Significance
Three stop codons are used as translation termination signals within the mRNA. Nonsense suppression is defined as the read through of stop codons in an mRNA by a class of mutant tRNAs called nonsense suppressor tRNAs. Although much is known about nonsense suppression in bacteria (e.g., Escherichia coli) and eukaryotes (e.g., yeast) and although suppressor tRNAs have served as invaluable tools in bacterial and bacterial viral genetics, very little is known about nonsense suppression in archaea, the third kingdom of life. We describe here for the first time, to our knowledge, the suppression of all three stop codons in Haloferax volcanii, a halophilic archaeon, by suppressor tRNAs derived from serine or tyrosine tRNA.
Keywords: nonsense suppression, suppressor tRNA, inducible tRNA expression, haloarchaea, Haloferax volcanii
Abstract
Bacterial strains carrying nonsense suppressor tRNA genes played a crucial role in early work on bacterial and bacterial viral genetics. In eukaryotes as well, suppressor tRNAs have played important roles in the genetic analysis of yeast and worms. Surprisingly, little is known about genetic suppression in archaea, and there has been no characterization of suppressor tRNAs or identification of nonsense mutations in any of the archaeal genes. Here, we show, using the β-gal gene as a reporter, that amber, ochre, and opal suppressors derived from the serine and tyrosine tRNAs of the archaeon Haloferax volcanii are active in suppression of their corresponding stop codons. Using a promoter for tRNA expression regulated by tryptophan, we also show inducible and regulatable suppression of all three stop codons in H. volcanii. Additionally, transformation of a ΔpyrE2 H. volcanii strain with plasmids carrying the genes for a pyrE2 amber mutant and the serine amber suppressor tRNA yielded transformants that grow on agar plates lacking uracil. Thus, an auxotrophic amber mutation in the pyrE2 gene can be complemented by expression of the amber suppressor tRNA. These results pave the way for generating archaeal strains carrying inducible suppressor tRNA genes on the chromosome and their use in archaeal and archaeviral genetics. We also provide possible explanations for why suppressor tRNAs have not been identified in archaea.
The availability of Escherichia coli strains carrying nonsense suppressor tRNA genes played a crucial role in much of the early work on bacterial (1) and bacterial viral genetics (2–4). For example, use of strains carrying amber suppressors enabled the isolation and propagation of T4 bacteriophage mutants defective in phage assembly and morphogenesis. Infection of E. coli strains not carrying an amber suppressor by the mutant T4 phages and biochemical and EM analyses of the phage lysates led to identification of the step at which phage morphogenesis was blocked in each of the mutants and provided a picture of the T4 phage genes involved in morphogenesis (5, 6). Similar approaches were used to identify genes involved in morphogenesis of phages P22 and λ and dissect their morphogenetic pathways (7, 8).
In eukaryotes as well, nonsense suppressor tRNAs have played important roles in the genetic analysis of yeast (9, 10) and worms (11, 12). Suppressor tRNAs have not been identified in flies and mammals. However, ectopic expression of suppressor tRNA genes has been used to identify and suppress nonsense (stop codon) mutations in Drosophila (13, 14) and mammalian cell lines or viruses (15). A mammalian cell line carrying an inducible amber suppressor tRNA gene has been used to propagate a Polio virus mutant carrying an amber mutation in the RNA replicase gene (16).
The availability of archaeal strains carrying suppressor tRNA genes would greatly facilitate archaeal and archaeviral genetics. Surprisingly, in contrast to bacteria and eukaryotes, little is known about genetic suppression in archaea (17), and there has been no characterization of suppressor tRNAs and identification of nonsense mutations in any of the archaeal genes. A few of the methanogenic archaea contain a tRNA that can read the stop codon UGA and insert the noncanonical amino acid selenocysteine at specific sites in a protein. These cases are, however, specialized, in that read through of the UGA codon requires a cis-acting structural element elsewhere in the mRNA (18). Some archaea, including Methanosarcinaceae, use the stop codon UAG to insert the noncanonical amino acid pyrrolysine into a few proteins (19, 20). It is possible, however, that, in this case, UAG has been or is being usurped as a sense codon (21) in much the same way as UGA as a sense codon for tryptophan in mitochondria and Mycoplasma and UAA and UAG as sense codons for glutamine in Tetrahymena (22, 23).
There could be several reasons why suppressor tRNAs have not been identified in archaea. First, suppression could be weak and difficult to detect. Second, suppressor tRNAs, particularly those expressed constitutively, may be inherently toxic to archaea. Third, in contrast to bacteria and eukaryotes, archaea contain very few tRNA genes that are redundant (24). Therefore, mutation of any archaeal tRNA gene to produce a suppressor could abrogate the normal function of the tRNA and therefore, be lethal. This scenario is reminiscent of the situation in E. coli, where isolation of an amber or ochre suppressor derived from the tryptophan tRNA (tRNATrp) gene, of which there is only a single copy in WT strains, required the generation of merodiploid strains carrying an additional copy of the tRNATrp gene (25, 26).
To distinguish between the possibilities mentioned above and with the eventual goal of generating archaeal strains carrying suppressor tRNA genes on the chromosome, we mutated the serine and tyrosine tRNA genes of the archaeon Haloferax volcanii to amber, ochre, and opal suppressors and studied their activities in suppression of amber, ochre, and opal stop codons using β-galactosidase (β-gal) genes carrying the corresponding mutations as reporters. We show that the suppressor tRNAs are active in suppression of the corresponding codons in H. volcanii. Using a promoter for tRNA expression regulated by concentrations of tryptophan in the medium, we have also demonstrated inducible and regulatable suppression of stop codons. Furthermore, we show that transformation of a ΔpyrE2 H. volcanii strain with plasmids carrying the genes for a pyrE2 amber mutant and the serine amber suppressor tRNA produced transformants that could grow on plates in minimal medium lacking uracil. In addition to providing the only example of suppression of amber, ochre, and opal stop codons in archaea, these results pave the way for generation of archaeal strains carrying inducible suppressor tRNA genes on the chromosome and their use in genetic analyses of archaea and archaeal viruses.
Results
Suppression of Nonsense Codons in H. volcanii WFD11.
We first investigated whether ectopically expressed suppressor tRNAs would be functional in archaea. Using a plasmid-based reporter for measuring the activity of amber, ochre, and opal suppressor tRNAs in H. volcanii, we mutated the codon for serine 184 of the Haloferax lucentensis (previously named Haloferax alicantei) β-gal gene in plasmid pMLH32 (27) to amber, ochre, and opal stop codons (Fig. 1A). Concurrently, the anticodon sequence GGA of one of the serine tRNA genes () of H. volcanii was mutated to CUA, UUA, or UCA (amber, ochre, or opal suppressor, respectively) (Fig. S1A). The choice of was based on the fact that recognition of by seryl-tRNA synthetase does not involve the anticodon sequence. Therefore, mutation of the anticodon sequence to produce suppressor tRNAs is unlikely to affect their aminoacylation by seryl-tRNA synthetase.
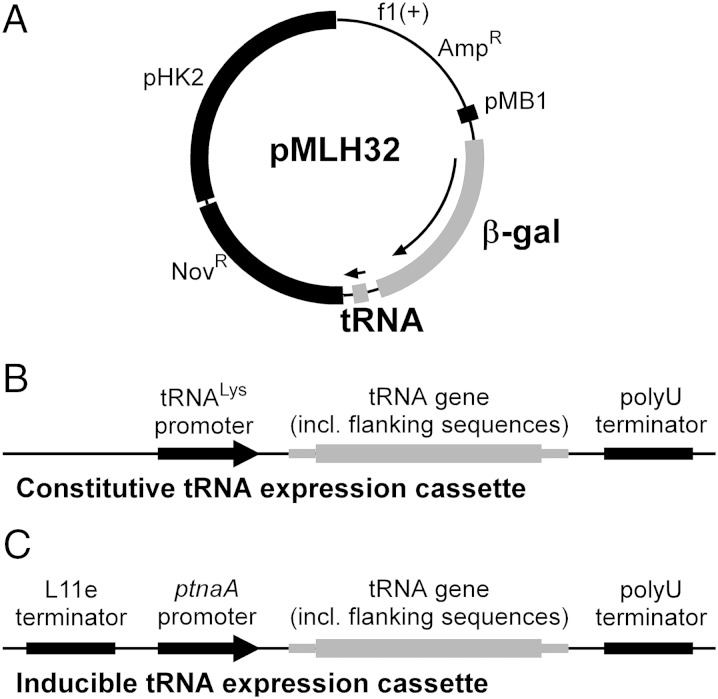
Fig. 1.
Schematic representation of plasmids used for expression of nonsense suppressor tRNAs in H. volcanii. (A) pMLH32-derived plasmids for coexpression of suppressor tRNA and β-gal reporter genes. (B) Constitutive tRNA expression cassette, in which the tRNA gene is placed under control of the promoter. (C) Inducible tRNA expression cassette, in which the tRNA gene is placed under control of the ptnaA promoter.
For expression of and mutants derived from it in H. volcanii, we first placed the tRNA coding sequences and 20 nt each of upstream and downstream flanking sequences under control of the strong constitutive promoter of plasmid pUCsptProM (28). The tRNA expression cassette (Fig. 1B) was subsequently transferred from pUCsptProM into pMLH32 downstream of the β-gal gene (Fig. 1A). The resulting pMLH32 derivatives contained the WT or mutant β-gal genes in various combinations with WT or mutant serine tRNA genes. A detailed description of plasmids is provided in SI Materials and Methods.
β-Gal activity was measured in total cell extracts of H. volcanii WFD11 (29) transformants carrying various pMLH32-derived plasmids using a luminescence-based assay and is given in relative luminescence units (RLU) per 1 s (Table 1 and Table S1). The activity in transformants carrying the combination of the WT β-gal gene and the WT gene ( 1.35 × 106 RLU/s) was set at 100%. In cells expressing mutant β-gal mRNAs, no β-gal activity was observed in the absence of suppressor tRNAs. Coexpression of the β-gal amber mRNA and the serine amber suppressor tRNA ( am indicates amber) yielded a β-gal activity of 0.32 × 106 RLU/s, corresponding to a suppression efficiency of 23.56% for the amber suppressor. The serine ochre and opal suppressors were significantly less active, with suppression efficiencies of 0.85% ( oc indicates ochre) and 0.24% ( op indicates opal), respectively. All suppressor tRNAs showed high specificity for their cognate codons, with the exception of the ochre suppressor tRNA, which could also read the amber codon to some extent ( 0.43%). The above results were also corroborated by spraying H. volcanii WFD11 transformants grown on solid media with X-gal (Fig. S2). To our knowledge, these data provide the first report of amber, ochre, and opal suppression in an archaeon.
Table 1.
Constitutive suppression of nonsense codons in H. volcanii WFD11 by serine suppressor tRNAs
| Plasmid | β-Gal | tRNA | β-Gal activity* (×103 RLU/s) | Suppression efficiency* (%) |
| WT | WT | 1,347.18 | 100.00 | |
| S184am | WT | 0.27 | 0.02 | |
| S184am | Amber | 317.35 | 23.56 | |
| S184am | Ochre | 5.83 | 0.43 | |
| S184am | Opal | 0.22 | 0.02 | |
| S184oc | WT | 0.10 | 0.01 | |
| S184oc | Amber | 0.30 | 0.02 | |
| S184oc | Ochre | 11.42 | 0.85 | |
| S184oc | Opal | 0.08 | 0.01 | |
| S184op | WT | 0.35 | 0.03 | |
| S184op | Amber | 0.45 | 0.03 | |
| S184op | Ochre | 0.58 | 0.04 | |
| S184op | Opal | 3.17 | 0.24 | |
| No plasmid† | — | — | <0.25 | N/A |
N/A, not available.
Values for β-gal activity and suppression efficiency represent the mean from two independent transformations (n = 6). SEM values are given in Table S1.
Untransformed WFD11 cells.
Expression of serine amber, ochre, and opal suppressor tRNAs in H. volcanii WFD11 was confirmed by acid urea PAGE and Northern blotting. Suppressor tRNAs were fully aminoacylated (Fig. S3A) and expressed well above the background originating from the endogenous (Fig. S3B). Thus, the substantial differences in translational efficiencies of amber, ochre, and opal suppressor tRNAs in H. volcanii WFD11 (Table 1 and Fig. S2) are not caused by large differences in expression or aminoacylation levels.
Inducible and Regulated Expression of an Amber Suppressor tRNA in H. volcanii WFD11.
Plasmid-based constitutive expression of suppressor tRNAs described above displayed signs of toxicity for the amber suppressor tRNA indicated by heterogeneity in colony size, longer lag periods, and generally, reduced growth rates (Fig. S3 C and D). Because toxicity of strong suppressor tRNAs that are constitutively expressed is an established phenomenon (30), we opted for an inducible and regulated promoter for expression of suppressor tRNAs in H. volcanii. We chose the tryptophan-inducible tryptophanase promoter (ptnaA) originally described for expression of protein genes in H. volcanii (31, 32). The serine amber suppressor tRNA gene was cloned under control of the ptnaA promoter of pTA1228 (33), and the newly generated inducible tRNA expression cassette was subsequently shuttled from pTA1228 into pMLH32 (Fig. 1C). The construct was designated (i indicates inducible).
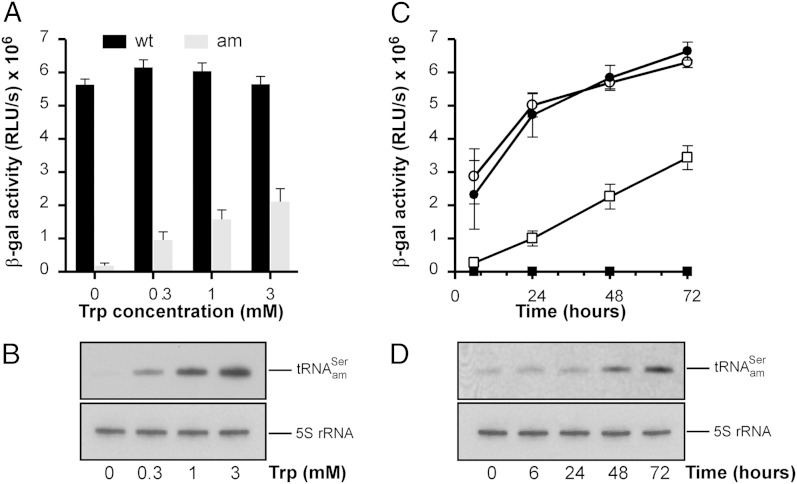
As a test of inducible and regulated expression of the serine amber suppressor in H. volcanii WFD11, we first analyzed the effect of changing the tryptophan concentration in the medium. Transformants carrying the plasmid were selected on solid H. volcanii media based on yeast extract–tryptone (Hvo-YT; Hvo indicates H. volcanii) or yeast extract–peptone–casamino acids (Hvo-YPC) that contained ∼0.3 or ∼0.15 mM tryptophan, respectively. Precultures of transformants were grown in a casamino acids-based medium (Hvo-Ca) lacking tryptophan for 4–5 d and then, diluted into fresh Hvo-Ca medium supplemented with 0, 0.3, 1, or 3 mM tryptophan; 48 h postinduction, cells were harvested and assayed for β-gal activity. Fig. 2A shows that β-gal activity increased with increasing amounts of tryptophan. The highest levels of β-gal activity were observed with 3 mM tryptophan, yielding 2.1 × 106 RLU/s, corresponding to an apparent suppression efficiency of 37.30%. We use the term apparent suppression efficiency to indicate the fact that the expression of WT β-gal mRNA used as a control is constitutive, and thus, β-gal activity stays constant (see below), whereas β-gal activity in cells expressing the β-gal amber mutant depends on induction of suppressor tRNA expression by tryptophan and hence, increases with time. β-Gal activity in the absence of tryptophan was 0.2 × 106 RLU/s (3.25%), indicating a small extent of leakiness before induction. Northern blot analysis of the serine amber suppressor tRNA using an anticodon-specific probe correlates well with tryptophan-dependent expression of the amber suppressor in H. volcanii WFD11 (Fig. 2B).
Fig. 2.
Inducible and regulatable expression of the serine amber suppressor tRNA in H. volcanii WFD11. (A and B) Tryptophan-dependent suppression of the amber codon in β-gal mRNA. (A) β-Gal activities were measured in total cell extracts obtained from H. volcanii WFD11 transformed with (black bars) or (gray bars) and grown in the presence of 0, 0.3, 1, or 3 mM tryptophan. β-Gal activities 48 h postinduction with tryptophan are given. (B) Northern blot analysis of the serine amber suppressor tRNA. Total RNA was separated on an 8% (wt/vol) polyacrylamide gel under denaturing conditions; the amber suppressor tRNA was visualized by Northern blot hybridization using a 5′–32P-labeled oligonucleotide complementary to nucleotides 31–45e that preferentially recognizes the amber suppressor tRNA over the endogenous ; 5S rRNA was used as an internal standard. (C and D) Time course of tryptophan-dependent amber suppression. β-Gal activities in H. volcanii WFD11 transformed with (open circles), (open squares), (closed circles), and (closed squares) and grown in the presence of 3 mM tryptophan for 6, 24, 48, and 72 h. (D) Total RNA was isolated and analyzed by Northern hybridization as described in B. Values in A and C represent means ± SEMs (n = 6). Trp, tryptophan.
Next, we monitored β-gal activity and expression levels of the amber suppressor tRNA at a fixed tryptophan concentration over time. Transformants were selected and precultured as before; tRNA expression was induced by transferring cells to fresh Hvo-Ca medium supplemented with 3 mM tryptophan, and β-gal activity was measured after 6, 24, 48, and 72 h. Significant levels of β-gal activity above background could be detected as soon as 6 h after induction with tryptophan; β-gal activity increased steadily over time, reaching 19.99%, 39.70%, and 54.50% of apparent suppression efficiency after 24, 48, and 72 h, respectively (Fig. 2C). These results correspond to levels of suppressor tRNA detected by Northern blot analysis (Fig. 2D).
Our results show that the ptnaA promoter works well for inducible and regulated expression of tRNAs in H. volcanii. Importantly, no significant toxicity was observed for transformants expressing the serine amber suppressor tRNA, because the initial selection of transformants and precultures was performed in media containing limited amounts of tryptophan (Hvo-YT or Hvo-YPC) or no tryptophan at all (Hvo-Ca), thereby limiting high-level expression of potentially toxic suppressor tRNAs to a time frame of 48–72 h.
Induced Suppression of Amber, Ochre, and Opal Codons in H. volcanii WFD11 and H1424 by Suppressor tRNAs Derived from .
Having established the suitability of a tryptophan-inducible tRNA expression system, the genes for the ochre and opal suppressors were also placed under control of the ptnaA promoter. H. volcanii WFD11 transformants were selected and precultured as before. tRNA expression was induced by addition of tryptophan to the medium to 3 mM, and β-gal activity was measured 48 h postinduction. Table 2 and Table S2, H. volcanii WFD11 show that the amber suppressor was again the strongest among the suppressors, with an apparent suppression efficiency of almost 50%, followed by ochre and opal suppressors, with apparent suppression efficiencies of 2.47% and 0.26%, respectively. We also note weak misreading of the amber codon in the β-gal mRNA by the serine ochre suppressor tRNA (0.38%). In all cases, β-gal activities before induction with tryptophan were negligible.
Table 2.
Inducible suppression of nonsense codons in H. volcanii by serine suppressor tRNAs
| Suppression efficiency* (%) | ||
| Preinduction | 48 h Postinduction | |
| H. volcanii WFD11 | ||
| 100.00 | 100.00 | |
| 2.31 | 48.51 | |
| 0.10 | 2.47 | |
| 0.02 | 0.26 | |
| 0.02 | 0.38 | |
| H. volcanii H1424 | ||
| 100.00 | 100.00 | |
| 0.43 | 39.01 | |
| 0.05 | 0.66 | |
| 0.06 | 0.10 | |
| 0.07 | 0.21 | |
Apparent suppression efficiency. Values are the mean from two independent transformations (n ≥ 9). Corresponding RLU per 1 s values (mean ± SEM) are given in Table S2.
The ptnaA promoter was used previously for expression of proteins in the H. volcanii strain H1424 (∆pyrE2 ∆hdrB Nph-pitA ∆mrr cdc48d-Ct) (31). This strain also carries a deletion of the orotate phosphoribosyl transferase gene (∆pyrE2), which allows chromosomal manipulation using well-established pyrE-based positive selection and counterselection systems (34), such as gene KOs or chromosomal integration of genes (for example, suppressor tRNA genes) at the pyrE2 locus. With the latter goal in mind, we studied the inducible expression of suppressor tRNAs in H. volcanii H1424. The results (Table 2 and Table S2, H. volcanii H1424) obtained with H. volcanii H1424 are similar to those obtained with H. volcanii WFD11.
Induced Suppression of Amber, Ochre, and Opal Codons in H. volcanii WFD11 and H1424 by Suppressor tRNAs Derived from .
To widen the scope of nonsense suppression in haloarchaea, we generated a different set of amber, ochre, and opal suppressor tRNAs derived from H. volcanii (Fig. S1B) and placed WT and mutant tRNA genes under control of the ptnaA promoter. We also mutated the codon for tyrosine 187 of the β-gal gene to amber, ochre, and opal stop codons. The resulting plasmids were used to transform H. volcanii WFD11 and H1424. β-Gal activity was determined before induction and 48 h postinduction with 3 mM tryptophan (Table 3 and Table S3). Results show that tyrosine suppressor tRNAs are active in suppression of nonsense codons and generally follow a pattern similar to that seen for serine suppressor tRNAs. There are five notable observations. (i) The tyrosine amber suppressor is much more active in reading the amber codon in the β-gal mRNA, with an apparent suppression efficiency as high as ∼75% in both strains 48 h postinduction. (ii) The tyrosine ochre suppressor tRNA also has much higher suppression efficiency with the cognate ochre codon (almost 30%) in H. volcanii WFD11. The suppression efficiency in H. volcanii H1424 is not quite as high (4.50%) but is still significantly higher than efficiencies obtained for the serine ochre suppressor tRNA in either WFD11 or H1424 (compare Table 2 with Table 3). The difference in suppression efficiency of the tyrosine ochre suppressor in WFD11 and H1424 is likely strain-specific. (iii) The opal suppressor tRNA is consistently the weakest among the tyrosine suppressors (6.36% in WFD11 and 2.94% in H1424). It is possible that this is partly caused by poor aminoacylation of the tyrosine opal suppressor tRNA (∼20% as estimated by PhosphorImager analysis), whereas tyrosine amber and ochre suppressor tRNAs are essentially quantitatively aminoacylated (Fig. S4). (iv) Despite the much improved activity of the tyrosine ochre suppressor tRNA, in particular, in H. volcanii WFD11, we do not see any significant misreading of the amber codon, suggesting that the tyrosine ochre suppressor is specific for the ochre codon. (v) Considering the high activities of the tyrosine amber and ochre suppressors in WFD11, it is not surprising that leaky transcription in medium lacking tryptophan leads to considerable levels of β-gal activity before induction with tryptophan (24.24% and 4.62%, respectively). Background activities before induction with tryptophan are significantly lower in H1424 (Table 3, compare H. volcanii WFD11 with H. volcanii H1424).
Table 3.
Inducible suppression of nonsense codons in H. volcanii by tyrosine suppressor tRNAs
| Suppression efficiency* (%) | ||
| Preinduction | 48 h Postinduction | |
| H. volcanii WFD11 | ||
| 100.00 | 100.00 | |
| 24.24 | 74.23 | |
| 4.62 | 29.98 | |
| 0.38 | 6.36 | |
| 0.19 | 1.33 | |
| H. volcanii H1424 | ||
| 100.00 | 100.00 | |
| 1.84 | 74.08 | |
| 0.14 | 4.50 | |
| 0.03 | 2.94 | |
| 0.01 | 0.03 | |
Apparent suppression efficiency. Values are the mean from two independent transformations (n ≥ 9). Corresponding RLU per 1 s values (mean ± SEM) are given in Table S3.
Genetic Suppression of in H. volcanii H1424.
As a prelude to investigating whether the plasmid-based suppression system described above can be used to genetically select for suppression of amber codons in chromosomal genes, we transformed the ∆pyrE2 H. volcanii strain H1424 with a mixture of plasmids and . The pTA1228 plasmid carried the gene for a pyrE2 mutant, in which the codon for serine 30 has been changed to an amber codon. As a control, H1424 was also transformed with and . The transformation mixtures were spread directly onto Hvo-Ca plates supplemented with 3 mM tryptophan without any preselection of transformants on novobiocin-containing plates. Strikingly, only transformants carrying the serine amber suppressor tRNA gene grew on plates lacking uracil (Fig. 3A), whereas transformants carrying the WT did not (Fig. 3B). With transformation mixes containing plasmids and spread directly onto Hvo-Ca plates not supplemented with tryptophan, we observed a much smaller number of tiny colonies, most likely from leaky transcription of the serine amber suppressor tRNA gene (Fig. 3C). Thus, complementation of the ∆pyrE2 phenotype of H. volcanii H1424 requires induction of the amber suppressor tRNA gene (Fig. S5).
Fig. 3.
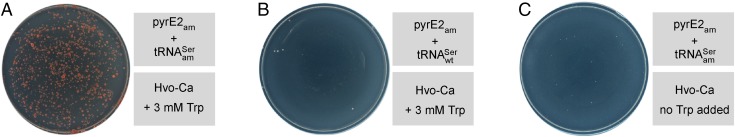
Genetic suppression of in H. volcanii H1424. Cells were transformed with (A and C) a mixture of plasmids pTA1228. and or (B) a mixture of plasmids pTA1228. and . Transformants were selected directly on Hvo-Ca medium supplemented with 3 mM tryptophan (Trp) as indicated.
Discussion
We have shown that plasmid-based expression of amber, ochre, and opal suppressor tRNAs derived from serine and tyrosine tRNAs can be used to suppress the corresponding stop codons in a reporter gene in H. volcanii. The suppressor tRNAs can be expressed constitutively (Table 1) or in an induced and regulatable manner (Tables 2 and 3). These results, combined with the finding that suppression of an auxotrophic amber mutation in the pyrE2 gene by an amber suppressor can be directly selected for using uracil-deficient medium (Fig. 3), pave the way for chromosomal insertion of suppressor tRNA genes using various approaches (35, 36) and isolation of H. volcanii strains expressing any suppressor tRNA gene of interest. The availability of such strains will facilitate archaeal and archaeviral genetics, and work along these lines is in progress.
Of two sets of H. volcanii suppressor tRNA genes generated in this work, the amber suppressor is, in both cases, the most active followed by the ochre suppressor and then, the opal suppressor, with the -derived ochre and opal suppressors being more active than the corresponding -derived suppressors. Also, in bacteria, such as E. coli, there is a range of suppressor tRNA activity depending on levels of expression of the tRNA, sequences in and around the anticodon loop that significantly affect activity of the suppressor tRNA (37–40) and mRNA codon contexts. For example, mutations in the anticodon loop region of the -derived suppressor tRNA can be used to increase the activity of the suppressor tRNAs in mammalian cells by between 36- and 200-fold (41). It is, therefore, possible that the weak activities, particularly of the H. volcanii serine ochre and opal suppressor tRNAs, can be increased by judicious mutations of sequences in the anticodon loop region.
A question of much interest is the specificity of the H. volcanii ochre suppressor tRNAs. In bacteria, ochre suppressors also suppress amber codons, whereas in eukaryotes, ochre suppressors are much more specific, particularly in mammalian cells (42, 43). For example, a human -derived ochre suppressor is specific for the ochre codon in CV1 and NIH 3T3 cells (44). Similarly, an E. coli -derived ochre suppressor expressed in mammalian HEK293 cells is specific for the ochre codon, whereas the same tRNA expressed in E. coli also suppresses the amber codon quite well (41). With the H. volcanii ochre suppressors developed here, the -derived suppressor reads the amber codon to a certain extent, although its overall activity is quite weak (Tables 1 and 2). In contrast, the H. volcanii -derived ochre suppressor, which has very high activity when expressed in H. volcanii WFD11, is much more ochre-specific and reads the amber codon very poorly (Table 3). Therefore, at least the H. volcanii -derived ochre suppressor may be specific for the ochre codon. However, to establish more definitively whether H. volcanii ochre suppressors are, in general, more bacteria-like, more eukaryote-like, or somewhere in between, it would be important to study the activity of other H. volcanii ochre suppressors, including anticodon loop mutants of -derived ochre suppressors with increased ochre suppression activity. The molecular mechanism of specificity of ochre suppressors in mammalian cells is not established and could be caused by differences in tRNA sequences, anticodon base modifications, and/or the ribosome.
Our success in generating suppressor tRNAs that function in H. volcanii raises, once again, the question of why suppressor tRNAs have not been identified before in archaea, with the rare exceptions of tRNAs that insert selenocysteine or pyrrolysine (18, 21, 45). Based on our results, it is unlikely that suppressor tRNAs are inherently toxic to archaea. The likely possibilities for absence of suppressor tRNAs could depend on whether the organism is euryarchaeal with a polyploid genome (for example, H. volcanii) or crenarchaeal with a monoploid genome (for example, Sulfolobus solfataricus) (46). In H. volcanii, with 12–20 copies of the chromosome, mutation of a single gene to a suppressor would produce very little suppressor tRNA and weak suppression activity. In the absence of any strong selection pressure, the mutant allele would probably be erased by gene conversion.
The possibility for absence of suppressor tRNAs in organisms, such as S. solfataricus, could be the lack of redundancy of tRNA genes in archaea compared with bacteria, such as E. coli, or eukaryotes, such as Saccharomyces cerevisiae. For example, the S. solfataricus, H. volcanii, E. coli K12, and S. cerevisiae genomes contain 46, 51, 86, and 286 tRNA genes, respectively, all coding for ∼44–46 different tRNA anticodons (24). Based on the number of tRNA genes in S. solfataricus, it would be impossible to generate an ochre or opal suppressor tRNA with the anticodon sequences UUA and UCA, respectively, through a single-nucleotide change in the anticodon, while maintaining the necessary and specific function of the parental tRNA. Five tRNAs with anticodons CAA, CGA, CUG, CUC, or CUU (reading codons UUG, UCG, CAG, GAG, and AAG, respectively) could, theoretically, be mutated to produce an amber suppressor with the anticodon CUA. These tRNAs are normally not considered essential, because for each of them, there is a corresponding tRNA, in which the first nucleotide C of the anticodon is replaced by a U. However, if these five tRNAs were essential for translation of specific mRNAs, mutation of their anticodon sequences to produce an amber suppressor would be strongly selected against. For example, it is known that although S. cerevisiae contains the gene for , which reads both UUA and UUG codons, and the gene for , which reads only the UUG codon, is still required for translation of mRNAs with multiple UUG codons (47).
The hypothesis of the ploidy state of the genome or the lack of tRNA gene redundancy limiting the generation of suppressor tRNAs in archaea would explain why there have been no suppressors isolated through the usual route of anticodon sequence mutations in tRNA genes, particularly in the absence of any strong genetic selection. It would also suggest that the approach used here is the only possible one for now to generate suppressors that function in archaea.
Materials and Methods
Strains and Plasmids.
A description of all strains and plasmids used in this work can be found in SI Materials and Methods and Table S4.
Transformation and Growth of H. volcanii Strains.
H. volcanii strains WFD11 (29) and H1424 (31) were routinely grown in rich medium based on Hvo-YT or Hvo-YPC or a casamino acids-based minimal medium (Hvo-Ca) (detailed composition in SI Materials and Methods). When necessary, novobiocin, thymidine, and uracil were added to a concentration of 0.2, 40, or 50 µg/mL, respectively. Transformation of H. volcanii mediated by PEG 600 was as described before (48).
Constitutive and Inducible Expression of Suppressor tRNAs in H. volcanii.
For constitutive expression of suppressor tRNAs in WFD11, transformants (single colonies) carrying the respective pMLH32-derived plasmids were grown at 37 °C in 5 mL Hvo-YT supplemented with novobiocin. Cells were harvested when cultures reached an A600 of ∼1. For inducible expression of suppressor tRNAs in WFD11 or H1424, transformants were first precultured at 37 °C for 4–5 d in 5 mL Hvo-Ca medium supplemented with novobiocin (WFD11) or novobiocin, thymidine, and uracil (H1424). Cells were then diluted to 0.1 A600 into fresh Hvo-Ca medium supplemented as before, and tryptophan was added to a final concentration of 0–3 mM as indicated. Cells were harvested 6–72 h postinduction.
β-Gal Assay.
β-Gal activity of H. volcanii transformants grown in liquid media was measured as follows. Cells corresponding to 0.2 A600 were resuspended in 400 µL β-gal assay buffer (2.5 M NaCl, 0.05 M Tris⋅HCl, pH 7.2, 10 µM MnCl2, 0.12% Triton X-100) and vortexed to lyse the cells; 1 µL cell lysate was diluted with 9 µL β-gal assay buffer, 10 µL Beta-Glo Reagent (Promega) was added, and the reaction was allowed to proceed at room temperature in the dark for 30 min. Measurement of β-gal activities was carried out in a Sirius Tube Luminometer (Berthold Detection Systems) using a 10-s premeasurement delay and a 15-s measurement period. β-Gal activities are given as RLU per 1 s. β-Gal activities represent the means ± SEMs of at least six individual colonies originating from at least two independent transformations. To detect β-gal activities of transformants grown on solid media, a solution of X-gal (20 mg/mL) in dimethylformamide was sprayed onto colonies, and the plates were incubated overnight at 37 °C for development of blue color.
Northern Blot Analysis of tRNA.
Extraction of total RNA, PAGE, and Northern blot hybridizations were carried out as described (48) with modifications (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Dr. Thorsten Allers for providing us with plasmids and strains and his most helpful advice. We also thank Dr. Mike Dyall-Smith for his comments and suggestions on the manuscript. This work was supported by National Institutes of Health Grant GM17151 (to U.L.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501558112/-/DCSupplemental.
References
- 1.Garen A, Siddiqi O. Suppression of mutations in the alkaline phosphatase structural cistron of E. coli. Proc Natl Acad Sci USA. 1962;48:1121–1127. doi: 10.1073/pnas.48.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benzer S, Champe SP. A change from nonsense to sense in the genetic code. Proc Natl Acad Sci USA. 1962;48:1114–1121. doi: 10.1073/pnas.48.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgar RS, Wood WB. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci USA. 1966;55(3):498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarabhai AS, Stretton AO, Brenner S, Bolle A. Co-linearity of the gene with the polypeptide chain. Nature. 1964;201:13–17. doi: 10.1038/201013a0. [DOI] [PubMed] [Google Scholar]
- 5.Epstein RH, et al. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harb Symp Quant Biol. 1963;28:375–394. [Google Scholar]
- 6.Epstein RH, Bolle A, Steinberg CM. Amber mutants of bacteriophage T4D: Their isolation and genetic characterization. Genetics. 2012;190(3):833–840. doi: 10.1534/genetics.112.138438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973;80(4):697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser D, Syvanen M, Masuda T. Processing and assembly of the head of bacteriophage lambda. J Supramol Struct. 1974;2(2-4):318–328. doi: 10.1002/jss.400020221. [DOI] [PubMed] [Google Scholar]
- 9.Liebman SW, Sherman F, Stewart JW. Isolation and characterization of amber suppressors in yeast. Genetics. 1976;82(2):251–272. doi: 10.1093/genetics/82.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherman F. Suppression in the yeast Saccharomyces cerevisiae. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces—Metabolism and Gene Expression. Cold Spring Harbor Laboratory Press; Plainview, NY: 1982. pp. 463–486. [Google Scholar]
- 11.Waterston RH, Brenner S. A suppressor mutation in the nematode acting on specific alleles of many genes. Nature. 1978;275(5682):715–719. doi: 10.1038/275715a0. [DOI] [PubMed] [Google Scholar]
- 12.Wills N, et al. The genes sup-7 X and sup-5 III of C. elegans suppress amber nonsense mutations via altered transfer RNA. Cell. 1983;33(2):575–583. doi: 10.1016/0092-8674(83)90438-5. [DOI] [PubMed] [Google Scholar]
- 13.Doerig RE, Suter B, Gray M, Kubli E. Identification of an amber nonsense mutation in the rosy516 gene by germline transformation of an amber suppressor tRNA gene. EMBO J. 1988;7(8):2579–2584. doi: 10.1002/j.1460-2075.1988.tb03107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laski FA, Ganguly S, Sharp PA, RajBhandary UL, Rubin GM. Construction, stable transformation, and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86(17):6696–6698. doi: 10.1073/pnas.86.17.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudziak RM, Laski FA, RajBhandary UL, Sharp PA, Capecchi MR. Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell. 1982;31(1):137–146. doi: 10.1016/0092-8674(82)90413-5. [DOI] [PubMed] [Google Scholar]
- 16.Sedivy JM, Capone JP, RajBhandary UL, Sharp PA. An inducible mammalian amber suppressor: Propagation of a poliovirus mutant. Cell. 1987;50(3):379–389. doi: 10.1016/0092-8674(87)90492-2. [DOI] [PubMed] [Google Scholar]
- 17.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rother M, Resch A, Gardner WL, Whitman WB, Böck A. Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol Microbiol. 2001;40(4):900–908. doi: 10.1046/j.1365-2958.2001.02433.x. [DOI] [PubMed] [Google Scholar]
- 19.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: Charging of a UAG-decoding specialized tRNA. Science. 2002;296(5572):1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 20.Polycarpo C, et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci USA. 2004;101(34):12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rother M, Krzycki JA. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea. Archaea. 2010;2010:453642. doi: 10.1155/2010/453642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitenberger CA, RajBhandary UL. Some highlights of mitochondrial research based on analyses of Neurospora crassa mitochondrial DNA. Trends Biochem Sci. 1985;10(12):478–483. [Google Scholar]
- 23.Hanyu N, Kuchino Y, Nishimura S, Beier H. Dramatic events in ciliate evolution: Alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986;5(6):1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soll L, Berg P. Recessive lethal nonsense suppressor in Escherichia coli which inserts glutamine. Nature. 1969;223(5213):1340–1342. doi: 10.1038/2231340a0. [DOI] [PubMed] [Google Scholar]
- 26.Yaniv M, Folk WR, Berg P, Soll L. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J Mol Biol. 1974;86(2):245–260. doi: 10.1016/0022-2836(74)90016-3. [DOI] [PubMed] [Google Scholar]
- 27.Holmes ML, Dyall-Smith ML. Sequence and expression of a halobacterial beta-galactosidase gene. Mol Microbiol. 2000;36(1):114–122. doi: 10.1046/j.1365-2958.2000.01832.x. [DOI] [PubMed] [Google Scholar]
- 28.Nieuwlandt DT, Daniels CJ. An expression vector for the archaebacterium Haloferax volcanii. J Bacteriol. 1990;172(12):7104–7110. doi: 10.1128/jb.172.12.7104-7110.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlebois RL, Lam WL, Cline SW, Doolittle WF. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci USA. 1987;84(23):8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian H, et al. Screening system for orthogonal suppressor tRNAs based on the species-specific toxicity of suppressor tRNAs. Biochimie. 2013;95(4):881–888. doi: 10.1016/j.biochi.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Stroud A, Liddell S, Allers T. Genetic and biochemical identification of a novel single-stranded DNA-binding complex in Haloferax volcanii. Front Microbiol. 2012;3:224. doi: 10.3389/fmicb.2012.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Large A, et al. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol Microbiol. 2007;66(5):1092–1106. doi: 10.1111/j.1365-2958.2007.05980.x. [DOI] [PubMed] [Google Scholar]
- 33.Brendel J, et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J Biol Chem. 2014;289(10):7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185(3):772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peck RF, DasSarma S, Krebs MP. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol Microbiol. 2000;35(3):667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- 36.Allers T, Mevarech M. Archaeal genetics - the third way. Nat Rev Genet. 2005;6(1):58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 37.Yarus M. Translational efficiency of transfer RNA’s: Uses of an extended anticodon. Science. 1982;218(4573):646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]
- 38.Yarus M, Cline S, Raftery L, Wier P, Bradley D. The translational efficiency of tRNA is a property of the anticodon arm. J Biol Chem. 1986;261(23):10496–10505. [PubMed] [Google Scholar]
- 39.Ericson JU, Björk GR. tRNA anticodons with the modified nucleoside 2-methylthio-N6-(4-hydroxyisopentenyl)adenosine distinguish between bases 3′ of the codon. J Mol Biol. 1991;218(3):509–516. doi: 10.1016/0022-2836(91)90697-5. [DOI] [PubMed] [Google Scholar]
- 40.Raftery LA, Egan JB, Cline SW, Yarus M. Defined set of cloned termination suppressors: In vivo activity of isogenetic UAG, UAA, and UGA suppressor tRNAs. J Bacteriol. 1984;158(3):849–859. doi: 10.1128/jb.158.3.849-859.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köhrer C, Sullivan EL, RajBhandary UL. Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: Concomitant suppression of three different termination codons in an mRNA in mammalian cells. Nucleic Acids Res. 2004;32(21):6200–6211. doi: 10.1093/nar/gkh959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner S, Beckwith JR. Ochre mutants, a new class of suppressible nonsense mutants. J Mol Biol. 1965;13(3):629–637. [Google Scholar]
- 43.Eggertsson G, Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988;52(3):354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capone JP, Sedivy JM, Sharp PA, RajBhandary UL. Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: Use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol Cell Biol. 1986;6(9):3059–3067. doi: 10.1128/mcb.6.9.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280(21):20740–20751. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 46.Soppa J. Ploidy and gene conversion in Archaea. Biochem Soc Trans. 2011;39(1):150–154. doi: 10.1042/BST0390150. [DOI] [PubMed] [Google Scholar]
- 47.Chan CT, et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandal D, et al. Identification and codon reading properties of 5-cyanomethyl uridine, a new modified nucleoside found in the anticodon wobble position of mutant haloarchaeal isoleucine tRNAs. RNA. 2014;20(2):177–188. doi: 10.1261/rna.042358.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.