius was smaller than 40 µm, cells at the front edge occasionally broke off and moved as single cells. Such events were more frequent at small radii (Movie S2 and Fig. 2A). After detaching from the monolayer, these cells moved back and forth at very high speeds (up to 100 µm⋅h−1) along the wire and/or around it. Because of this erratic movement, the progressing monolayer eventually caught up with these maverick cells, which then readily reincorporated in the monolayer (Fig. 2B). These individually migrating cells switched between elongated and retracted morphologies. In particular, they displaced the fastest when they took a characteristic “rounded up” shape by which they minimized their contact area with the wire (Fig. 2C).
Fig. 2.
(A) Frequencies of detachment of a cell at the front edge. The “frequency of detachment” is the proportion of experiments for which detachment was observed in 22 h. Track measurements were concatenated for 50 µm < w < 200 µm. Measurements in presence of HGF were conducted on 10-µm wires. Error bars are SDs (n > 17 for each box). (B) Sequence illustrating the detachment of a cell at the front edge. Note the change of direction and the reintegration of this cell in the epithelium. R = 5.3 µm. The line is a guide for the eye. (C) Close-up of a detached cell in its fastest migration mode (similar to the cell marked with an empty triangle on B). Note the rounded shape. Superimposition of phase contrast and fluorescence images (LifeAct cells). R = 10 µm.
Experiments performed on the adhesive tracks showed that, at equivalent confinement, cell detachment was seldom observed in tracks of widths larger than 50 µm (equivalent radius R = w/2π ≈ 8 µm) (Fig. 2A). In the few outlying cases where cells did break off on tracks, they remained close to the front or stretched along the edge of the track before being reincorporated into the monolayer. Therefore, we conclude that the unique phenotype of the detached cells seen on wires is due to curvature and not to confinement.
As long as the wire radius was larger than 5 µm, several cells were needed to circle the wire. Above this radius, cells maintained the classical 2D arrangement observed on flat surfaces, and on the largest radii, migration fingers preceded by leader cells were present (26, 27). In contrast, single cells could wrap around thin wires (R < 5 µm) and formed chains in which adhesions were maintained between the back edge of a cell and the front edge of its follower (Fig. 1E and Movie S2).
Cell Architecture on Cylindrical Fibers.
Since we observed a change of behavior of the cells as a function of the wire radius, we turned toward their polarity and their cytoskeleton organization. The cells were fixed and imaged after the front had progressed for 48 h. Staining the monolayer on the wire for ezrin, an apical marker (28), confirmed that the apical surface of the cells was facing the medium and their basal plane contacted the wire (see SI Appendix, Fig. S3). Thus, the tissue polarity seen on flat substrates was conserved on the curved surfaces.
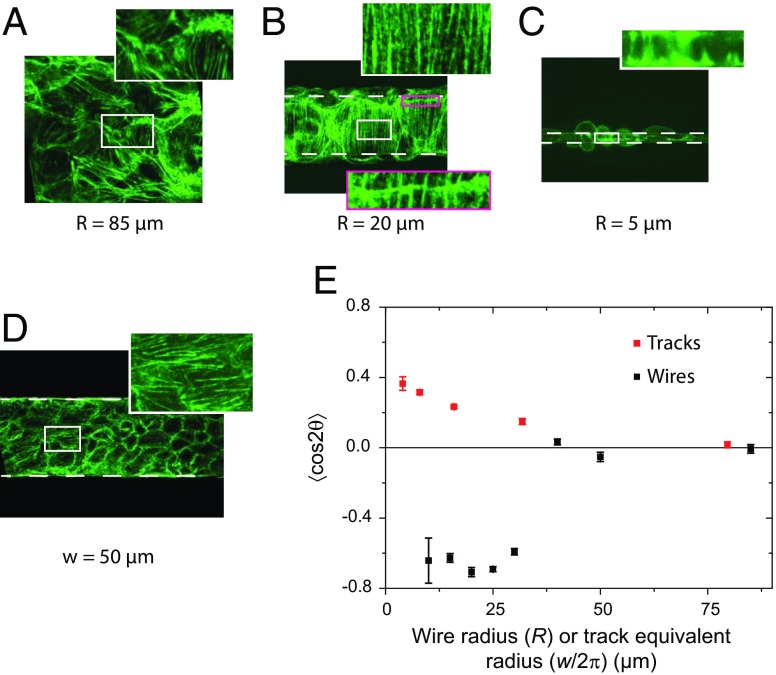
The cell architecture was probed by labeling the actin cytoskeleton. Actin at the basal plane in contact with the wire was primarily in the form of fibers (see SI Appendix, Fig. S4). We found that the actin orientation across the tissue was random for large radii, as it is for flat substrates (Fig. 3A; see SI Appendix, Fig. S5). In contrast, the actin fibers were highly oriented perpendicular to the wire’s longitudinal axis below R ≈ 40 µm (Fig. 3 B and C; see SI Appendix, Fig. S5). It is worth noticing that the same transverse actin alignment was observed even for wires whose radius was small enough to be fully wrapped around by a single cell (Fig. 3C). The same organization of the actin fibers was also observed on uncoated glass wires (see SI Appendix, Fig. S6A) and with another epithelial cell line [retinal pigmented epithelial (RPE1) cells] but not with NIH 3T3 fibroblasts that tended to orient in the direction of the wires (see SI Appendix, Fig. S6 B and C). These results indicate that this organization is not caused by surface composition and could be general to epithelial tissues.
Fig. 3.
(A−C) Orientation of the actin stress fibers for wires of various radii. On the 85-µm wire, the actin fibers are disordered. Below R = 40 µm, actin fibers take a circumferential orientation. Note the continuity of the actin fibers visible across a cell boundary (B, Inset), demonstrating the mechanical continuity in these pluricellular assemblies. C shows circumferential orientation even though the wire is wrapped up by only one cell. (D) The actin fibers on tracks of similar confinement are oriented along the tracks. (E) The order parameter , quantifies these two opposite trends (different signs of S) at small radii/widths. Note the common horizontal scale used to compare wires and tracks. On wires, the transition between not ordered (R > 40 µm) and ordered (R < 40 µm) is very abrupt. Error bars are SDs. (A, B, and D) Phalloidin-Alexa488 labeling. (C) LifeAct cells.
Very interestingly, we observed continuous fibers from cell to cell (Fig. 3B, Inset). This observation implies a mechanical relay at the cell−cell adhesion sites (29). It is a clear signature of local mechanical continuity.
The orientation of these actin fibers was further quantified by using the apolar order parameter , where θ is the angle between the actin fiber and the wire or track axis (Fig. 1A) (30, 31). S = 0 corresponds to a random orientation, S = 1 corresponds to a perfect alignment with the main axis, and S = −1 corresponds to a perfect orientation perpendicular to the main axis. Fig. 3E clearly shows the tendency of the fibers to orient themselves perpendicular to the wire below R ≈ 40 µm.
Experiments performed on micropatterned tracks showed that the actin fibers tended to orient parallel to the tracks at small widths (24) (Fig. 3D). Of note, the order was more pronounced on wires at small confinement. Further, the order parameter measured in tracks decreased smoothly to zero with decreasing confinement (larger width), unlike the situation on wires that shows a sharp transition around R = 40 µm (Fig. 3E). Note that in this plot, S is plotted against R or w/2π to allow comparison between wires and tracks at equivalent confinement. Moreover, on tracks, actin cables on the boundary cells lined the limit of the adhesive track. These boundary effects were not present on the wires where cells were laterally confined but did not experience physical borders. We therefore conclude that the transverse actin orientation on wires is a consequence of curvature and not confinement.
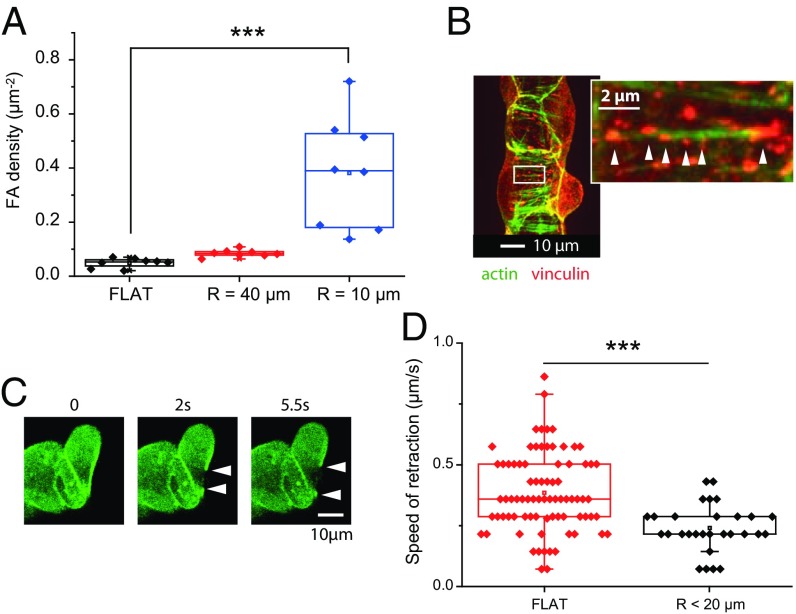
We then characterized the adhesions of the cells on the wires. Fig. 4A shows the density of FAs on the wires for different radii. On wires of radius R < 10 µm, we measured an increase in the density of FAs, as well as a decrease in their size compared with the flat control (0.7 ± 0.9 µm2 on flat substrates vs. 0.3 ± 0.5 µm2 on wires of radius 10 µm; P = 3×10−12). Furthermore, many FAs were found to be spaced along the same fibers (Fig. 4B). The situation on wires is therefore very different from flat surfaces where stress fibers are anchored to the surface via only two large, well-defined FAs at their extremities (see SI Appendix, Fig. S7).
Fig. 4.
(A) Influence of the radius of curvature on the density of FAs. FAs are more numerous on thin wires. (B) At a more local scale, the FAs decorate the circumferential actin fibers; vinculin immunofluorescence and phalloidin-Alexa488 staining. (C) Succession of images of the front-edge actomyosin cable after laser ablation (R = 10 µm); MDCK-LifeAct cells. (D) Initial retraction speeds of the actin cable after photoablation.
Strikingly, local laser photoablation did not trigger a significant retraction of these fibers (retraction smaller than 0.5 µm for R < 20 µm). This observation is consistent with the high density of FAs and their localization along the fibers. It is in sharp contrast to the control experiments on flat surfaces where we routinely observed retractions of several microns, as expected (32). Furthermore, the leading edge presented a clearly defined circumferential pluricellular actin “cable” similar to the contractile cable present at the edge of a classical 2D closing wound (27, 33, 34) (see SI Appendix, Fig. S8). On the wires, the number of adhesions was much less along this cable than along the cells’ stress fibers and retraction after the cut was larger (up to several micrometers), which is a clear signature of the elastic tension stored in this cable (Movie S3 and Fig. 4C). Upon ablation, the initial retraction speed was significantly smaller on small radii wires compared with flat surfaces (0.2 ± 0.1 µm⋅s−1 vs. 0.4 ± 0.2 µm⋅s−1, P = 10−5), this last value being consistent with previous measurements (27, 33) (Fig. 4D).
Collective Migration Speed Is Controlled by Confinement.
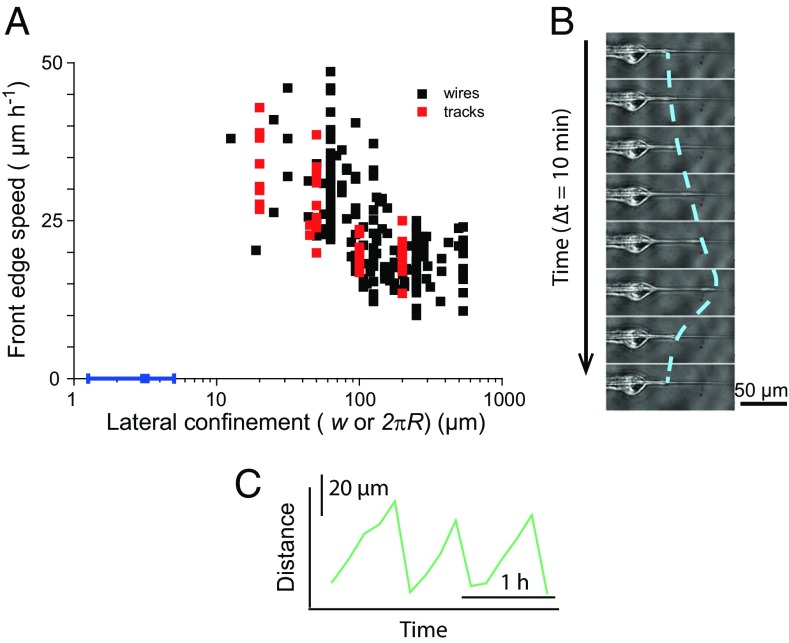
We then turned our attention to the impact of the wire radius on the speed of collective migration, as cell dynamics is related to cytoskeletal organization and adhesion (35). Cells on wires with a radius below R = 20 µm (corresponding to a confinement 2πR = 126 µm) migrated collectively faster than cells on thicker wires, which had velocities comparable to those on a flat surface (Fig. 5A, black points).
Fig. 5.
(A) Front velocity as a function of lateral confinement. Confinement is either the width w of the tracks (red points) or the perimeter 2πR of the wires (black points). Confinement controls the velocity. The blue point shows the arrest of migration on submicron wires. (B) The migration stops at very small radii, at which point the leading cell sends out periodic protrusions. The line is a guide for the eye. (C) Position of the extremity of the protrusion as a function of time. Note the asymmetric shape of these protrusion−retraction cycles.
The cell fronts on wires above R = 10 µm (confinement larger than 63 µm) migrated with a constant speed, as evident from the kymographs (see SI Appendix, Fig. S9A). For smaller radii, the average progression was faster (up to 35 ± 3 µm/h), but it was also less regular (see SI Appendix, Fig. S9B), because of cell divisions that interfered with the forward motion on the thin wires. This effect, also present on small-width tracks (24), became noticeable when the number of cells around the wire was small.
Surprisingly, despite the very different cytoskeletal organization on wires compared with the one on tracks, the velocity−confinement relationship of the front progression in these two cases was very similar (Fig. 5A). We therefore conclude that this velocity increase results from confinement and not curvature. Similarly, the oscillatory behavior at small radii is also observed with small-width tracks (24) and is therefore the consequence of the division events that become more noticeable in situations of high confinement.
Cells Cannot Migrate Collectively on Submicron Wires.
We now focus on the dynamics of the cells at extremely small radii. To explore situations characterized by radii smaller than 1 µm, we had to slightly alter our system, since submicron wires cannot be easily handled. Here, we used tapered glass cones, drawn from a pipette puller, with a submicron radius at their tips. Cones with a range of tapers from 30° down to 1.5° were tested.
With the exception of the detachments of the front cells discussed above, collective cell migration in the form of a multicellular streaming speared by the wire was conserved down to submicron radii. For thinner wires, however, the migrating front stopped before the tip of the cone, at a submicron radius (see SI Appendix, Fig. S10A, and Fig. 5A, blue point). This observation was reproduced for all taper angles tested, meaning that the critical radius that causes the arrest of the migrating front is not dependent on the taper angle in this range and can be extrapolated to straight wires.
At the front edge of the monolayer, the leading cell continued to send out protrusions during 20–30 h that were remarkably periodic, with a period of typically 1 h (Movie S4 and Fig. 5 B and C; see SI Appendix, Fig. S10A, Inset). The shape of the protrusion−retraction cycles was clearly asymmetric, with progressions on the wire 6 times slower than the retraction (Fig. 5C), demonstrating the active process by which the cells’ lamellipodia continued to extend but failed to drag the cell body as they would on large radii wires.
At later stages, as the monolayer became denser by proliferation, oscillating behavior spread to the entire monolayer that then experienced large-scale compression-spreading cycles (Movie S5). As a result of these compressions, the monolayer eventually detached from the wire and buckled, forming a cell-free cavity between the monolayer and the wire, in the form of a hollow cyst that opened further into a cylindrical lumen (see SI Appendix, Fig. S10B).
Active Migration and Proliferation.
Both active migration and proliferation must be considered in propelling front migration for the long-timescale experiments performed here. Therefore, we probed these two contributions with different drugs.
Inhibition of myosin II by blebbistatin resulted in a radius-independent migration velocity (see SI Appendix, Fig. S11). The same trend has been also reported on tracks (24). Interestingly, preventing lamellipodium protrusions by Rac1 inhibition with NSC23766 did not affect the velocity at large radii but slowed down the front for R = 10 µm (see SI Appendix, Fig. S11). Rac1 being classically correlated with protrusive activity (36), we conclude that the velocity increase at small radius/high confinement is controlled by protrusions. On thick wires, the migration mode is different and controlled by contractility, highlighting the role of leader cells in migration at these large confinements (27).
Finally, inhibiting proliferation with mitomycin-C resulted in a drastic slowdown of the bulk migration, while the front velocity was largely unaffected, in the 12-h timescale of the experiments (before the drug became toxic) (see SI Appendix, Fig. S12).
Discussion
We have shown in the present study that growing cells on wires had several consequences that can originate from curvature itself or from the intrinsic lateral confinement of the cells.
Curvature caused cells at the leading edge to detach from the monolayer and progress individually on the wires. Because of their strong cell−cell adhesions, MDCK monolayers are prototypical of cohesive epithelial sheets. This cell detachment is therefore particularly surprising. This effect was further explored by adding Hepatocyte Growth Factor (HGF), which is a well-known scatter factor, inducer of the Epithelial−Mesenchymal Transition (EMT) of MDCK cells (37–39). In the present case, it was used at a subscattering concentration. In the presence of HGF, we measured enhanced velocities at the leading edge independent of the wire radius (see SI Appendix, Fig. S13). Quantitatively, this velocity (∼40 µm⋅h−1) is the one attained in control experiments at the smallest radii (see SI Appendix, Fig. S11). Furthermore, even though the migration remained collective, we observed increased cell detachment events in the presence of HGF (Fig. 2A). We hypothesize here that these cell detachments at the front edge on thin wires could be the signature of a curvature-induced EMT. More experiments are clearly needed to fully prove or disprove this hypothesis, for example, by labeling mesenchymal markers such as vimentin (40). We note that cells from tumors often wrap around and migrate along lymphatic vessels or muscle fibers (41, 42) in vivo. As cells undergo EMT, this initial collective mode of migration then turns into a more individual one where detached single cells are guided by collagen bundles (43). Our observations may therefore mimic this behavior. Interestingly, other mechanical cues have been identified as EMT effectors, such as substrate rigidity (44) or in-plane curvature (40). Out-of plane curvature is therefore another mechanical parameter potentially impacting this transition.
Below a wire radius of typically 5 µm, we found that a single cell can fully wrap the wire. This observation of cells migrating in chains while individually wrapping around the wire recapitulates a mode of migration actually already observed in vivo or in in vitro 3D studies as cells experience an intermediate situation between single-cell and collective migration (45). In these multicellular streaming arrangements, cells have been reported to be only weakly engaged in adhesive junctions with their neighbors. Our experiments provide evidence that this is indeed the case at the leading edge where leading cells split away (Fig. 2).
Regarding the actin distribution for wire radii smaller than 40 µm, we observed a circumferential orientation, induced by curvature and independent of the surface coating, that was reproduced with another epithelial cell line. A similar distribution of actin has also been previously observed for single epithelial cells (16) and for confluent microvascular brain endothelial cells (19) plated on wires. The same orientation is also observed in vivo during the morphogenesis of the trachea of the Drosophila embryo that develops on a cylindrical chitin template appearing at the same stage (46, 47). In that case, the spacing between actin fibers is of the same order of magnitude as in our observations (typically 1–2 µm) for a comparable radius (47). These observations suggest that substrate geometry could play a role in organizing actin in this in vivo situation as well. Of note, the apical basal polarity in the trachea is opposite to the one adopted by cells on wires, suggesting that actin is primarily aligned by substrate geometry rather than polarity cues.
Actin fibers in the Drosophila trachea also appear continuous from cell to cell, implying a local mechanical continuity at the cell−cell boundary. Similar observations have been reported in endothelial monolayers where adherens junctions have been shown to be instrumental in this mechanical continuity via the cadherin/catenin complex that can anchor to the stress fibers extremities, independently of the FAs (29). Because of this continuity condition, the theoretical arguments invoking a balance between shear stress and the anisotropic bending stiffness of the stress fibers that were used to describe perpendicular alignment of the stress fibers at the single-cell level may be transposable to the present study (16, 18). In this context, the decoration of the individual stress fibers by the FAs that appears to correlate with the circumferential orientation is likely to be an important contribution, as it would limit the impact of the bending anisotropy of the stress fibers themselves that tends to orient the fibers along the wire axis.
The monolayer exhibits a tensile actin cable at its front edge. Similar supracellular structures are commonly observed in the healing of circular wounds on flat surfaces, but the purse-string function of these contractile cables has been recently downplayed, the migration being driven by lamellipodial protrusions at the free edge (33, 34). The tissue organization on wires is directly comparable to this closing of circular wounds; they differ by a topological transformation in which the cell-free region within the closing wound is the equivalent of the wire cross section. Obviously, on wires, the purse-string mechanism cannot be productive to the migration; however, it is, as on flat surfaces, efficient to maintain the regularity of the front edge during migration. The slower retraction of the actin cable on R ≤ 20-µm wires can be attributed to either a decreased tension or to an enhanced friction with the surface compared with cables on flat substrates. The increase in the density of FAs on highly curved surfaces suggests that the actin meshwork inside the cell is more pinned down to the wire than when on a flat surface. We hypothesize that the change in the substrate attachment of the cell's actin meshwork may act as an added source of friction, resisting cable retraction.
Velocity of migration in the bulk has been shown to be under control of proliferation, cell division replenishing the monolayer as it spreads and covers the free area. As for the front edge, Rac1-controlled protrusions are important to the cell migration on thin but not thick wires. This highlights the role of migration fingers that can be at play at large radii but could not develop at small radii, triggering an alternative migration behavior in a similar way to what has been observed for different wound sizes (27, 33). Myosin-II inhibition recapitulated on wires the response observed in tracks (confinement-independent velocity), evidencing further that the velocity on small radii wires is controlled by the confinement. A similar velocity−confinement relationship is also present on micrometer-size tracks at the single-cell level (48) and was attributed to a decrease in adhesion area. In the present case, the increased density of FAs overcomes this geometrical factor. However, these smaller adhesions are likely to have different dynamics and therefore contribute differently to migration. As the wire geometry allows the cells to migrate in a confined but boundary-free situation, this increase in migration speed is truly a confinement effect and is not due to contact guidance by the edges of the tracks.
Finally, we have identified a limiting diameter below which cells don’t migrate. The available surface is then too small to allow cells’ lamellipodium to develop sufficient adhesions, leading to the unstable protrusions/retraction cycles (49, 50). As it becomes denser, the whole monolayer then undergoes strong morphogenetic movements that eventually lead to cyst formation at the cone tips. Collective pulsations and formation of 3D structures have previously been identified with the same cell type in other confined environments (51). It appears, therefore, rather general that epithelial tissues encountering a boundary develop new strategies to continue their growth in the form of 3D bulging.
Materials and Methods
Experimental Substrates.
Glass wires were manufactured by heating and pulling glass capillaries by hand or with a pipette puller (P-2000, Sutter Instruments). We used, respectively, Pasteur pipets (Volac; Fisher Scientific) and glass capillaries (WPI Inc.). Also, R = 40-µm and R = 85-µm wires were purchased (CM Scientific). In all three cases, the initial material was borosilicate glass. Wires were cut to size and aligned at the edge of a silicone slab on a coverslip. They were then held in place with a heat-curable silicone elastomer (Sylgard 184; Dow Corning). The entire structure was cured at 65 °C, cleaned with ethanol and water, oxidized inside an air plasma cleaner (Harrick Plasma), and used as is or coated with fibronectin (Sigma) at 50 µg⋅mL−1 in PBS. Wires’ surface uniformity was characterized by SEM (Zeiss Ultra 55; Plateforme de l’Institut des Matériaux de Paris Centre), after gold metallization.
Nonadhesive Surface Treatment.
Tracks of varying widths were micropatterned onto cleaned glass coverslips using a highly repellent surface treatment of polyacrylamide and polyethyleneglycol (52). The treatment was subsequently coated with a layer of positive photoresist (S1813; Microchem), which was patterned by UV exposure through a mask (Selba). After development, air plasma was used to eliminate the protein-repellent layer on the exposed areas. Finally, the undeveloped photoresist was dissolved with acetone.
Cell Culture and Reagents.
MDCK, RPE1 or NIH 3T3 cells were maintained in Dulbecco’s Modified Eagle’s Medium (Gibco) with 10% (vol/vol) FBS (Sigma), 1% penicillin (10,000 U/mL) streptomycin (10 mg⋅mL−1) (Gibco), and 1% 2-mM l-glutamine (Gibco) at 5 vol% CO2, 95% relative humidity, and 37 °C. The LifeAct-GFP transfected MDCK cells (33) were cultured under the same conditions with medium supplemented with 400 µg⋅mL−1 geneticin (Invitrogen). Cells were seeded on the PDMS supports and allowed to adhere for 2 h. The substrates were then washed with PBS, and medium was added to the entire dish. Cells were cultured for 3–5 d, allowing them to divide and migrate onto the wires before being put under the microscope. Blebbistatin (Sigma) was used at a concentration of 50 µM. HGF (Sigma) was used at 10 ng⋅mL−1, a subscattering concentration. NSC23766 (Tocris Bioscience) was used at 50 µM. Mitomycin C (Sigma), an inhibitor of division, was used at a concentration of 5 µg⋅mL−1.
Immunofluorescence Labeling and Staining.
Cells were fixed in 4% (wt/vol) paraformaldehyde, permeabilized in 0.1% Triton X-100, and blocked in 10% (vol/vol) FBS in PBS. Ezrin was labeled with an anti-ezrin produced in rabbit (1:300; from Monique Arpin UMR 144, Institut Curie, Paris) followed by an Alexa 488 chicken anti-rabbit (1:500; Life Technologies). Vinculin labeling was performed with a mouse monoclonal anti-vinculin antibody (1:500; Sigma) followed by Alexa 546 goat anti-mouse (1:500; Life Technologies). Actin was labeled using Alexa 488 phalloidin or Alexa 546 phalloidin (1:1,000; Life Technologies). Myosin was labeled with rabbit anti-phospho myosin light chain (1:100; Ozyme) followed by Alexa 488 chicken anti-rabbit (1:1,000; Life Technologies). Hoescht 33342 (1:10,000; Sigma) was used to mark the nuclei. The wires were stored in PBS and imaged within 24 h.
Microscopy.
Time-lapse experiments were conducted at 10× and 20× magnification under an automated inverted Olympus IX-71 phase contrast video microscope with temperature, humidity, and CO2 regulation (Life Imaging Service). Fluorescently marked cells were observed under an upright Imager Z2 spinning disk microscope (Zeiss) equipped with a 63× water immersion objective. Due to optical limitations and, in particular, the finite numerical aperture of the microscope objectives, accuracy on the measurement of the radius of the glass wires was limited to ±1 µm.
Laser Photoablation.
Photoablation experiments were performed on an LSM 710 NLO (Zeiss) microscope equipped with a two-photon MaiTai laser and a 63× water objective. The two-photon laser was used at 85% power and at a wavelength of 890 nm. Retraction speed of the fibers was measured over the first 3.8 s after ablation.
Image Analysis.
Images were processed with the ImageJ software (53) or with Matlab (MathWorks) routines. Further analysis was occasionally performed on Origin (OriginLab). In particular, local orientation analysis was performed in Matlab by a Fourier Transform technique (54). The order parameter, S, was computed by averaging cos2θ over the images (around 120 subwindows for the smallest radii and up to 10,000 for the largest ones) and wires (at least four) for the same radius.
Statistical Analysis.
Unless otherwise specified, error bars in the figures represent the SDs. Two-sample t test was used to compare distributions. The number of replicates was always larger than eight, over at least four independent experiments.
Supplementary Material
Acknowledgments
We gratefully thank A. Buguin, Y. Chen, J. Condeelis, S. Coscoy, N. Gov, S. Li, P. Marcq, P. Recho, and the members of the “biology-inspired physics at mesoscales” group for discussions and advice. We also thank L. Malaquin and F. Pillier for their help with the SEM. The “biology inspired physics at mesoscales” group is part of the CelTisPhyBio Labex. We acknowledge the Cell and Tissue Imaging Platform (member of France-Bioimaging) of the Genetics and Developmental Biology Department (UMR3215/U934) of Institut Curie, and in particular O. Renaud and O. Leroy, for help with the microscopes. H.G.Y. thanks the Fondation Pierre-Gilles de Gennes for financial support. We thank the Association pour la Recherche sur le Cancer for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418857112/-/DCSupplemental.
References
- 1.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Martínez E, Engel E, Planell JA, Samitier J. Effects of artificial micro- and nano-structured surfaces on cell behaviour. Ann Anat. 2009;191(1):126–135. doi: 10.1016/j.aanat.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Théry M. Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci. 2010;123(Pt 24):4201–4213. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- 5.Wang CC, Jamal L, Janes KA. Normal morphogenesis of epithelial tissues and progression of epithelial tumors. Wiley Interdiscip Rev Syst Biol Med. 2012;4(1):51–78. doi: 10.1002/wsbm.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubarsky B, Krasnow MA. Tube morphogenesis: Making and shaping biological tubes. Cell. 2003;112(1):19–28. doi: 10.1016/s0092-8674(02)01283-7. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro C, Neumann M, Affolter M. Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol. 2004;14(24):2197–2207. doi: 10.1016/j.cub.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen JP, English K, Tenlen JR, Priess JR. Notch signaling and morphogenesis of single-cell tubes in the C. elegans digestive tract. Dev Cell. 2008;14(4):559–569. doi: 10.1016/j.devcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci USA. 2012;109(24):9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiddes LK, et al. A circular cross-section PDMS microfluidics system for replication of cardiovascular flow conditions. Biomaterials. 2010;31(13):3459–3464. doi: 10.1016/j.biomaterials.2010.01.082. [DOI] [PubMed] [Google Scholar]
- 11.Weiss P. Experiments on cell and axon orientation in vitro; the role of colloidal exudates in tissue organization. J Exp Zool. 1945;100:353–386. doi: 10.1002/jez.1401000305. [DOI] [PubMed] [Google Scholar]
- 12.Curtis AS, Varde M. Control of cell behavior: Topological factors. J Natl Cancer Inst. 1964;33:15–26. [PubMed] [Google Scholar]
- 13.Dunn GA, Heath JP. A new hypothesis of contact guidance in tissue cells. Exp Cell Res. 1976;101(1):1–14. doi: 10.1016/0014-4827(76)90405-5. [DOI] [PubMed] [Google Scholar]
- 14.Fisher PE, Tickle C. Differences in alignment of normal and transformed cells on glass fibres. Exp Cell Res. 1981;131(2):407–410. doi: 10.1016/0014-4827(81)90244-5. [DOI] [PubMed] [Google Scholar]
- 15.Abercrombie M. The Croonian Lecture 1978: The crawling movement of metazoan cells. Proc R Soc B. 1980;207(1167):129–147. [Google Scholar]
- 16.Svitkina TM, Rovensky YA, Bershadsky AD, Vasiliev JM. Transverse pattern of microfilament bundles induced in epitheliocytes by cylindrical substrata. J Cell Sci. 1995;108(Pt 2):735–745. doi: 10.1242/jcs.108.2.735. [DOI] [PubMed] [Google Scholar]
- 17.Hwang CM, et al. Controlled cellular orientation on PLGA microfibers with defined diameters. Biomed Microdevices. 2009;11(4):739–746. doi: 10.1007/s10544-009-9287-7. [DOI] [PubMed] [Google Scholar]
- 18.Biton YY, Safran SA. The cellular response to curvature-induced stress. Phys Biol. 2009;6(4):046010. doi: 10.1088/1478-3975/6/4/046010. [DOI] [PubMed] [Google Scholar]
- 19.Ye M, et al. Brain microvascular endothelial cells resist elongation due to curvature and shear stress. Sci Rep. 2014;4:4681. doi: 10.1038/srep04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander S, Weigelin B, Winkler F, Friedl P. Preclinical intravital microscopy of the tumour-stroma interface: Invasion, metastasis, and therapy response. Curr Opin Cell Biol. 2013;25(5):659–671. doi: 10.1016/j.ceb.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. Intravital. 2014;1(1):32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugassy C, Barnhill RL. Angiotropic melanoma and extravascular migratory metastasis: A review. Adv Anat Pathol. 2007;14(3):195–201. doi: 10.1097/PAP.0b013e31805048d9. [DOI] [PubMed] [Google Scholar]
- 23.Montesano R, Matsumoto K, Nakamura T, Orci L. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell. 1991;67(5):901–908. doi: 10.1016/0092-8674(91)90363-4. [DOI] [PubMed] [Google Scholar]
- 24.Vedula SRK, et al. Emerging modes of collective cell migration induced by geometrical constraints. Proc Natl Acad Sci USA. 2012;109(32):12974–12979. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marel A-K, et al. Flow and diffusion in channel-guided cell migration. Biophys J. 2014;107(5):1054–1064. doi: 10.1016/j.bpj.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poujade M, et al. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci USA. 2007;104(41):15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reffay M, et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat Cell Biol. 2014;16(3):217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 28.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105(Pt 4):1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 29.Millán J, et al. Adherens junctions connect stress fibres between adjacent endothelial cells. BMC Biol. 2010;8:11. doi: 10.1186/1741-7007-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reffay M, et al. Orientation and polarity in collectively migrating cell structures: Statics and dynamics. Biophys J. 2011;100(11):2566–2575. doi: 10.1016/j.bpj.2011.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemkemer R, Jungbauer S, Kaufmann D, Gruler H. Cell orientation by a microgrooved substrate can be predicted by automatic control theory. Biophys J. 2006;90(12):4701–4711. doi: 10.1529/biophysj.105.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, et al. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys J. 2006;90(10):3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cochet-Escartin O, Ranft J, Silberzan P, Marcq P. Border forces and friction control epithelial closure dynamics. Biophys J. 2014;106(1):65–73. doi: 10.1016/j.bpj.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anon E, et al. Cell crawling mediates collective cell migration to close undamaged epithelial gaps. Proc Natl Acad Sci USA. 2012;109(27):10891–10896. doi: 10.1073/pnas.1117814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125(7):1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 36.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 37.Lee K, Nelson CM. New insights into the regulation of epithelial-mesenchymal transition and tissue fibrosis. Int Rev Cell Mol Biol. 2012;294:171–221. doi: 10.1016/B978-0-12-394305-7.00004-5. [DOI] [PubMed] [Google Scholar]
- 38.Weidner KM, Behrens J, Vandekerckhove J, Birchmeier W. Scatter factor: Molecular characteristics and effect on the invasiveness of epithelial cells. J Cell Biol. 1990;111(5 Pt 1):2097–2108. doi: 10.1083/jcb.111.5.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridley AJ, Comoglio PM, Hall A. Regulation of scatter factor/hepatocyte growth factor responses by Ras, Rac, and Rho in MDCK cells. Mol Cell Biol. 1995;15(2):1110–1122. doi: 10.1128/mcb.15.2.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez EW, Chen QK, Gjorevski N, Nelson CM. Tissue geometry patterns epithelial-mesenchymal transition via intercellular mechanotransduction. J Cell Biochem. 2010;110(1):44–51. doi: 10.1002/jcb.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: A modified skin-fold chamber model. Histochem Cell Biol. 2008;130(6):1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 42.Gritsenko PG, Ilina O, Friedl P. Interstitial guidance of cancer invasion. J Pathol. 2012;226(2):185–199. doi: 10.1002/path.3031. [DOI] [PubMed] [Google Scholar]
- 43.Gligorijevic B, Bergman A, Condeelis J. Multiparametric classification links tumor microenvironments with tumor cell phenotype. PLoS Biol. 2014;12(11):e1001995. doi: 10.1371/journal.pbio.1001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilghman RW, et al. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS ONE. 2010;5(9):e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 46.Devine WP, et al. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc Natl Acad Sci USA. 2005;102(47):17014–17019. doi: 10.1073/pnas.0506676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matusek T, et al. The Drosophila formin DAAM regulates the tracheal cuticle pattern through organizing the actin cytoskeleton. Development. 2006;133(5):957–966. doi: 10.1242/dev.02266. [DOI] [PubMed] [Google Scholar]
- 48.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184(4):481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10(1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 50.Xue N, Bertulli C, Sadok A, Huang YYS. Dynamics of filopodium-like protrusion and endothelial cellular motility on one-dimensional extracellular matrix fibrils. Interface Focus. 2014;4(2):20130060. doi: 10.1098/rsfs.2013.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deforet M, Hakim V, Yevick HG, Duclos G, Silberzan P. Emergence of collective modes and tri-dimensional structures from epithelial confinement. Nat Commun. 2014;5:3747. doi: 10.1038/ncomms4747. [DOI] [PubMed] [Google Scholar]
- 52.Tourovskaia A, Figueroa-Masot X, Folch A. Long-term microfluidic cultures of myotube microarrays for high-throughput focal stimulation. Nat Protoc. 2006;1(3):1092–1104. doi: 10.1038/nprot.2006.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rasband WS (1997−2012) ImageJ ( Natl Inst Health, Bethesda), Version 1.46b.
- 54.Bosveld F, et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science. 2012;336(6082):724–727. doi: 10.1126/science.1221071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.