Significance
Microorganisms are key players in emissions of the greenhouse gas (GHG) methane from anoxic carbon-rich peat soils of the Arctic permafrost region. Although available data and modeling suggest a significant temperature-induced increase of GHG emissions from these regions by the end of this century, the controls of and interactions within the underlying microbial networks are largely unknown. This temperature-gradient study of an Arctic peat soil using integrated omics techniques reveals critical temperatures at which microbial adaptations cause changes in metabolic bottlenecks of anaerobic carbon-degradation pathways. In particular taxonomic shifts within functional guilds at different levels of the carbon degradation cascade enable a fast adaptation of the microbial system resulting in high methane emissions at all temperatures.
Keywords: microorganisms, methane, SOC mineralization, temperature, Arctic peat
Abstract
Arctic permafrost soils store large amounts of soil organic carbon (SOC) that could be released into the atmosphere as methane (CH4) in a future warmer climate. How warming affects the complex microbial network decomposing SOC is not understood. We studied CH4 production of Arctic peat soil microbiota in anoxic microcosms over a temperature gradient from 1 to 30 °C, combining metatranscriptomic, metagenomic, and targeted metabolic profiling. The CH4 production rate at 4 °C was 25% of that at 25 °C and increased rapidly with temperature, driven by fast adaptations of microbial community structure, metabolic network of SOC decomposition, and trophic interactions. Below 7 °C, syntrophic propionate oxidation was the rate-limiting step for CH4 production; above this threshold temperature, polysaccharide hydrolysis became rate limiting. This change was associated with a shift within the functional guild for syntrophic propionate oxidation, with Firmicutes being replaced by Bacteroidetes. Correspondingly, there was a shift from the formate- and H2-using Methanobacteriales to Methanomicrobiales and from the acetotrophic Methanosarcinaceae to Methanosaetaceae. Methanogenesis from methylamines, probably stemming from degradation of bacterial cells, became more important with increasing temperature and corresponded with an increased relative abundance of predatory protists of the phylum Cercozoa. We concluded that Arctic peat microbiota responds rapidly to increased temperatures by modulating metabolic and trophic interactions so that CH4 is always highly produced: The microbial community adapts through taxonomic shifts, and cascade effects of substrate availability cause replacement of functional guilds and functional changes within taxa.
Microorganisms are key players in the turnover of soil organic carbon (SOC) in the large carbon storages of the Arctic permafrost region (1). These soils contribute about 3–10% of the global emissions of the potent greenhouse gas (GHG) CH4 (2). By the end of this century, the surface temperatures in the Arctic are predicted to increase by 2–11 °C in winter and 1–6 °C in summer (3). As a consequence, CH4 and CO2 production from SOC decomposition are expected to increase, possibly causing a positive feedback to climate change (4).
Estimates of current CH4 release from boreal and tundra biomes differ substantially (5–8). Predicting future emissions is even more difficult, because the complexity of the soil microbiota limits the understanding of temperature effects on SOC decomposition (4). Temperature-related CH4 emission will be greatly affected by the active anoxic Arctic soil layer. In these soils, low temperature, phenolic compounds, and recalcitrant organic matter limit decomposition rates (4, 9), and the proximate drivers of organic matter transformations are the microbial communities (10, 11). As temperatures increase, higher GHG emissions from Arctic soils are expected because of direct effects on microbial enzymes, but temperature might have indirect effects on microbial communities, altering the effect on GHG emissions (12, 13).
In anoxic peat, plant polymers are degraded through several hydrolysis and fermentation steps involving at least four functionally distinct types of microorganisms: primary and secondary fermenters and two groups of methanogens (14, 15). A rate-limiting step is polysaccharide hydrolysis (16–18); syntrophic oxidation of organic acids and alcohols, which produce little energy (19), also might be rate limiting, particularly at low temperature (17). High in situ concentrations of fermentation intermediates have been detected in Arctic (20), sub-Arctic (21, 22), boreal (23), and temperate peat (24). Formate, H2/CO2, and acetate are considered the major substrates for methanogenesis in most environments (25).
CH4 emissions can be mitigated by microbial CH4 oxidation, constituting the biological CH4 filter in soils. In continental ecosystems, CH4 oxidation is primarily aerobic and is performed by Proteobacteria (26) and Verrucomicrobia (27). Proteobacterial methanotrophs closely related to the aerobic Methylobacter are characteristic for circum-Arctic soils (28–30). Stable isotope signature studies indicate that anaerobic CH4 oxidation is a sink for CH4 in peat soils (31), but the oxidants, enzymes, and organisms involved are currently unknown.
Anaerobic degradation of SOC to CH4 and CO2 involves metabolic interactions between microorganisms. Temperature is expected to affect both this metabolic network and the trophic network (the microbial foodweb). However, no integrated system-level study has yet addressed these metabolic fluxes or pathways in combination with the activity and identity of the associated microorganisms.
Here we studied the effect of temperature on the Arctic anoxic peat soil microbiota. This ecosystem is characterized by permafrost soil with a high organic content, thawed topsoil during summer, and an active growth season of 60–70 d with soil temperatures mostly below 10 °C. The topsoil temperature fluctuates to a large extent with the air temperature because of sparse vegetation.
We aimed at identifying system-level changes in metabolic and trophic interactions of microorganisms during anaerobic SOC degradation to CH4 and CO2 along a temperature gradient. We used metatranscriptomic, metagenomic, and targeted metabolic profiling to assign specific microorganisms to their respective function in the metabolic network. The insights acquired were investigated further by direct process measurements and specific inhibition assays. This investigation enabled us to identify distinct temperature-sensitive mechanisms that modulate the CH4 emissions from the Arctic peat microbiota in response to warming.
Results
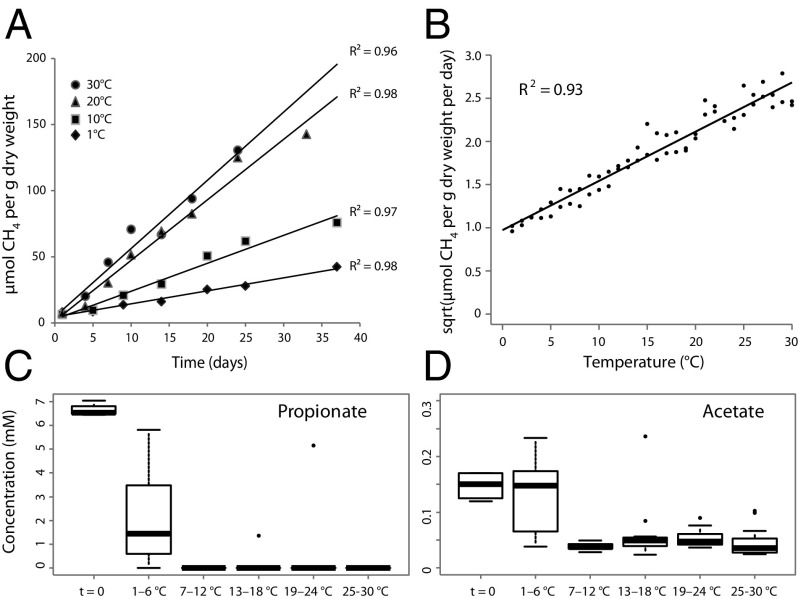
When Arctic peat soil was incubated in anoxic microcosms at 1–30 °C for 26–39 d, CH4 concentration increased linearly with time at all temperatures (Fig. 1A; see SI Appendix, Fig. S1 for details of the experimental set-up), indicating that the microbiota adapted rapidly even to drastic temperature shifts. As expected, CH4 production rates increased with temperature (Fig. 1B), exhibiting average temperature coefficient (Q10) values of 2.1 along the entire temperature gradient and a high CH4 production rate even at 4 °C (25% of that at 25 °C). A generalized additive model fitted to the Arrhenius function for the temperature dependence of CH4 production indicated nonlinearity (effective degrees of freedom = 2.6) and thus deviated from the expected linearity of a chemical reaction, suggesting that the activation energy changed with temperature. Thus, the Arrhenius law did not adequately describe the temperature dependence of CH4 production. The Ratkowsky model (32), however, explained the temperature dependence of both CH4 (R2 = 0.93) (Fig. 1B) and CO2 (R2 = 0.92) production rates (SI Appendix, Fig. S2). This result suggests that the relationship between temperature and CH4 production was not explained solely by the kinetics of a single enzyme-catalyzed reaction but rather by several reactions and possibly was related to changes in the microbiota along the temperature gradient.
Fig. 1.
(A) Linear regression of CH4 production in Arctic peat soil under anoxic conditions over time at 1, 10, 20, and 30 °C. (B) Linear regression of the plotted Ratkowsky function (square root of the CH4 production rate vs. temperature) for two replicate temperature gradients. (C and D) Boxplots of propionate (C) and acetate (D) concentrations (expressed in millimoles) in soils incubated at temperature windows of 6 °C ranging from 1–30 °C. Measurements from the temperature gradient series A and B are included. Whiskers indicate the most extreme values within 1.5 multiplied by the interquartile range; values outside that range are represented by dots. Box: 25% quartile; median, 75% quartile.
We next assayed the potential activities of hydrolytic enzymes responsible for degradation of the two major SOC components, cellulose and hemicellulose (SI Appendix, Fig. S3), as markers for the initial degradation step of SOC. As with CH4 production, the potential activities were high at 4 °C (i.e., between 50 and >100% of that at 15 °C) and were highest at 25 °C for all enzymes.
We wanted to know whether an increase in biomass with temperature could explain the increased CH4 production. Therefore we extracted nucleic acids as biomass indicators from soil slurries incubated in three temperature ranges that defined the low (3–5 °C), medium (14–16 °C), and high (24–26 °C) temperature windows. However, the amount of RNA and DNA in soil (expressed in micrograms per gram of dry weight soil) at these temperature windows did not differ, showing that the bulk (DNA) and active (RNA) microbial biomass did not increase with temperature (SI Appendix, Fig. S4).
The determination of fermentation intermediates revealed that butyrate and ethanol concentrations were mostly below the detection limits (SI Appendix, Fig. S5), and the H2 partial pressure was low throughout the experiment at all temperatures (0.8–1.3 Pa, n = 472). In contrast, the pools of the major fermentation intermediates propionate and acetate were nonlinearly affected by temperature (Fig. 1 C and D).
Although the concentrations of propionate and acetate were high between 1 and 6 °C (median concentrations of 1.5 mM and 150 µM, respectively), the propionate pool was completely depleted above 6 °C, and acetate concentrations were much lower (median concentrations, 50 µM). This biphasic behavior of propionate and acetate concentrations suggests a strong shift in anaerobic fermentative pathways in the peat microbiota at around 7 °C. Also, at 1–6 °C, propionate and acetate concentrations were inversely related with a strong negative correlation (gradient A: r = −0.92; gradient B: r = −0.97).
To assess possible shifts in microbiota structure and function related to these observations, we generated metatranscriptomes and metagenomes from RNA and DNA coextracted from soils incubated at the three temperature windows (SI Appendix, Table S1).
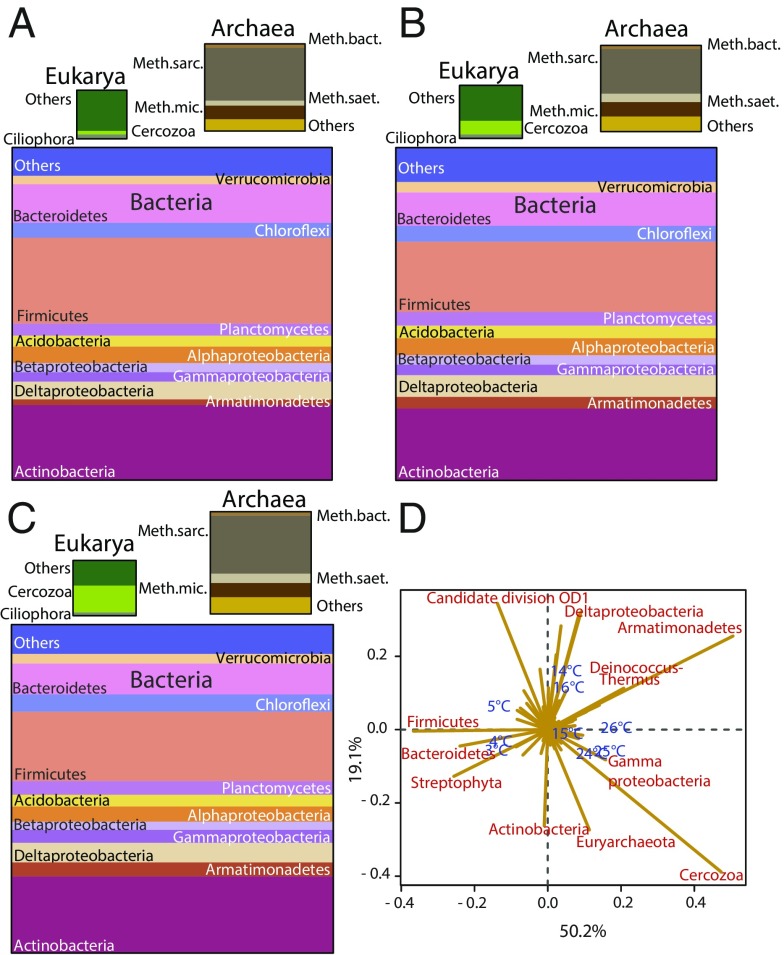
First we used the small subunit (SSU) rRNA transcripts for broad three-domain community profiling. In the three temperature windows, Bacteria dominated with 88–91% of the SSU rRNA, Archaea comprised 7–9%, and Eukarya comprised 2–3% (Fig. 2 A–C). Correspondence analysis (CA) of SSU rRNA (Fig. 2D) and its gene (SI Appendix, Fig. S6) revealed taxonomic shifts with temperature. The most prominent changes were in the Firmicutes and Bacteroidetes; their relative abundance decreased with increasing temperature. The relative abundance of the bacterial phylum Armatimonadetes and the eukaryotic Cercozoa increased with temperature (Fig. 2). All observed patterns stem from read abundances that were high enough to be above the data noise. A constrained CA showed that temperature alone explained 43.8 and 33.6% of total inertia (variation in the CA space) for rRNA and its gene, respectively. With only two exceptions, the changes in relative abundance of SSU rRNA and its gene correlated for all taxa (SI Appendix, Fig. S7).
Fig. 2.
(A–C) Phylum-based (class-based for proteobacteria) taxonomic proportions of SSU rRNA in soil at 3–5 °C (A), 14–16 °C (B), and 24–26 °C (C). The sizes of the boxes correspond to the relative abundance of SSU rRNA. The relative abundances are calculated as the average of the three temperatures in each temperature window, e.g., 3, 4, and 5 °C. (D) Biplot of CA of phylum-based (class-based for proteobacteria) taxonomic proportions in soils incubated at the various temperatures. Sample profiles are shown in blue; taxa responsible for the majority of the differences between sample profiles are shown in red. The x axis represents the first dimension, which explained 50.2% of the inertia; the y axis represents the second dimension, which explained 19.1% of the inertia. The distance between the sample profiles indicates the difference in taxonomic composition. The length of the line connecting the taxon to the center of the plot is equivalent to the weight of this taxon profile in the final solution of the samples, given the inertia explained by the dimensions it crosses (i.e., the longer the line, the larger is the part of the inertia it explains). The direction of the line indicates the sample orientation of its weight (e.g., if a line points toward higher temperatures, the relative abundance of that taxon is highest at high temperatures).
CA of gene-expression profiles showed that the relative abundances of transcripts in several functional categories differed among the three temperature windows (SI Appendix, Fig. S8). Temperature explained most of the inertia (38.4%). A temperature-constrained CA showed that transcription related to fermentation, intracellular carbon storage, and CH4 cycling [e.g., formate metabolism, polyhydroxybutyrate (PHB) metabolism, methanogenesis, CH4 oxidation, and glycine degradation] was particularly responsive.
Next we monitored the relative abundances of mRNAs relevant for the anaerobic SOC degradation cascade. Taxonomic assignments of mRNAs were used to identify replacements of taxa within functional guilds.
Transcripts of Hydrolytic Enzymes.
We wanted to know whether any population and/or transcriptional changes were associated with the temperature-dependent increase in potential activities of cellulases and hemicellulases. No significant increases in the relative abundance of the respective mRNAs or genes were found (SI Appendix, Figs. S9 and S10). In addition, no taxon replacements were identified, suggesting that the same taxa, mainly Firmicutes, Bacteroidetes, and Actinobacteria, were responsible for these functions throughout the temperature gradient (SI Appendix, Fig. S11). Interestingly, however, the remaining transcripts were assigned to more than 10 additional phyla, and 10–20% of all identified hydrolase transcripts could not be assigned to any taxon beyond the domain level.
Transcripts for Fermentative Metabolisms.
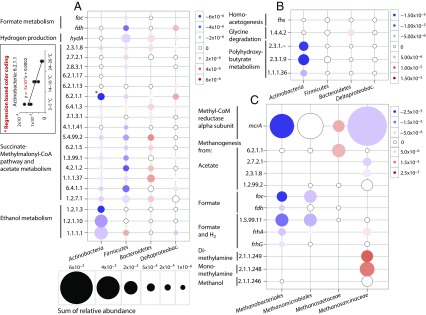
We studied the fermentation and CH4-cycling pathways in detail (Fig. 3 and SI Appendix, Fig. S12). Transcripts for the succinate-methylmalonyl-CoA (SMC) pathway for propionate fermentation and for formate, hydrogen, acetate, and ethanol metabolism were relatively highly abundant, but changes related to temperature were small and could not explain the observed biphasic pattern of propionate and acetate concentrations. However, taxonomic annotation of the transcripts revealed a taxon replacement within functional guilds (Fig. 3). Within the Firmicutes, the relative abundance of transcripts for the SMC pathway was much lower at the middle and high temperature windows than at 3–5 °C. The reduced abundance corresponded to a decline in transcripts for acetate metabolism, formate dehydrogenase, formate transporters, and [FeFe] hydrogenase (Fig. 3A), all stemming from the family Peptococcaceae within Clostridiales. The correlating decrease in gene expression in all these pathway steps with temperature indicated declining activity of a Peptococcaceae population that oxidized propionate to acetate, formate, and H2, as described recently (Fig. 3A and SI Appendix, Fig. S13) (19). Interestingly, transcripts encoding ethanol dehydrogenase from Firmicutes increased with temperature; however, these Firmicutes transcripts were assigned to the family Eubacteriaceae within Clostridiales. Although the abundance of SMC pathway transcripts of Deltaproteobacteria did not change, transcripts of Bacteroidetes (including transcripts of acetate metabolism and [FeFe] hydrogenases; Fig. 3A) increased with temperature. This finding suggested that the Bacteroidetes replace Clostridiales as dominant propionate oxidizers (to acetate and H2). The relative abundance of transcripts for formyltetrahydrofolate synthetase (gene fhs), a key enzyme in the acetyl-CoA pathway for homoacetogenesis (33), was consistently low and did not change with temperature for any taxon, indicating that homoacetogenesis is not an important source of acetate at any temperature.
Fig. 3.
Changes in the relative abundance of transcripts for key functions (y axes) in fermentation (A and B) and methanogenesis (C) within specific taxa (x axes) with temperature. Red circles indicate increases in the relative abundance, and blue circles indicate decreases in the relative abundance of transcripts with increasing temperature. The size of the circles indicates the sum of relative abundance across all temperatures. The color-coded schemes in each panel show a linear translation of the slope of regression lines to a color gradient. The regression lines were fitted to the relative abundance of transcripts for the functions assigned to taxa across the temperature windows 3–5 °C, 14–16 °C, and 24–26 °C as exemplified with the data for EC 6.2.1.1 assigned to Actinobacteria (A, Inset). Each point within the temperature windows was treated as a replicate of the median temperature. The y axis labels are EC designations (e.g., 2.3.1.8), or gene names (e.g., fdh). (A) foc, formate channel; fdh, formate dehydrogenase; hydA, [FeFe] hydrogenase subunit a; 2.3.1.8, phosphate acetyltransferase; 2.7.2.1, acetate kinase; 2.8.3.1, propionate-CoA transferase; 6.2.1.17, propionate-CoA ligase; 6.2.1.13, acetate-CoA ligase (ADP forming); 6.2.1.1, acetate-CoA ligase (AMP forming); 6.4.1.3, propionyl-CoA carboxylase; 2.1.3.1, methylmalonyl-CoA carboxytransferase; 4.1.1.41, methylmalonyl-CoA decarboxylase; 5.4.99.2, methylmalonyl-CoA mutase; 6.2.1.5, succinate-CoA ligase (ADP forming); 1.3.99.1, succinate dehydrogenase; 4.2.1.2, fumarate hydratase; malate dehydrogenase; 6.4.1.1, pyruvate carboxylase; 1.2.7.1, pyruvate synthase; 1.2.1.3, aldehyde dehydrogenase (NAD+); 1.2.1.10, acetaldehyde dehydrogenase (acetylating); 1.1.1.1, alcohol dehydrogenase. (B) fhs, formyltetrahydrofolate synthetase; 1.4.4.2, glycine dehydrogenase (aminomethyl transferring); 2.3.1.-, polyhydroxybutyrate polymerase; 2.3.1.9, acetyl-CoA acetyltransferase; 1.1.1.36, acetoacetyl-CoA reductase. (C) mcrA, methyl-CoA reductase alpha subunit; 6.2.1.1, acetate-CoA ligase (AMP forming); 2.7.2.1, acetate kinase; 2.8.3.1, propionate-CoA transferase; 1.2.99.2, carbon monoxide dehydrogenase; foc, formate channel; fdh, formate dehydrogenase; 1.5.99.11, 5,10-methylenetetrahydromethanopterin reductase; frhA/frhG, coenzyme F420 hydrogenase alpha and gamma subunits; 2.1.1.249, dimethylamine methyltransferase; 2.1.1.248, monomethylamine methyltransferase; 2.1.1.246, methanol methyltransferase.
High relative abundances of transcripts for the metabolism of ethanol, propionate, and acetate were assigned to Actinobacteria (Fig. 3A), whereas the relative abundance of transcripts of formate dehydrogenase (gene fdh) and [FeFe] hydrogenase alpha subunit (gene hydA) assigned to this taxon was low. Members of Actinobacteria thus appear to be the most important players in the oxidation of ethanol to propionate and acetate, a pathway proposed previously (Fig. 3A and SI Appendix, Fig. S14) (34). Transcripts related to ethanol, propionate, acetate, and PHB metabolism assigned to Actinobacteria decreased with increasing temperature, indicating that this group became less important. The results suggest that they were partly replaced by other ethanol oxidizers, but it is not clear from the results whether the pathway of ethanol oxidation was the same (Fig. 3 and SI Appendix, Fig. S15). The temperature dependence and relative importance of taxa other than those displayed in Fig. 3 are shown in SI Appendix, Fig. S15.
Transcripts for Syntrophy, Methanogenesis, and CH4 Oxidation.
At low temperature, transcripts associated with methanogenesis from formate and H2/CO2 were highly abundant and assigned mostly to Methanobacteriales (Fig. 3C). The steep decrease in these transcripts from low to high temperatures correlated with the decrease in transcripts of propionate oxidizers/formate-producing Peptococcaceae within Firmicutes (Fig. 3A). In fact, relative transcript abundances of formate dehydrogenase (Fdh), the key enzyme of both pathways, were highly correlated (r = 0.88) between Firmicutes and Methanobacteriales, suggesting a syntrophic relationship between these taxa for the oxidation of propionate to acetate and CH4, with formate as intermediate. In bacterial syntrophs grown in cocultures with different methanogens, the expression of genes for key enzymes in syntrophic production and transfer of H2 and formate has been shown to depend on the metabolism of the methanogenic partner (35).
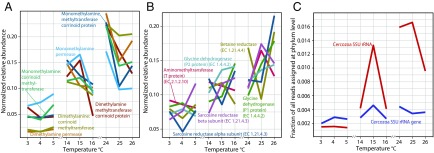
The relative abundance of transcripts for acetotrophic methanogenesis within the family Methanosaetaceae [key enzyme: acetate-CoA ligase (Kyoto Encyclopedia of Genes and Genomes Ligand Database for Enzyme Nomenclature EC 6.2.1.1)] increased with temperature, consistent with increases in SSU rRNA (r = 0.86) and mcrA (r = 0.96) transcripts assigned to Methanosaetaceae. In contrast, transcripts of the acetotrophic methanogenesis of Methanosarcinaceae (key enzyme: acetate kinase, E.C. 2.7.2.1) decreased (Fig. 3C), indicating a functional replacement of the latter by Methanosaetaceae. All transcripts for methanogenesis from methylamines were assigned to Methanosarcinaceae (Fig. 3C). Remarkably, the relative abundance of transcripts for monomethylamine- and dimethylamine-dependent methanogenesis increased three- to fourfold from 4 °C to 24 °C (Fig. 4A), whereas transcripts for trimethylamine-dependent methanogenesis were similarly abundant at all temperatures.
Fig. 4.
(A) Changes in the relative abundance of transcripts for methanogenesis from dimethylamines and monomethylamines with temperature. (B) Changes in the relative abundance of transcripts for enzymes involved in the degradation of betaine (betaine reductase), sarcosine (sarcosine reductase), and glycine (aminomethyltransferase, glycine dehydrogenase). These enzymes are proposed to be involved in the formation of methylamines. (C) Changes in the relative abundance of SSU rRNA sequence tags assigned to the eukaryotic phylum Cercozoa with temperature. The relative abundances in A and B were normalized to the sum of relative abundances across temperatures for each function.
Methylamines are products of the degradation of glycine and possibly other amino acids (from proteins or peptidoglycan), compatible solutes, and lipids. Indeed, transcripts for the glycine-cleavage system were most abundant, followed by transcripts for sarcosine reductase and betaine reductase, and all increased twofold, on average, from the lowest to the highest temperature window (Fig. 4B). The temperature-dependent increase in the relative abundance of SSU rRNA and SSU rRNA genes for predatory protists of the phylum Cercozoa corresponded to the increase of transcripts for methanogenesis from methylamine, and of sarcosine reductase, betaine reductase, and the glycine-cleavage system (Fig. 4C).
Transcripts and genes of methanotrophic Methylococcales were relatively abundant (SI Appendix, Fig. S16) and were most similar to genes from Methylobacter tundripaludum, e.g., the aerobic pathway of CH4 oxidation to CO2, identified by transcripts of particulate CH4 monooxygenase (pMMO) and methanol and formate dehydrogenases (SI Appendix, Table S2). The relative abundance of pMMO transcripts was ∼100-fold higher than that of methanol and formate dehydrogenases, which catalyze the downstream steps of CH4 oxidation.
Metabolic Networks and Mass Balances.
We aimed at a high-resolution understanding of the carbon flow from SOC through fermentation intermediates to CH4 and CO2. For this purpose we added an inhibitor of acetotrophic methanogenesis (36, 37), methylfluoride (CH3F), to one series of the temperature gradient to cause the accumulation of substrates and thereby allow the study of upstream processes (38). CH3F inhibition resulted in the accumulation of acetate, propionate, ethanol, and very low amounts of butyrate (SI Appendix, Fig. S17), whereas smaller amounts of CH4 and CO2 were produced than by noninhibited microcosms (SI Appendix, Fig. S2). This experiment revealed that the minimum fraction of CH4 originating from H2 and formate comprised ∼35% at all temperatures; thus a maximum of 65% of formed CH4 could be attributed to methanogenesis from acetate. Throughout the inhibition experiment the median partial pressure of H2 (1–18 °C: 1.6–1.9 Pa; n = 144; and 18–30 °C: 3.1–3.6 Pa; n = 96) was higher than in the noninhibited control microcosms.
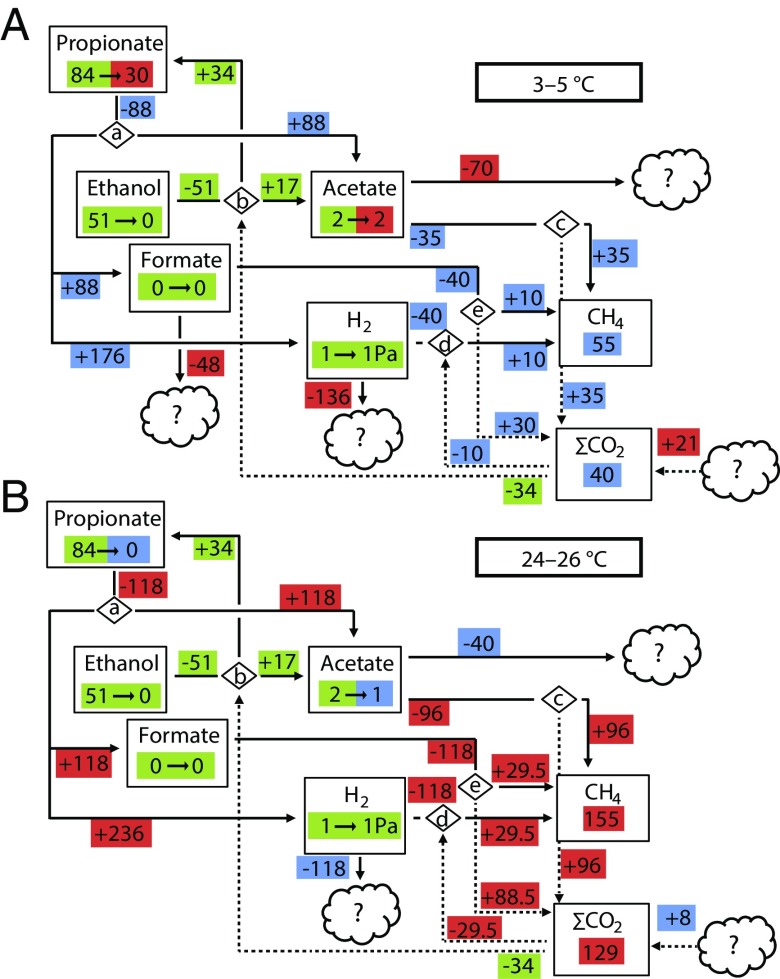
We then calculated mass balances from the beginning and final concentrations of fermentation products. We combined the mass balances with the ratio between hydrogenotrophic and acetotrophic methanogenesis to construct system-level metabolic maps of carbon flow for low (2–6 °C; Fig. 5A) and high (23–27 °C; Fig. 5B) temperatures.
Fig. 5.
Mass balances for two temperature windows, 2–6 °C (incubated for 39 d) (A) and 23–27 °C (incubated for 26 d) (B). Boxes show pool sizes at start → pool sizes at end point; masses are expressed in micromoles per gram dry weight. Zeros correspond to the respective detection limits. Hydrogen partial pressures are expressed in pascals. The pools of CH4 and CO2 are the masses that accumulated during the incubation period. Negative and positive masses associated with arrows represent masses going out from and masses going into pools during the incubation time, respectively; masses are expressed in micromoles per gram dry weight per incubation time. Green color indicates that pools or mass flows are the same in both temperature windows. Red indicates a higher value, and blue indicates a lower value in that particular pool or flow compared with the other temperature window. ∑CO2 is the sum of all CO2 species, gaseous and dissolved. All flows of ∑CO2 are indicated by dotted arrows. All other mass flows are indicated by solid arrows. Diamonds indicate reactions constrained to the following based on metatranscriptomic analysis: a: propionate fermentation to acetate, formate, and hydrogen [CH3CH2COO− + 2H2O = CH3COO− + HCOO− + 2H2 + H+]; b: ethanol fermentation to acetate and propionate [3(CH3CH2OH) + 2HCO3− = CH3COO− + H+ + 2(CH3CH2COO)− + 3H2O]; c: acetoclastic methanogenesis [CH3COO− + H+ = CH4 + CO2]; d: methanogenesis from hydrogen and carbon dioxide [4H2 + CO2 = CH4 + 2H2O]; e: methanogenesis from formate [4HCOO− + H+ + H2O = CH4 + 3HCO3−]. The amount of CH4 produced from H2 + CO2 and the amount CH4 produced from formate have been set as equal, because the data do not allow the ratio to be determined.
The most important fermentation intermediates were ethanol, propionate, and acetate. Metatranscriptomic data (Fig. 3A) were used to constrain the metabolite flux to the pathway reactions ethanol fermentation to propionate and acetate (34) and propionate oxidation to acetate, formate, and H2 (39). Subsequent thermodynamic calculations showed that ethanol oxidation was exergonic along the entire temperature gradient (SI Appendix, Fig. S18). Propionate oxidation to formate and H2 also was exergonic except in the temperature range 7–12 °C. The alternative propionate oxidation pathway producing acetate and H2, but no formate, was exergonic along the entire temperature gradient (SI Appendix, Fig. S18). Acetate was the primary substrate for methanogenesis at all temperatures (ca. 65%; see above). Although formate was not detected, the presence of transcripts for formate metabolism suggested that the pathway was active (Fig. 3A). Because CH3F inhibition resulted in the accumulation of upstream intermediates, e.g., propionate (a substrate for syntrophic oxidation), the production of H2 and/or formate also could have been affected. Thus, we might have underestimated methanogenesis from formate and H2.
Interestingly, not all consumption of acetate, H2, and formate could be explained, as indicated by the unknown sinks in Fig. 5. Furthermore, the mass balances could not fully explain the sources of the accumulated pool of total CO2 (gaseous + aqueous: ∑CO2), as indicated by the unknown source in Fig. 5. The ∑CO2 proportion of total mineralized carbon (CH4 + ∑CO2) increased with temperature (average at 1–10 °C: 40%; average at 11–20 °C: 44%; average at 21–30 °C: 45%), indicating a shift in the carbon flow with temperature.
An Integrated System-Level Model for Anaerobic SOC Decomposition in Arctic Peat Soil.
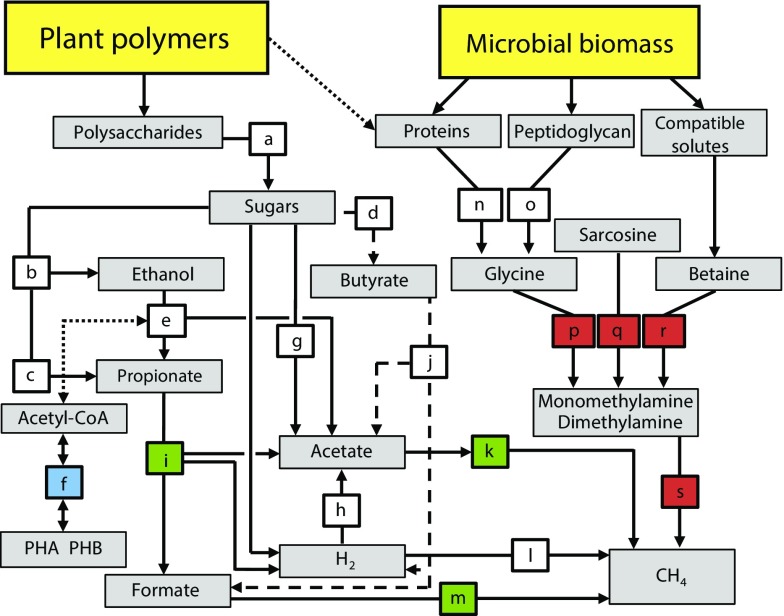
This integrative metatranscriptomic, metagenomic, and targeted metabolite profiling allowed the development of a system-level model of the predominant metabolic pathways and the key temperature effects on anaerobic SOC decomposition in the Arctic peat soil investigated (Fig. 6). Three steps in polysaccharide degradation were temperature sensitive: syntrophic propionate oxidation, acetotrophic methanogenesis, and methanogenesis from formate. There were taxon replacements within the functional guilds for these pathways, with different taxa dominating at 3–5 °C and at 14–16 °C and 24–26 °C. The taxon replacements corresponded to shifts from high concentrations of propionate and acetate below 7 °C to low concentrations at higher temperatures, again pointing to syntrophic oxidation of propionate as the limiting step for SOC mineralization below 7 °C. Transcription related to PHB metabolism decreased substantially with temperature, suggesting a decrease in the utilization of intracellular storage compounds. A large increase in the relative abundance of transcripts for monomethylamine and dimethylamine methanogenesis corresponded to an increase in transcripts for the degradation of known sources of methylamines: glycine (peptidoglycan and proteins), sarcosine (intermediate in glycine metabolism), and glycine betaine (compatible solute). This observation might be explained by the increase in relative abundance of the predatory protist phylum Cercozoa, which indicated an accelerated microbial loop causing an increase in the turnover of bacterial cells, sources of proteins, peptidoglycan, and compatible solutes.
Fig. 6.
Conceptual model displaying the decomposition network of SOM in high-Arctic peat soils. SOM input is from two main sources: plants and microbial cells. The selection of network components is based on the combined information presented in the study, including inhibition studies; measurements of gases, organic acids, and ethanol; enzyme assays; metatranscriptomics; and metagenomics. Each colored box represents a biochemical transformation of a substrate (origin of arrow) to a product (end point of arrow). Dashed lines indicate a less prominent pathway (i.e., butyrate oxidation). The dotted line indicates putative links between metabolisms/pools (ethanol oxidation and PHB metabolism; availability of plant cell wall proteins). The color coding is based on the identified temperature responses. Blue: decrease in transcription; red: increase in transcription; green: a switch in the taxa responsible, with consequences for propionate and acetate metabolisms; white: no clear change. Pathways are indicated by letters a–s: a, hydrolysis; b, c, and d, primary fermentations; e, ethanol fermentation to propionate and acetate or to acetate and H2; f, PHB metabolism; g and h, acetogenesis; i, propionate fermentation to acetate, formate, and H2 or to acetate and H2; j, butyrate fermentation to acetate, formate, and H2 or to acetate and H2; k, methanogenesis from acetate; l, methanogenesis from H2 and CO2; m, methanogenesis from formate; n, proteolysis; o, penta-glycine proteolysis; p, glycine cleavage/degradation; q, sarcosine reduction; r, betaine reduction; s, methanogenesis from methylamines.
Discussion
Low-Temperature Adaptation.
We investigated the temperature response of microorganisms responsible for anaerobic degradation of SOC and CH4 production in Arctic peat soil. The relatively high CH4 production rate at 4 °C (25% of the rate at 25 °C) implies that the Arctic peat microbiota investigated here is well adapted to low temperature. At lower latitudes, the CH4 production shows a stronger response to increasing temperature: In temperate peat soils, the rate at 4 °C is <1% of the rate at 25 °C (40); in temperate lake sediments, the rate is 10% of that at 25 °C (41); and in sub-Arctic peat soils, the rate is 17% of that at 27 °C (22). Also methane fluxes in wetland, rice paddy, and aquatic ecosystems located within a broad range of latitudes have a higher temperature sensitivity; the average CH4 emissions increases by 57-fold from 1 °C to 30 °C (42). The results show that this methane-producing Arctic ecosystem has a lower temperature sensitivity than temperate/sub-Arctic ecosystems.
The proportion of hydrogenotrophic methanogenesis in the Arctic peat soil was high (35%), in contrast to lake sediments (41, 43–45) and rice soil (46, 47), where only a minor proportion or no CH4 originates from hydrogenotrophic methanogenesis at low temperature. This finding is in accordance with studies of sub-Arctic (22) and boreal peat soil (21), where the proportion of hydrogenotrophic methanogenesis is 30 and 80%, respectively. In lake and tundra ecosystems at low temperatures, homoacetogens are important H2 consumers, and acetate is the major precursor for methanogenesis (17, 44, 45, 48). However, homoacetogens were not important in this Arctic peat soil, as shown by the low relative abundance of fhs transcripts and the high and constant proportion of hydrogenotrophic methanogenesis at all temperatures.
Temperature-Dependent, Rate-Limiting Steps for CH4 Production.
The step in anaerobic SOC decomposition with the lowest rate defines the rate-limiting step for CH4 production. In rice soils, polysaccharide hydrolysis is the rate-limiting step for CH4 formation (16, 18). In our study, the accumulation of propionate and acetate below 7 °C indicated that terminal processes rather than upstream polysaccharide hydrolysis were rate limiting for CH4 production. Terminal processes also might be rate limiting in situ, considering that 7 °C is within the range of current Arctic summer soil temperatures, and high concentrations of fermentation intermediates have been measured in summer (20). In our study, propionate was syntrophically fermented at low temperature by propionate oxidizers within the Firmicutes in association with formate- and H2-using methanogens within the Methanobacteriales, thus keeping the formate and H2 concentrations in the microcosms low. However, the high and variable concentrations of acetate and propionate at low temperature, together with the prevalence of low-affinity acetate-using Methanosarcinaceae over high-affinity Methanosaetaceae indicated that acetotrophic methanogenesis and propionate oxidation were not syntrophically associated below 7 °C. However, the two processes were still connected, as shown by the nearly perfect negative correlation between acetate and propionate concentrations. These findings suggest that below 7 °C the relationship is cyclic, with opposite cycles of activity of propionate oxidizers and acetotrophic methanogens, increasing the Gibbs free-energy change of the reactions of both groups. Because of the high propionate and low acetate concentrations in several microcosms below 7 °C, propionate oxidation was exergonic, with a Gibbs free-energy change around the minimum found to sustain life in propionate-oxidizing bacteria (−6 to 12 kJ/mol) (49). Above 7 °C, the propionate pool was depleted and was associated with a shift in the functional guild responsible for propionate oxidation, with Firmicutes replaced by Bacteroidetes and Deltaproteobacteria. Calculations showed that between 7 °C and 12 °C, propionate oxidation to acetate, H2, and formate was endergonic, whereas the alternative pathway of propionate oxidation to acetate and H2 was exergonic. This finding suggests that propionate oxidizers might alter their pathways to enable energy conservation depending on the conditions, as discussed previously (19). The metatranscriptomic indications that at higher temperatures Bacteroidetes oxidize propionate to acetate and H2 but not to formate support this interpretation. Also, the concentration of acetate decreased with increasing temperature, in line with the increased abundance and activity of high-affinity acetotrophic methanogenic Methanosaetaceae (50), suggesting that more efficient acetate utilization was associated with more efficient propionate oxidation. A similar association was seen in a triculture of a high-affinity acetotrophic methanogen, a propionate oxidizer, and a formate-using methanogen (51), where low acetate and formate concentrations increased the efficiency of propionate oxidation over that of dicultures without the acetotrophic methanogen. The same relationship between acetate concentrations and propionate oxidation has been observed in wastewater treatment at low temperatures (52) and in anaerobic degradation in taiga pond sediment (17). The removal of the terminal bottleneck above 7 °C resulted in hydrolysis of polysaccharides becoming the rate-limiting step for CH4 production.
Microbial Loop Alters Methanogenesis Pathways.
The most pronounced temperature-related taxonomic shift was a tenfold increase in SSU rRNA of the predatory protist phylum Cercozoa (53). This shift and the overall low abundance of protists indicated a low grazing pressure at in situ temperatures and substantially higher grazing pressure at higher temperatures. The difference in grazing pressures is supported further by increased activity and substrate turnover not resulting in a larger microbial biomass. The increase in Cercozoa correlated with the increased abundance of transcripts for methanogenesis from mono- and dimethylamines. Sources of methylamines include glycine-betaine, sarcosine, and glycine (54, 55). Glycine comprises up to 60% of some proteins (54). Several plant cell-wall structural proteins contain long stretches of glycine (56), and Gram-positive bacterial cell walls contain pentaglycine bridges that make up a substantial fraction of peptidoglycan (57). The increase in the relative abundance of transcripts of proteins involved in glycine betaine, sarcosine, and glycine degradation suggested that these compounds are among the primary sources of methylamines in Arctic soils. Glycine from undigested cell walls or amino acids could be released by Cercozoa during grazing on bacteria. Indeed, nitrogen (58), sometimes as amino acids and peptides (59), is released during protist grazing on bacteria.
Furthermore, the compatible solute glycine-betaine, used as an intracellular osmo- and thermoprotectant in psychrophiles, could have been released during the shift from low to high temperature. However, the relative abundance of transcripts for proteins involved in glycine-betaine reduction was low. Taken together, this circumstantial evidence illustrates how higher-level trophic interactions of the soil microbial loop can indirectly impact the substrate availability for an important guild of GHG-producing microorganisms.
The storage compound for excess carbon and energy, PHB, can provide organisms with a crucial advantage under fluctuating carbon and/or limiting nutrient availability (60, 61). The high abundance of transcripts for PHB metabolism, mainly assigned to Actinobacteria, suggested that PHB is an important storage compound at low temperature. The decrease with increasing temperature suggested that the advantage of intracellular carbon storage in this taxon is restricted to low temperature. Both methanogenesis from methylamines (25) and fermentation of glycine (54) release ammonium (NH4+). Thus, the higher availability of nitrogen might explain the indicated decrease in PHB metabolism. However, other explanations are possible also.
Taxonomic and Metabolic Shifts Enable Rapid Thermal Adaptation of Microbiota.
A remarkable effect of temperature change was the taxonomic switches within functional guilds and the functional switches within taxa. A taxonomic switch within the functional guild for syntrophic propionate oxidation occurred with increasing temperature, where Firmicutes were replaced by Bacteroidetes and to some extent by Deltaproteobacteria. Other taxonomic switches occurred with increasing temperature: Within the guild for methanogenesis from H2/CO2 and formate, Methanomicrobiales replaced Methanobacteriales; within the guild for acetotrophic methanogenesis, Methanosaetaceae replaced Methanosarcinaceae; Methanosarcinaceae in turn changed their metabolism from acetotrophic methanogenesis to methylotrophic methanogenesis, thus exhibiting remarkable metabolic flexibility during thermal adaptation. Such taxon and metabolic shifts correspond well to the temperature dependence of microbial community metabolism in aquatic ecosystems modeled by Hall et al. (62). These authors propose that organisms adapt physiologically to be competitive for substrates within specific temperature ranges, and that this adaptation results in changes in their relative contribution to community metabolism. We suggest that similar mechanisms are triggered in the Arctic peat ecosystem studied, indicating that part of the complexity found in microbial communities is attributable to their flexibility across environmental gradients such as temperature. The switch from acetotrophic to methylotrophic methanogenesis of Methanosarcinaceae likely is a direct result of increased substrate availability, possibly from a more pronounced microbial loop, but also might have been triggered by more competitive Methanosaetaceae having a higher affinity to acetate. All observed effects are in the terminal steps of anaerobic decomposition, i.e, syntrophic oxidation of organic acids and methanogenesis, indicating that the temperature effect, which is primarily thermokinetic in upstream metabolism, becomes systemic (via changes in pathways and taxa) in downstream and terminal metabolism. The results agree with those of previous studies showing that thermal adaptation is the switch from cold-adapted taxa to warm-adapted taxa (63). However, in the tightly connected anaerobic SOC degradation network of this Arctic peat soil, additional mechanisms resulting in altered substrate concentrations are as important as thermal adaptation in shaping the community response to temperature increases.
Missing Sinks and Sources.
Mass balance calculations indicated that there were missing sinks and sources of carbon and reducing equivalents. The putative sink of acetate can be explained by either assimilation (64, 65) or adsorption to solids (66), whereas the unknown redox kinetics of both solid and dissolved humic matter (67) might have been a sink for reducing equivalents (H2 and formate). Among the putative sources of CO2 are primary fermentation of hexose or pentose sugars, anaerobic respiration, and/or CH4 oxidation. Because sulfate, nitrate, and nitrite were not detected, alternative electron acceptors must be considered. The CO2 proportion of mineralized carbon was below 50%, indicating that the carbon source deviated from C6H12O6, as is consistent with the indications for methanogenesis from methylamines originating from the degradation of proteins or fats, both of which would yield a CO2 proportion of mineralized carbon below 50%. However, the increase in CO2 proportion with temperature suggested that other processes, such as CH4 oxidation, might have affected the proportions of terminal products. High numbers of transcripts and SSU rRNA were assigned to CH4-oxidizing bacteria within the Methylococcales. The assignment of the majority of the transcripts to M. tundripaludum (68, 69) indicated that this CH4-oxidizing bacterium might be important not only in oxic soil layers (20, 70) but also under anoxic conditions. However, the data generated in this study are not suitable to test this hypothesis.
Conclusions
By the end of this century, summer temperatures in the Arctic possibly will increase by 1–6 °C, with substantial spatial and temporal variations. Our results showed that within a short period (30 ± 10 d), reflecting the Arctic active summer season, the peat microbiota quickly adapted to large temperature shifts. CH4 production below 10 °C was higher than in lower latitude ecosystems and increased rapidly even with small increases in temperature. Temperature changes were associated with changes in the microbial community and metabolic network and affected pathways of SOC decomposition (including CH4 production), metabolic bottlenecks, and the microbial loop. The threshold temperature of 7 °C—a temperature that might be reached in many high-latitude anoxic peat systems periodically within the next century—defined the switch between two different system balances. At temperatures below 7 °C, syntrophic oxidation of propionate was the rate-limiting step for CH4 production, depending on the efficiency of acetotrophic methanogens. Above 7 °C, hydrolysis of polysaccharides was the rate-limiting step. Temperatures higher than 7 ° C thus might relieve limitations inherent to terminal mineralization, including syntrophic fermentation and methanogenesis. Particularly striking was the plasticity of the microbial community involved in terminal mineralization, because cascade effects of temperature altered substrate availability in terminal steps.
This work has pointed out the remarkable ability of anaerobic Arctic peat microbiota to adapt to a wide range of temperatures within a short time, reflecting their response to fluctuating temperatures during the short Arctic summer. The consequence of increased temperatures is a large increase in CH4 and CO2 production, which potentially is of crucial importance to the global carbon cycle in a warming world.
Materials and Methods
Study Sites and Sampling.
Samples were obtained from active layers at 10–15 cm depth (thawed during summer) of a representative High Arctic peat soil from Svalbard, Knudsenheia (78°56.544 N, 11°49.055 E) in August 2010 at the growing season peak. Average monthly temperatures range from −14 to 6 °C, but the vegetation surface can experience up temperatures up to 25 °C (71). The site is located ca. 5 km northwest of the research settlement Ny Ålesund. The dominant peat-forming species is the moss Calliergon richardsonii. Heavy grazing by Barnacle Geese (Branta leucopsis) limits growth of grasses (Dupontia pelligera) and the formation extensive root networks. The peat layers had a pH of 5.3–5.8 when measured after KCl extraction (20), whereas the pH in the sampled peat layer was 7.1 when measured directly in pore water at the time of sampling. The peat is ∼40 cm deep. The site and soil characteristics have been described in more detail previously (20). Five 20 cm × 30 cm × 20 cm peat blocks from the site (designated “a”–“e”) were cut in close proximity and were treated as biological replicates of the same site and same in situ condition. The upper 4 cm with vegetation was removed before the blocks were placed in plastic boxes and covered with water from the site to prevent oxygen access. The sealed plastic boxes were transported to the Max Planck Institute for Terrestrial Microbiology, Marburg, Germany, over 3 d with cooling (ca. 4 °C), and blocks were processed immediately upon arrival. Samples of peat from each block were diluted 1:1 (vol/vol) with O2-free autoclaved distilled water and mixed. The experimental set-up is shown in SI Appendix, Fig. S1. The resulting slurries of peat blocks a–d and e were placed into sterile 100-mL test bottles and16-mL tubes, respectively, which were closed with butyl rubber stoppers and capped. A total of 20 bottles, each containing 50 mL of slurry from each of the batches a–d, and 124 tubes, each containing 8 mL of slurry from batch e, were set up. All handling was performed in a glove box under a N2 atmosphere.
Preincubation.
All bottles and tubes of all five slurry batches (a–e) were preincubated for 171 d at 4 °C to allow the system to equilibrate. All incubations were conducted in the dark to prevent photosynthesis. Two replicate bottles from each of slurry batches a–d (eight bottles total) were used to monitor the development of the system (SI Appendix, Fig. S19). CH4, CO2, organic acids, and ethanol in these two replicates were measured at regular intervals throughout the preincubation period. The CH4 concentration increased steadily (SI Appendix, Fig. S19). At four time points during the preincubation, organic acids and ethanol concentrations were measured. Average concentrations of these compounds in the slurries at the beginning and end of the preincubation, respectively, were ethanol: 2.25 and 0.08 mM; propionate: 1.5 and 1.3 mM; acetate: 3.4 and 0.94 mM; and butyrate: 0.09 and 0.25 mM. (See SI Appendix, Fig. S19 for a complete overview of the concentrations measured.) Slurry batches a, b, and d were not used for further experiments.
Temperature Gradient Experiment.
The 124 tubes of the fifth slurry batch (e) were used for the temperature-gradient experiment. First, four tubes of batch e were used (destructive sampling) for HPLC analyses before the temperature experiment was started to provide time 0 concentrations and masses of organic acids and ethanol. Four series of 30 tubes (designated series A, B, C, and D; a total of 120 tubes) were flushed with N2 and incubated at 1–30 °C in increments of 1 °C; i.e., one tube from each series was incubated at each temperature, as follows: 1–10 °C for 39 d, 11–20 °C for 35 d, and 21–30 °C for 26 d (see SI Appendix, Fig. S1 for the experimental set-up). To the tubes of series C, CH3F was added at a final mixing ratio of 0.5%, which was sufficient to inhibit acetotrophic methanogenesis. At higher concentrations (>0.5%), CH3F also inhibits methanogenesis from methylamines and methanol (72). CH4, CO2, and H2 were measured in the tubes of series A, B, and C on day zero and at intervals varying between 2 and 4 d until the termination of the experiment. After termination of the experiment pore water from the tubes in series A, B, and C was sampled for HPLC analyses to provide end-point concentrations and masses of organic acids and ethanol, pH was measured, and water content was estimated by drying peat overnight at 120 °C and reweighing the samples. The end-point samples had a pH between 6.5 and 7; the pH of samples of series A and B (without CH3F) increased slightly with increasing temperature, and the pH of samples of series C (with CH3F) decreased slightly with increasing temperature. Tubes of series D were shock frozen in liquid nitrogen at the end of the incubation period, stored at −80 °C, and used for extraction of RNA and DNA.
GC, HPLC, and Ion Chromatography Measurements.
Series A, B, and C of slurry batch e were sampled for simultaneous analysis of CH4, CO2, and H2 with a gas chromatograph equipped with a flame ionization detector with an RT-Q-Bond capillary column, a methanizer for CO2 measurements, a helium ionization detector with a capillary molecular sieve steel column (SRI-9300; SRI Instruments), and a thermal conductivity detector (SRI-9300; SRI Instruments), with N2 as the carrier gas. We used pressure lock syringes (Vici; Precision Sampling, Inc.), which ensured the transfer of all molecules to the column. We sampled 250 µL per analysis, adding up to a maximum of 2 mL. We considered the temperature and pressure of the calibration gas in the calculations of the mass of a species in the injected sample. Columns were calibrated with certified standards (Messer-Griesheim). Pore-water samples were filtered through 0.2-mm membrane filters (Schleicher and Schuell) and stored at −20 °C until analyzed. Organic acids (lactate, formate, acetate, propionate, and butyrate) and ethanol were measured by HPLC as described previously (21). All detected peaks corresponded to known substances for which we had included standards for quantification. Ion chromatography (IC) analysis was carried out as described (73); all IC measurements were below the detection limit (5 µM).
Calculations.
Moles of gas were calculated using the ideal gas law and by comparison with standards of known concentration under defined pressure, temperature, and volume, yielding the masses of the measured gases in the headspace mixture. H2 partial pressures were calculated by comparison with a defined standard (detection limit: 10 ppm range or 0.3–1 Pa; for further calculations, H2 measurements below the detection limit were set to 1 Pa). CH4 and CO2 measurements were corrected for the mass removed during sampling for gas measurements; the removed mass was calculated and added to the measured values. Dissolved CH4 was calculated from the temperature dependence constant and Henry’s law solubility constant, yielding total CH4 (74). Dissolved CO2 at different temperatures was calculated from the thermodynamic solubility and dissociation constants at the measured pH, yielding the mass of dissolved CO2 species and the corresponding ∑CO2 (75). Final values were normalized to the soil dry weight. We calculated the CH4 and CO2 production rates from the difference between the concentration at the start and the amount accumulated at the end. CH4 production rate Q10 values were calculated for all 10-°C increments (1–10 °C for series A and 20–30 °C, for series B), and the average of these values was presented. Fatty acid and ethanol concentrations were determined by comparing 1-mL samples and 1-mL standards run in parallel. The mass of each compound (expressed in moles) was calculated from the slurry volume and normalized to dry weights. We calculated the mass balances from the difference between start and end concentrations of fermentation intermediates and used the balances to estimate mass flow during incubation. The reactions were determined from the metatranscriptomic data, based on known pathways from model organisms. Potential CH4 production from hydrogenotrophic methanogenesis was estimated by assuming the CH4 produced in tubes with CH3F originates only from H2/CO2 and/or formate. When the amount of CH4 accumulated in tubes with CH3F is subtracted from the amount of CH4 accumulated in tubes without CH4, the values obtained are the minimum possible values for hydrogenotrophic methanogenesis and the maximum possible values for combined acetotrophic and methylotrophic methanogenesis. The putative sinks for acetate, hydrogen, and formate along the temperature gradient were determined as the difference between masses at the start and end of the experiment. The unknown source of CO2 was calculated as the discrepancy between calculated flow of CO2 and ∑CO2 at the end point. We assumed a 1:1 relationship between substrates and products for all reaction steps leading to acetotrophic methanogenesis (see the legend of Fig. 5 for reaction balances) and that the amount of CH4 produced under CH3F inhibition represents the CH4 production from hydrogen and formate. The standard Gibbs free energies (ΔG°r) of the reactions were calculated from the standard Gibbs free energies of formation (ΔG°f) of the reactants and products (76). The actual Gibbs free energies (ΔGr) under experimental conditions were calculated, replacing ΔG°r with ΔG°T that was corrected for the actual temperatures (39). Calculations were performed using the measured concentrations and partial pressures (H2) of all involved compounds except dissolved CO2 and bicarbonate, for which the calculated dissolved fractions were used. All activity coefficients (γ) were assumed to be equal to 1 because no ions were detected (≤5 µM).
Nucleic Acid Extraction, Linear Amplification, and Illumina Sequencing.
The frozen samples from series D of batch e previously incubated at 3, 4, 5, 14, 15, 16, 24, 25, and 26 °C were homogenized separately with mortar and pestle in liquid nitrogen. Coextraction of RNA and DNA from peat soil was performed as previously described (20). Briefly, a cetrimonium bromide-containing lysis buffer and phenol:chloroform:isoamylalcohol (25:24:1) were added to all peat samples in lysis matrix E tubes (MP Biomedicals) containing silica beads and exposed to 30 s of vigorous shaking in a FastPrep machine (MP Biomedicals) for the extraction of nucleic acids. After PEG precipitation, ethanol washing and dissolution of pellets in nuclease-free water, nucleic acids were treated with DNase or RNase before metatranscriptome and metagenome generation, respectively. Total RNA was amplified using the MessageAmp II-Bacteria Kit (Ambion Life Technologies) following the kit protocol, except that the linear amplification step was carried out for 14 h. Paired-end 101-bp reads were sequenced using the Illumina HighSeq2000 sequencer (Illumina) at the Norwegian Sequencing Centre (University of Oslo, Oslo, Norway).
Enzyme Assays.
After preincubation and at the time when the temperature-gradient experiment began, three 50-mL bottles of batch c slurry were incubated at temperatures of 4, 15, and 25 °C (for a total of nine bottles). After the last samples of the temperature-gradient experiment were processed, i.e., after 39 d of incubation, the bottles were incubated for another 36 d before enzyme assays were initiated using the samples incubated at 4, 15, and 25 °C mentioned here. The contents of the bottles then were distributed in 3-mL aliquots into 16-mL tubes, each aliquot corresponding to one reaction (see below). The potential activity of cellulolytic enzymes was determined according to the method of Boschker and Cappenberg (77) modified for soil (18), using fluorogenic 4-methylumbelliferone (MUF)-labeled substrates. The substrates for exo-β-1,4-glucanase, β-1,4-glucosidase, β-1,4-xylosidase, and α-l-arabinofuranosidase were 4-MUF-β-d-cellobioside (MUFCel), 4-MUF-β-d-glucopyranoside (MUFGlu), 4-MUF-β-d-xylopyranoside (MUFXyl), and MUF-α-l-arabinofuranoside (MUFAra) (Sigma-Aldrich), respectively.
To determine saturating concentrations, an experiment was carried out with two replicates and one negative control (see below) with 2, 4, 6, and 7 mM of all four substrates at 25 °C (three tubes × four concentrations × four substrates at one temperature = 48 tubes total). To compare the potential enzyme activities at 4, 15, and 25 °C, experiments were carried out with two replicates and one negative control for each temperature and each substrate at saturated concentration (6 mM). The data generated for the 6-mM concentration at 25 °C above were compared with new assays at 4 °C and 15 °C (three tubes × one concentration × four substrates × two temperatures = 24 tubes total). Aliquots (3 mL) of the preincubated soil slurry were mixed with 0.7 mL sodium-phosphate buffer (50 mM, pH 7.0) and 2.3 mL MUF-labeled substrate (6 mM final concentration). With MUFCel as substrate, 0.3 mL gluconolactone (500 mM in 0.1 M potassium phosphate buffer; pH 7.0; inhibition of glucosidases) and 2 mL of MUFCel (6 mM final concentration) were used. Hydrolysis reactions were carried out at the preincubation temperature and were stopped after 15 min (MUFGlu, MUFXyl, MUFAra) or 90 min (MUFCel) by adding 1.8 mL of the slurry to 5 mL ethanol (96% vol/vol). Negative controls were terminated immediately after the addition of substrate. After thorough mixing, samples were centrifuged (20 min, 3,000 × g, 4 °C), and 4 mL of the supernatant was mixed with 1 mL of NaOH-glycine buffer (1 M, pH 11.0). Fluorescence was measured in a fluorescence microplate reader (excitation: 365 nm, emission: 450 nm; gain: 81) (Tecan Safire; Tecan Group).
Sequence Analyses.
Briefly, metatranscriptomic and metagenomic sequence reads were processed and analyzed as described previously (20, 78). A detailed description of sequence analyses can be found in SI Appendix, Sequence Analyses.
Statistical and Quantitative Data Analyses.
A detailed description of statistical and quantitative data analyses can be found in SI Appendix, Statistical and Quantitative Data Analyses.
Supplementary Material
Acknowledgments
We thank Alexandra Hahn (Marburg) for HPLC and IC analyses and Stephanie Hainbuch (Marburg) for GC and HPLC analyses during sample preincubation; Frøydis Strand for assistance with figure design and preparation; Karen A. Brune for linguistic revision of the manuscript; and the Norwegian High Performance Computing Consortium (NOTUR) for access to computational resources. This work was funded by The Research Council of Norway Grant 191696/V49.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive (SRA) database, www.ncbi.nlm.nih.gov/sra (accession nos. SRX885604, SRX885607, SRX885609, SRX885611, and SRX885615–SRX885628).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420797112/-/DCSupplemental.
References
- 1.Tarnocai C, et al. Soil organic carbon pools in the northern circumpolar permafrost region. Global Biogeochem Cycles. 2009;23:GB2023. [Google Scholar]
- 2.Olefeldt D, Turetsky MR, Crill PM, McGuire AD. Environmental and physical controls on northern terrestrial methane emissions across permafrost zones. Glob Change Biol. 2013;19(2):589–603. doi: 10.1111/gcb.12071. [DOI] [PubMed] [Google Scholar]
- 3.van Oldenborgh GJ, et al. In: Intergovernmental Panel on Climate Change: Annex I: Atlas of Global and Regional Climate Projections in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Stocker TF, et al., editors. IPCC; Cambridge, UK: 2013. [Google Scholar]
- 4.Davidson EA, Janssens IA. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature. 2006;440(7081):165–173. doi: 10.1038/nature04514. [DOI] [PubMed] [Google Scholar]
- 5.Koven CD, et al. Permafrost carbon-climate feedbacks accelerate global warming. Proc Natl Acad Sci USA. 2011;108(36):14769–14774. doi: 10.1073/pnas.1103910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter BP, Heimann M, Matthews E. Modeling modern methane emissions from natural wetlands: 1. Model description and results. J Geophys Res Atmos. 2001;106(D24):34189–34206. [Google Scholar]
- 7.Mikaloff Fletcher SE, Tans PP, Bruhwiler LM, Miller JB, Heimann M. CH4 sources estimated from atmospheric observations of CH4 and its C-13/C-12 isotopic ratios: 1. Inverse modeling of source processes. Global Biogeochem Cycles. 2004;18(4):17. [Google Scholar]
- 8.Bousquet P, et al. Source attribution of the changes in atmospheric methane for 2006–2008. Atmos Chem Phys. 2011;11(8):3689–3700. [Google Scholar]
- 9.Fenner N, Freeman C. Drought-induced carbon loss in peatlands. Nat Geosci. 2011;4(12):895–900. [Google Scholar]
- 10.Schimel JP, Gulledge J. Microbial community structure and global trace gases. Glob Change Biol. 1998;4(7):745–758. [Google Scholar]
- 11.Allison VJ, Yermakov Z, Miller RM, Jastrow JD, Matamala R. Using landscape and depth gradients to decouple the impact of correlated environmental variables on soil microbial community composition. Soil Biol Biochem. 2007;39(2):505–516. [Google Scholar]
- 12.Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: Terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8(11):779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- 13.Weedon JT, et al. Temperature sensitivity of peatland C and N cycling: Does substrate supply play a role? Soil Biol Biochem. 2013;61:109–120. [Google Scholar]
- 14.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61(2):262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schink B, Stams AM. Syntrophism among prokaryotes. In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. 4th Ed. Springer; Berlin: 2013. pp. 471–493. [Google Scholar]
- 16.Fey A, Conrad R. Effect of temperature on the rate limiting step in the methanogenic degradation pathway in rice field soil. Soil Biol Biochem. 2003;35(1):1–8. [Google Scholar]
- 17.Kotsyurbenko OR. Trophic interactions in the methanogenic microbial community of low-temperature terrestrial ecosystems. FEMS Microbiol Ecol. 2005;53(1):3–13. doi: 10.1016/j.femsec.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Glissmann K, Conrad R. Saccharolytic activity and its role as a limiting step in methane formation during the anaerobic degradation of rice straw in rice paddy soil. Biol Fertil Soils. 2002;35(1):62–67. [Google Scholar]
- 19.Müller N, Worm P, Schink B, Stams AJM, Plugge CM. Syntrophic butyrate and propionate oxidation processes: From genomes to reaction mechanisms. Environ Microbiol Rep. 2010;2(4):489–499. doi: 10.1111/j.1758-2229.2010.00147.x. [DOI] [PubMed] [Google Scholar]
- 20.Tveit A, Schwacke R, Svenning MM, Urich T. Organic carbon transformations in high-Arctic peat soils: Key functions and microorganisms. ISME J. 2013;7(2):299–311. doi: 10.1038/ismej.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metje M, Frenzel P. Effect of temperature on anaerobic ethanol oxidation and methanogenesis in acidic peat from a northern wetland. Appl Environ Microbiol. 2005;71(12):8191–8200. doi: 10.1128/AEM.71.12.8191-8200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metje M, Frenzel P. Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ Microbiol. 2007;9(4):954–964. doi: 10.1111/j.1462-2920.2006.01217.x. [DOI] [PubMed] [Google Scholar]
- 23.Duddleston KN, Kinney MA, Kiene RP, Hines ME. Anaerobic microbial biogeochemistry in a northern bog: Acetate as a dominant metabolic end product. Global Biogeochem Cycles. 2002;16(4):1063. [Google Scholar]
- 24.Drake HL, Horn MA, Wüst PK. Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ Microbiol Rep. 2009;1(5):307–318. doi: 10.1111/j.1758-2229.2009.00050.x. [DOI] [PubMed] [Google Scholar]
- 25.Whitman WB, Bowen TL, Boone DR. The methanogenic bacteria. In: Dworkin M, Falkow S, Rosenberg E, Schliefer K-H, editors. Prokaryotes. 3rd Ed. Vol 2. Springer; New York: 2006. pp. 165–207. [Google Scholar]
- 26.Bowman J. The methanotrophs — the families Methylococcaceae and Methylocystaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. 3rd Ed. Vol 2. Springer; New York: 2006. pp. 266–289. [Google Scholar]
- 27.Dunfield PF, et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature. 2007;450(7171):879–882. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 28.Wartiainen I, Hestnes AG, Svenning MM. Methanotrophic diversity in high arctic wetlands on the islands of Svalbard (Norway)—denaturing gradient gel electrophoresis analysis of soil DNA and enrichment cultures. Can J Microbiol. 2003;49(10):602–612. doi: 10.1139/w03-080. [DOI] [PubMed] [Google Scholar]
- 29.Liebner S, Rublack K, Stuehrmann T, Wagner D. Diversity of aerobic methanotrophic bacteria in a permafrost active layer soil of the Lena Delta, Siberia. Microb Ecol. 2009;57(1):25–35. doi: 10.1007/s00248-008-9411-x. [DOI] [PubMed] [Google Scholar]
- 30.Martineau C, Whyte LG, Greer CW. Stable isotope probing analysis of the diversity and activity of methanotrophic bacteria in soils from the Canadian high Arctic. Appl Environ Microbiol. 2010;76(17):5773–5784. doi: 10.1128/AEM.03094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smemo KA, Yavitt JB. Evidence for Anaerobic CH4 oxidation in freshwater peatlands. Geomicrobiol J. 2007;24(7-8):583–597. [Google Scholar]
- 32.Ratkowsky DA, Olley J, McMeekin TA, Ball A. Relationship between temperature and growth rate of bacterial cultures. J Bacteriol. 1982;149(1):1–5. doi: 10.1128/jb.149.1.1-5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drake H, Küsel K, Matthies C. Acetogenic prokaryotes. In: Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Springer; Berlin: 2013. pp. 3–60. [Google Scholar]
- 34.Schink B, Kremer DR, Hansen TA. Pathway of propionate formation from ethanol in pelobacter-propionicus. Arch Microbiol. 1987;147(4):321–327. [Google Scholar]
- 35.Meyer B, Kuehl JV, Deutschbauer AM, Arkin AP, Stahl DA. Flexibility of syntrophic enzyme systems in Desulfovibrio species ensures their adaptation capability to environmental changes. J Bacteriol. 2013;195(21):4900–4914. doi: 10.1128/JB.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frenzel P, Bosse U. Methyl fluoride, an inhibitor of methane oxidation and methane production. FEMS Microbiol Ecol. 1996;21(1):25–36. [Google Scholar]
- 37.Daebeler A, Gansen M, Frenzel P. Methyl fluoride affects methanogenesis rather than community composition of methanogenic archaea in a rice field soil. PLoS ONE. 2013;8(1):e53656. doi: 10.1371/journal.pone.0053656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chidthaisong A, Conrad R. Specificity of chloroform, 2-bromoethanesulfonate and fluoroacetate to inhibit methanogenesis and other anaerobic processes in anoxic rice field soil. Soil Biol Biochem. 2000;32(7):977–988. [Google Scholar]
- 39.Conrad R, Wetter B. Influence of temperature on energetics of hydrogen metabolism in homoacetogenic, methanogenic, and other anaerobic bacteria. Arch Microbiol. 1990;155(1):94–98. [Google Scholar]
- 40.Freitag TE, Prosser JI. Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl Environ Microbiol. 2009;75(21):6679–6687. doi: 10.1128/AEM.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glissman K, Chin KJ, Casper P, Conrad R. Methanogenic pathway and archaeal community structure in the sediment of eutrophic Lake Dagow: Effect of temperature. Microb Ecol. 2004;48(3):389–399. doi: 10.1007/s00248-003-2027-2. [DOI] [PubMed] [Google Scholar]
- 42.Yvon-Durocher G, et al. Methane fluxes show consistent temperature dependence across microbial to ecosystem scales. Nature. 2014;507(7493):488–491. doi: 10.1038/nature13164. [DOI] [PubMed] [Google Scholar]
- 43.Nozhevnikova AN, et al. Influence of temperature and high acetate concentrations on methanogenesis in lake sediment slurries. FEMS Microbiol Ecol. 2007;62(3):336–344. doi: 10.1111/j.1574-6941.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- 44.Schulz S, Conrad R. Influence of temperature on pathways to methane production in the permanently cold profundal sediment of Lake Constance. FEMS Microbiol Ecol. 1996;20(1):1–14. [Google Scholar]
- 45.Schulz S, Matsuyama H, Conrad R. Temperature dependence of methane production from different precursors in a profundal sediment (Lake Constance) FEMS Microbiol Ecol. 1997;22(3):207–213. [Google Scholar]
- 46.Fey A, Conrad R. Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl Environ Microbiol. 2000;66(11):4790–4797. doi: 10.1128/aem.66.11.4790-4797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin KJ, Conrad R. Intermediary metabolism in methanogenic paddy soil and the influence of temperature. FEMS Microbiol Ecol. 1995;18(2):85–102. [Google Scholar]
- 48.Kotsyurbenko OR, Nozhevnikova AN, Soloviova TI, Zavarzin GA. Methanogenesis at low temperatures by microflora of tundra wetland soil. Antonie van Leeuwenhoek. 1996;69(1):75–86. doi: 10.1007/BF00641614. [DOI] [PubMed] [Google Scholar]
- 49.Scholten JC, Conrad R. Energetics of syntrophic propionate oxidation in defined batch and chemostat cocultures. Appl Environ Microbiol. 2000;66(7):2934–2942. doi: 10.1128/aem.66.7.2934-2942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elferink S, Visser A, Pol LWH, Stams AJM. Sulfate reduction in methanogenic bioreactors. FEMS Microbiol Rev. 1994;15(2-3):119–136. [Google Scholar]
- 51.Dong X, Plugge CM, Stams AJM. Anaerobic degradation of propionate by a mesophilic acetogenic bacterium in coculture and triculture with different methanogens. Appl Environ Microbiol. 1994;60(8):2834–2838. doi: 10.1128/aem.60.8.2834-2838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lettinga G, et al. High-rate anaerobic treatment of wastewater at low temperatures. Appl Environ Microbiol. 1999;65(4):1696–1702. doi: 10.1128/aem.65.4.1696-1702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bass D, et al. Phylogeny and classification of Cercomonadida (Protozoa, Cercozoa): Cercomonas, Eocercomonas, Paracercomonas, and Cavernomonas gen. nov. Protist. 2009;160(4):483–521. doi: 10.1016/j.protis.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Andreesen JR. Glycine metabolism in anaerobes. Antonie van Leeuwenhoek. 1994;66(1-3):223–237. doi: 10.1007/BF00871641. [DOI] [PubMed] [Google Scholar]
- 55.Smith EA, Macfarlane GT. Dissimilatory amino Acid metabolism in human colonic bacteria. Anaerobe. 1997;3(5):327–337. doi: 10.1006/anae.1997.0121. [DOI] [PubMed] [Google Scholar]
- 56.Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- 57.Schneider T, et al. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol Microbiol. 2004;53(2):675–685. doi: 10.1111/j.1365-2958.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- 58.Bonkowski M. Protozoa and plant growth: The microbial loop in soil revisited. New Phytol. 2004;162(3):617–631. doi: 10.1111/j.1469-8137.2004.01066.x. [DOI] [PubMed] [Google Scholar]
- 59.Nagata T, Kirchman DL. Release of dissolved free and combined amino-acids by bacterivorous marine flagellates. Limnol Oceanogr. 1991;36(3):433–443. [Google Scholar]
- 60.Budde CF, Mahan AE, Lu J, Rha C, Sinskey AJ. Roles of multiple acetoacetyl coenzyme A reductases in polyhydroxybutyrate biosynthesis in Ralstonia eutropha H16. J Bacteriol. 2010;192(20):5319–5328. doi: 10.1128/JB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Escapa IF, García JL, Bühler B, Blank LM, Prieto MA. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida. Environ Microbiol. 2012;14(4):1049–1063. doi: 10.1111/j.1462-2920.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- 62.Hall EK, Neuhauser C, Cotner JB. Toward a mechanistic understanding of how natural bacterial communities respond to changes in temperature in aquatic ecosystems. ISME J. 2008;2(5):471–481. doi: 10.1038/ismej.2008.9. [DOI] [PubMed] [Google Scholar]
- 63.Bradford MA. Thermal adaptation of decomposer communities in warming soils. Front Microbiol. 2013;4:333. doi: 10.3389/fmicb.2013.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hori T, Noll M, Igarashi Y, Friedrich MW, Conrad R. Identification of acetate-assimilating microorganisms under methanogenic conditions in anoxic rice field soil by comparative stable isotope probing of RNA. Appl Environ Microbiol. 2007;73(1):101–109. doi: 10.1128/AEM.01676-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chidthaisong A, Rosenstock B, Conrad R. Measurement of monosaccharides and conversion of glucose to acetate in anoxic rice field soil. Appl Environ Microbiol. 1999;65(6):2350–2355. doi: 10.1128/aem.65.6.2350-2355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hordijk CA, Kamminga H, Cappenberg TE. Kinetic-studies of acetate in fresh-water sediments - use of stable isotopic tracers. Geochim Cosmochim Acta. 1994;58(2):683–694. [Google Scholar]
- 67.Bauer M, Heitmann T, Macalady DL, Blodau C. Electron transfer capacities and reaction kinetics of peat dissolved organic matter. Environ Sci Technol. 2007;41(1):139–145. doi: 10.1021/es061323j. [DOI] [PubMed] [Google Scholar]
- 68.Wartiainen I, Hestnes AG, McDonald IR, Svenning MM. Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78 degrees N) Int J Syst Evol Microbiol. 2006;56(Pt 1):109–113. doi: 10.1099/ijs.0.63728-0. [DOI] [PubMed] [Google Scholar]
- 69.Svenning MM, et al. Genome sequence of the Arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol. 2011;193(22):6418–6419. doi: 10.1128/JB.05380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Graef C, Hestnes AG, Svenning MM, Frenzel P. The active methanotrophic community in a wetland from the High Arctic. Environ Microbiol Rep. 2011;3(4):466–472. doi: 10.1111/j.1758-2229.2010.00237.x. [DOI] [PubMed] [Google Scholar]
- 71.Bengtson SA. Terrestrisk liv på Svalbard. In: Bengtson SA, Mehlum F, Severinsen T, editors. Svalbardtundraens økologi. Norsk Polar Institutt, Polarmiljøsenteret N-9296; Tromsø, Norway: 1999. pp. 21–33. [Google Scholar]
- 72.Penger J, Conrad R, Blaser M. Stable carbon isotope fractionation by methylotrophic methanogenic archaea. Appl Environ Microbiol. 2012;78(21):7596–7602. doi: 10.1128/AEM.01773-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isaksen MF, Bak F, Jorgensen BB. Thermophilic sulfate-reducing bacteria in cold marine sediment. FEMS Microbiol Ecol. 1994;14(1):1–8. [Google Scholar]
- 74.Lide DR, Frederikse HPR. In: CRC Handbook of Chemistry and Physics. 76th Ed Lide DR, editor. CRC; Boca Raton, FL: 1995. [Google Scholar]
- 75.de Vries JJ. 2001 Chapter 9, Chemistry of carbonic acid in water. Principles and applications, introduction: Theory, methods, review. Environmental Isotopes in the Hydrological Cycle, ed Mook WG (Free University, Amsterdam), Vol 1. Available at www.hydrology.nl/images/docs/ihp/Mook_I.pdf. Accessed July 17, 2014.
- 76.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tveit AT, Urich T, Svenning MM. Metatranscriptomic analysis of arctic peat soil microbiota. Appl Environ Microbiol. 2014;80(18):5761–5772. doi: 10.1128/AEM.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boschker HTS, Cappenberg TE. A sensitive method using 4-methylumbelliferyl-beta-cellobiose as a substrate to measure (1,4)-beta-glucanase activity in sediments. Appl Environ Microbiol. 1994;60(10):3592–3596. doi: 10.1128/aem.60.10.3592-3596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.