ABSTRACT
The p160/steroid receptor coactivator (SRC) family comprises three pleiotropic coregulators (SRC-1, SRC-2, and SRC-3; otherwise known as NCOA1, NCOA2, and NCOA3, respectively), which modulate a wide spectrum of physiological responses and clinicopathologies. Such pleiotropy is achieved through their inherent structural complexity, which allows this coregulator class to control both nuclear receptor and non-nuclear receptor signaling. As observed in other physiologic systems, members of the SRC family have recently been shown to play pivotal roles in uterine biology and pathobiology. In the murine uterus, SRC-1 is required to launch a full steroid hormone response, without which endometrial decidualization is markedly attenuated. From “dovetailing” clinical and mouse studies, an isoform of SRC-1 was recently identified which promotes endometriosis by reprogramming endometrial cells to evade apoptosis and to colonize as endometriotic lesions within the peritoneal cavity. The endometrium fails to decidualize without SRC-2, which accounts for the infertility phenotype exhibited by mice devoid of this coregulator. In related studies on human endometrial stromal cells, SRC-2 was shown to act as a molecular “pacemaker” of the glycolytic flux. This finding is significant because acceleration of the glycolytic flux provides the necessary bioenergy and biomolecules for endometrial stromal cells to switch from quiescence to a proliferative phenotype, a critical underpinning in the decidual progression program. Although studies on uterine SRC-3 function are in their early stages, clinical studies provide tantalizing support for the proposal that SRC-3 is causally linked to endometrial hyperplasia as well as with endometrial pathologies in patients diagnosed with polycystic ovary syndrome. This proposal is now driving the development and application of innovative technologies, particularly in the mouse, to further understand the functional role of this elusive uterine coregulator in normal and abnormal physiologic contexts. Because dysregulation of this coregulator triad potentially presents a triple threat for increased risk of subfecundity, infertility, or endometrial disease, a clearer understanding of the individual and combinatorial roles of these coregulators in uterine function is urgently required. This minireview summarizes our current understanding of uterine SRC function, with a particular emphasis on the next critical questions that need to be addressed to ensure significant expansion of our knowledge of this underexplored field of uterine biology.
Keywords: decidualization, endometriosis, implantation, metabolism, placentation, steroid receptor coactivators, uterus
INTRODUCTION
Estrogen and progesterone are indispensable for the establishment and maintenance of pregnancy, whereas dysregulation of these hormone signals is associated with a broad spectrum of adverse reproductive outcomes and clinicopathologies, a subset of which is caused by a dysfunctional uterus [1]. To gain mechanistic insight into normal and abnormal hormone responses of the uterus at the molecular level, significant effort has been expended to identify the key downstream genes, pathways, and networks that mediate a specific uterine response to steroid hormone. These responses can be modulated at multiple levels that regulate normal physiology but can also contribute to disease when they become dysfunctional. Although tremendous progress has been attained in disclosing crucial steroid hormone signaling pathways in uterine physiologic and pathophysiologic contexts; reviewed previously [2–5], the identity of the pivotal coregulators (coactivators or corepressors) that operate in these signaling scenarios remains elusive. Given coregulators in general are potent modulators of a tissue's transcriptional response to both intracellular and extracellular physiologic signals [6, 7], identifying which of these transcriptional coregulators is required for a given uterine response constitutes a critical underpinning for future advances in our mechanistic understanding of hormone-dependent uterine biology and pathobiology. From a clinical perspective, advancing our knowledge in this area promises to provide a broader conceptual framework for understanding the etiopathogenesis of common reproductive morbidities that are causally linked with an abnormal steroid hormone response of the uterus; these include recurrent implantation failure, early miscarriage due to placental insufficiency, increased time to pregnancy, and endometriosis.
Remarkably, only a small number of coregulators (from nearly 450 identified to date [8]; http://www.nursa.org) have been operationally linked to male and female reproductive function and dysfunction [9–19]. Of these, the p160/steroid receptor coactivator (SRC) family, which is a subset of the nuclear coactivator (NCOA) family, has emerged as a preeminent class of reproductive coregulators, with the potential for being potent molecular descriptors and/or targets for diagnosing and/or therapeutically treating a dysfunctional uterus in the future. We focus this minireview on the role of SRC family members in uterine function and dysfunction, and highlight the invaluable contributions the genetically engineered mouse has made to our understanding of this new field of uterine coregulator biology.
THE p160/STEROID RECEPTOR COACTIVATOR FAMILY: MULTIFUNCTIONAL INTEGRATORS OF DIVERSE CELLULAR SIGNALS
A Multifunctional Domain Structure Drives Regulatory Complexity
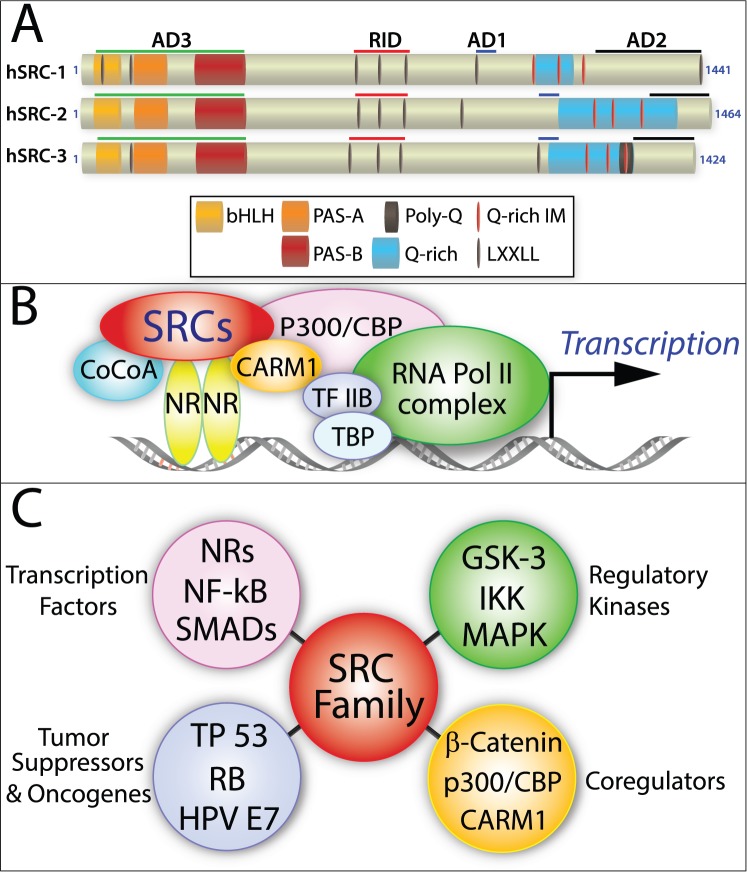
The SRC family consists of three evolutionary conserved coregulators of transcription: SRC-1 (NCOA-1, RIP160 [20]), SRC-2 (NCOA-2, TIF-2, GRIP-1 [21, 22]), and SRC-3 (NCOA-3, AIB-1, TRAM-1, pCIP [23–25]; Fig. 1A). The SRCs were first recognized as primary coactivators that transmit the transcriptional activation signal from DNA-bound nuclear receptors (NRs) to secondary coregulators, which in turn relay this signal to the downstream transcriptional machinery to enhance target gene transcription [20] (Fig. 1B). For each SRC, however, amino acid sequence analysis reveals a complex functional domain organization comprising numerous diverse protein-protein interaction surfaces (Fig. 1A). These findings suggested that members of this coregulator family possess the potential to act as multifunctional integrators of a wide spectrum of signaling cues that extend far beyond NR biology (Fig. 1C).
FIG. 1.
The multidomain structure of SRCs underpins their multifunctionality. A) Protein functional domain organization of human SRC-1, SRC-2, and SRC-3: the activation domains 1–3, receptor interaction domain, basic helix-loop-helix domain, the Per/ARNT/Sim domains A and B, leucine-X-X-leucine-leucine (X = any amino acid), glutamine-rich/interaction motif, and polyglutamine sequence are denoted by AD1-3, RID, bHLH, PAS A and B, LXXLL, Q-rich/IM, and poly-Q, respectively. B) Model of SRC-mediated transactivation of NRs: coiled-coil transcriptional coactivator A, CREB binding protein, coactivator-associated arginine methyltransferase 1, transcription factor IIB, transcription factor binding protein, and RNA polymerase II are indicated by CoCoA, CBP, CARM1, TF IIB, TBP, and RNA Pol II, respectively. C) The SRC family integrates signals beyond NR biology: NFKB [135], SMADs [136], TP53 [33], RB [137], HPV E7 [138], GSK-3 [139], IKK [140], and MAPK [141, 142] denote nuclear factor kappa-light-chain-enhancer of activated B cells, a contraction or pormanteau of Sma (Caenorhabditis elegans sma gene for small body size) and MAD (mothers against decapentaplegic), tumor protein 53, retinoblastoma protein, human papillomavirus E-7, glycogen synthase kinase 3, inhibitor of kappa B kinase, and mitogen-activated protein kinase, respectively.
The three members of the SRC family belong to a broader class of transcriptional regulators (transcription factors and coregulators) that harbor the basic helix-loop-helix-Per-Arnt-Sim (bHLH-PAS) domain [26]. Identified in 19 mammalian bHLH-PAS proteins—which include such pleiotropic factors as hypoxia-inducible factor-1α and hypoxia-inducible factor-2α (HIF-1A and HIF-2A/EPAS1), circadian locomotor output cycles kaput (CLOCK), and aryl hydrocarbon receptor (AHR)—the bHLH-PAS domain consists of a bHLH sequence contiguously followed by tandem PAS domains (PAS-A and PAS-B) [26] (Fig. 1A). Because of its structural complexity, this large composite domain mediates diverse functions, from homodimerization and heterodimerization of proteins, to DNA binding, to signal sensing and transduction. Located within the first 380 amino acids of each SRC, the bHLH-PAS domain is the most conserved region (60% amino acid identity) shared between the three coregulator homologs (Fig. 1A) and acts as a critical protein interface for binding not only secondary coregulators (i.e., coiled-coil coactivator A [CoCoA/ Calcoco1] [27]; GRIP1-associated coactivator 63 [GAC 63/SLC30A9] [28]; and Flightless-I [Flii] [29]) but also transcription factors, such as members of the signal transducer and activator of transcription (STAT) family [30–32], TP53 [33], myocyte enhancer factor-2C (MEF2C) [34], and transcriptional enhancer factor-4 (TEF-4/TEAD2) [35]. Although the bHLH region within bHLH-PAS proteins ordinarily mediates protein-protein dimerization via its HLH region and directs DNA binding through its four to six basic amino acids, SRCs have not been shown to directly bind DNA, despite possessing this region.
The juxtaposed tandem PAS domains (each ∼110 amino acid residues in length) form an antiparallel β-sheet flanked by three α-helices that generate an evolutionarily conserved hydrophobic groove [36], which allows binding of proteins containing an amphipathic helical LXXLL motif (L = leucine, X = any amino acid) [37]. Because SRCs also contain LXXLL motifs, SRC homodimerizations and heterodimerizations can potentially occur through binding of their respective LXXLL and PAS motifs [38, 39]. However, it is important to note that because the bHLH-PAS domain also binds structurally dissimilar proteins that lack LXXLL motifs (i.e., Calcoco1 and Flii [27, 29]), the specificity of this domain is significantly broader than initially suspected. Given the large size of the bHLH-PAS domain and complexity of its interaction surfaces, it is believed that many signal inputs that interface with this domain have yet to be determined. Because the bHLH-PAS domain acts synergistically with two previously identified C-terminal activation domains in SRCs to ensure target gene transcription, the SRC bHLH-PAS domain is often referred to as activation domain 3 (AD3 [27]; Fig. 1A).
The central region of SRCs contains the receptor interaction domain (RID), which enables direct interaction of SRCs with members of the NR superfamily. The RID contains three LXXLL motifs [37], which bind to a hydrophobic pocket within the ligand-binding domain of NRs (the activation function-2 [AF-2] region); the AF-2 is formed following a ligand-induced conformational change of the NR [40]. Therefore, the LXXLL motifs not only enable physical recruitment of SRCs as primary coactivators to the transcriptional complex but represent the first protein interface that receives the NR transcriptional activation signal before transmission to secondary coregulators.
Activation domains 1 and 2 (AD-1 and AD-2) in the C-terminal portion of SRCs recruit secondary coregulators to the nucleating transcriptional complex that locally remodel transcriptionally inactive or repressed chromatin through posttranslational modifications (PTMs), as well as recruit and interact with components of the RNA polymerase II transcriptional preinitiation complex at target promoters. The AD-1 domain binds p300 and cyclic AMP-response element binding (CREB) protein binding protein (CBP), both of which are potent histone acetyltransferases (HATs) that unwind chromatin to allow access and rapid formation of hierarchical protein assemblies, which form the transcription preinitiation complex [41, 42]. Of note, SRC-1 and SRC-3 also exhibit weak intrinsic HAT activity in their C-terminal region, which may contribute to PTM of components comprising the downstream transcriptional machinery [43, 44]. The AD-1 domain also can bind other members of the bHLH-PAS family (i.e., AHR and ARNT/HIF-1β [45]) and transcription factors (NFKB1 [46]). In concert with the AD-1 domain, the AD-2 domain recruits protein arginine N-methyltransferase (PRMT) family members: coactivator-associated arginine methyltransferase-1 (CARM1) [47] and PRMT1 [48], which methylate guanidino nitrogens of arginyl residues of histone proteins and other chromatin-associated proteins. Therefore, SRCs act as the “molecular go-between” for DNA-bound receptors and secondary coactivators in which the latter exhibit potent epigenetic enzymatic activities required to modulate the rate of transcription.
Posttranslational Modifications Expand the Functional Diversity of the SRC Family
Along with their complex domain structure, the functional scope of SRCs is further extended and modulated by dynamic sets of PTMs, which include phosphorylation, ubiquitination, sumoylation, and methylation of discrete amino acid residues located throughout each coregulator [49]. Termed a PTM code, specific permutations of PTMs confer distinct functional readouts, from “fine-tuning” coactivator potency, to regulating assembly or disassembly with other proteins, to increasing coregulator stability or instability, to controlling intracellular localization [50].
Pleiotropic Coregulators of Diverse Physiology and Pathology
Although their distinct structural architecture and their regulation by numerous PTMs enables SRCs to act as pleiotropic coregulators in myriad physiological systems, SRCs also serve as causative agents in many disease processes [51, 52]. Aberrant gene amplification and unscheduled “ramp-up” (or abnormal persistence) of SRC transcription were first recognized in early studies as common mechanisms by which dysregulation of this coregulator family leads to cancer [23, 52, 53]. Alteration of the fixed-domain structure of SRCs, which generates fusion proteins or coregulator isoforms, has also been linked to a number of pathologies. For example, chromosomal translocation at each SRC locus has been shown to generate fusion “oncoproteins” that contribute to a subset of aggressive leukemias and other malignancies [54–59]. The fusion oncoprotein in these clinical cases contains a truncated form of the SRC, which includes the RID, AD-1, Q-rich region, and AD-2 (Fig. 1A). Alternative splicing and proteolytic processing are additional mechanisms by which SRCs escape their fixed-domain structure to produce truncated variants or isoforms with entirely different functions and subcellular locations compared with their full-length precursors. These SRC variants have been shown to exert diverse pathological responses, from promoting mammary gland metastasis [60, 61] to driving endometriosis (the latter will be discussed in more detail in the next section).
STEROID RECEPTOR COACTIVATOR-1
A Modulator of Steroid Hormone Responsiveness in Reproductive Tissues
Two decades ago, a yeast two-hybrid screen isolated SRC-1 as the first identified SRC family member [20]. Because the C-terminal region of human progesterone receptor (PR) was used as bait, almost immediately this coregulator—and by extension other members of the family—was proposed to have a crucial role in steroid hormone-dependent reproductive biology. Soon after, this proposal was confirmed in the mouse, in which absence of SRC-1 leads to a significant decrease in steroid hormone responsiveness in all major target tissues associated with male or female reproductive biology [62]. In the case of the female, a marked partial resistance to estrogen and progesterone exposure was observed in the uterus and the mammary gland [62]. Without SRC-1, the murine endometrium fails to exhibit a full decidual response when exposed to a standard estrogen and progesterone hormone regimen and deciduogenic stimulus [62]. Furthermore, the murine endometrium requires SRC-1 to display a full uterotrophic response when exposed to unopposed estrogen [62]. As a corollary, more recent studies using combinatorial knockout mouse models indicate that SRC-1 may collaborate with SRC-3 in the embryo to ensure normal placental morphogenesis [63]; a more detailed description of these findings is provided later in this minireview.
The Long and the Short of SRC-1′s Role in Endometriosis
An estrogen-dependent, proinflammatory disease, endometriosis, is defined as the survival, colonization, and growth of endometrial tissue at anatomic sites outside the uterus, primarily the pelvic peritoneal cavity and ovaries [64, 65]. Although its etiology is uncertain, Sampson [66] proposed more than 90 years ago that endometriosis is caused by retrograde menstruation of endometrial cells through the fallopian tubes into the peritoneal cavity. Up to 10% of reproductive-age women in the United States chronically suffer from the symptoms of endometriosis, which include infertility and severe pelvic pain [67]. Indeed, estimates of endometriosis incidence as high as a third of all patients diagnosed with infertility and nearly half of all patients with persistent pelvic pain have been reported [67].
For the majority of endometriosis cases, small rather than large endometriotic lesions are detected at inflamed ectopic sites, indicating that cell survival rather than unbridled cell division is initially favored by endometriotic cells following their entry into the hostile inflammatory environment of the pelvic peritoneal cavity [68]. In support of this proposal, endometriotic cells are known to be extremely recalcitrant to apoptosis and can survive in a foreign environment in which proinflammatory cytokines would trigger apoptosis in normal endometrial cells [69]. Therefore, a number of groups have attempted to identify the distinguishing molecular features of these endometriotic cells with a view to developing more effective prognostic, diagnostic, and/or treatment strategies in the clinical management of this debilitating disease.
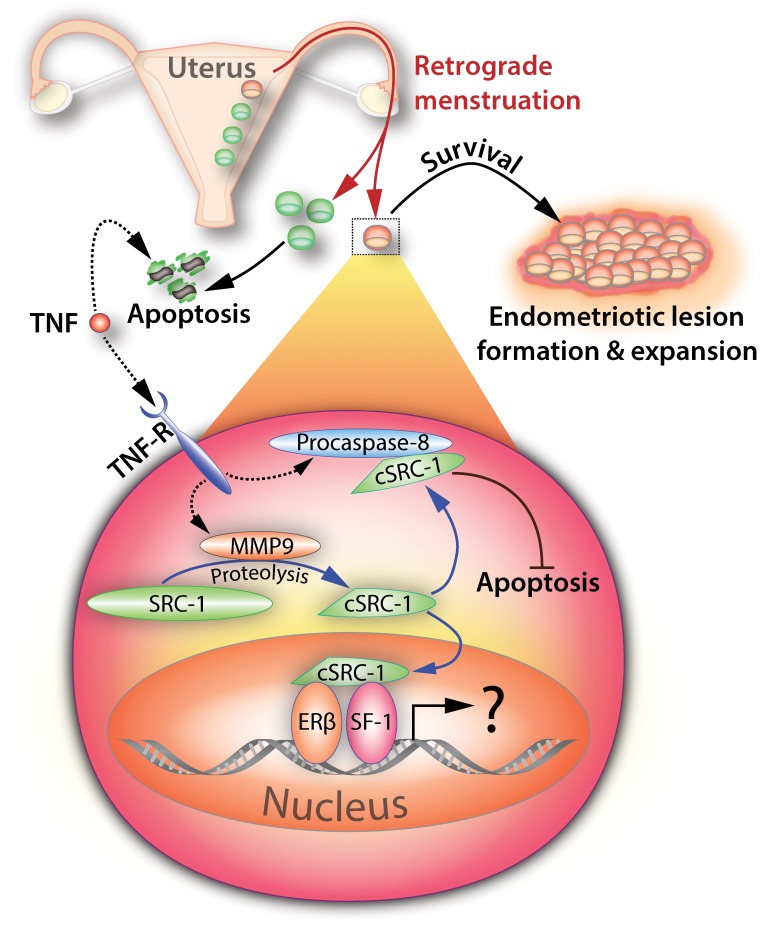
In this regard, the O'Malley group recently discovered a specific isoform of SRC-1 that is selectively expressed at high levels in human and mouse ectopic endometriotic lesions [70]. Molecular analyses revealed that a 70-kDa SRC-1 C-terminal isoform is proteolytically generated from full-length SRC-1 by matrix metalloproteinase-9 (MMP9; Fig. 2), a proteinase that is known to be upregulated in endometriotic cells during their migration and implantation into distant anatomic sites [71, 72]. Intriguingly, MMP9 is induced by activated tumor necrosis factor-α (TNFα), a cytokine that is secreted from activated peritoneal leukocytes as part of the proinflammatory response to ectopic menstrual effluents [73, 74]. Importantly, this newly identified TNFα-MMP9-SRC-1 isoform signaling axis was shown to promote the pathogenic progression of endometriosis by preventing TNFα-mediated apoptosis in human endometriotic cells, thereby promoting survival (Fig. 2). This nongenomic mechanism of action entails the interaction of the SRC-1 isoform with cytosolic caspase 8 to block TNFα-induced apoptosis (Fig. 2).
FIG. 2.
Endometriosis is driven by a short isoform of SRC-1. Retrograde menstruation from the uterus (red arrows) is hypothesized to release a subpopulation of endometrial cells (red) that is predisposed to causing endometriosis. Although the majority of the endometrial cell effluent (green) undergoes normal apoptosis in the peritoneal cavity in response to proinflammatory signals (i.e., tumor necrosis factor), the disease-forming subset of endometrial cells (red) evades apoptosis by generating a cleaved isoform of SRC-1 (cSRC-1) through a nongenomic mechanism of action. In response to proinflammatory cytokines, matrix metalloproteinase-9 proteolytically cleaves full-length SRC-1 to a ∼70-kDa cleaved isoform (cSRC-1) in the cytoplasm. Binding to procaspase-8, the cSRC-1 blocks programmed cell death, thereby allowing the endometrial cell to survive and colonize as an endometriotic lesion in the hostile environment of the pelvic peritoneal cavity. Because the cSRC-1 isoform still retains the ability to enter the nucleus and interact with NR family members, it is conceivable that this isoform also promotes NR-mediated (or genomic) signals that accelerate endometriosis progression. Tumor necrosis factor, tumor necrosis factor receptor, steroid receptor coactivator-1, cleaved steroid receptor coactivator-1, matrix metalloproteinase-9, estrogen receptor-β, and steroidogenic factor-1 are indicated by TNF, TNF-R, SRC-1, cSRC-1, MMP9, ER-β, and SF-1, respectively.
Therefore, these recent findings support the proposal that endometrial cells from a subset of women at high risk for endometriosis have evolved not only to evade but to co-opt the systemic proinflammatory response that normally clears ectopic endometrial cellular effluents from the peritoneal cavity. By co-opting the TNFα proinflammatory response, these endometrial cells generate a truncated isoform of SRC-1 that acts through a nongenomic pathway in the cytoplasm to suppress the TNFα-induced programmed cell death pathway (Fig. 2).
Apart from providing new insights into the molecular pathogenesis of endometriosis, the discovery of a new molecular signaling paradigm raises a number of important questions to be addressed in the future. 1) Because the SRC-1 isoform also distributes to the nucleus and retains its NR interaction function [70], does the isoform modulate the transactivational function of estrogen receptor-β (ER-β) and/or the orphan NR, steroidogenic factor-1 (SF-1/NR5A1)? Expressed at high levels in endometriotic lesions, ER-β (rather than ER-α) is considered a key NR mediator of estradiol-driven endometriosis [75–77]. Similarly, SF-1 is highly expressed in endometriotic cells and is primarily responsible for the local production of estradiol [78]. 2) What are the initial molecular triggers that activate the TNFα-MMP9-SRC-1 isoform axis? Epigenetic dysregulation of MMP expression is one answer recently put forward [79, 80]. 3) Is this signaling axis operational in the etiopathogenesis of other target tissues in which proinflammatory, steroid hormone, and coregulator signals intersect? For example, the association between inflammation and cancer progression is now well recognized [81]. In particular, induction of MMP9 has been linked to the progression of a number of cancers [82]. Whether cancer cells similarly use the TNFα-MMP9-SRC-1 (or other SRC isoforms) to evade apoptosis is an important question for future investigation.
STEROID RECEPTOR COACTIVATOR-2
Establishment of the Gravid Uterus Requires SRC-2
Soon after the discovery of SRC-1, the human (TIF-2) and murine (Grip-1) orthologs of SRC-2 were identified by the Chambon/Gronemeyer and Stallcup groups, respectively [21, 22]. The close homology between SRC-1 and SRC-2 was the first indication of a new family of transcriptional modulators with overlapping and distinct coregulator properties [21]. As with SRC-1, in vitro studies demonstrated that SRC-2 acts as a coactivator for many members of the NR superfamily, including NRs required for reproductive biology and dysfunction. Confirming predictions drawn from these early studies, ablation of SRC-2 in the mouse demonstrated the functional importance of this coregulator to both male and female reproductive function [83]. In the case of the female, Chambon's group revealed that a global knockout of murine SRC-2 results in an infertility defect in which absence of maternal SRC-2 leads to significant placental hypoplasia [83]. With decreased numbers of trophoblastic trabeculae in the labyrinth zone caused by delayed development of the chorioallantoic placenta, pregnancy failure occurs at midgestation in dams devoid of SRC-2. However, using an alternative murine genetic approach in which SRC-2 function is abrogated postnatally and specifically in cells expressing PR, Mukherjee et al. [84] demonstrated that the requirement for endometrial SRC-2 occurs much earlier, specifically during the peri-implantation period. Absence of SRC-2 in uterine cells positive for PR expression results in embryo implantation failure due to an early block in progesterone-dependent endometrial stromal cell (ESC) decidualization. As a critical cellular transformation process that regulates the depth and orientation of embryo invasion prior to chorioallantoic placentation, ESC decidualization is indispensable for establishing the maternofetal interface. Of clinical significance, SRC-2 also is critical for progesterone-driven decidualization of primary human ESCs in culture [85], supporting an evolutionarily conserved role for this endometrial coregulator in the reproductive success of the hemochorial placental mammal. Having demonstrated the functional importance of SRC-2 in endometrial decidualization, tandem studies on primary human ESCs and the mouse would illuminate a novel metabolic signaling mechanism by which SRC-2 elaborates the endometrial decidual response.
Metabolic Reprogramming by SRC-2 Underpins Endometrial Decidualization
Prior to decidualization, the endometrium progresses from a “prereceptive” to a “receptive” state within a narrow time frame (the window of receptivity), during which the luminal epithelium of the endometrium is transiently responsive to embryo apposition, attachment, and trophoblast invasion [2]. Even with normal embryo development, embryo implantation will fail if the endometrium does not transition to the receptive state. Of clinical importance, the inability to diagnose or therapeutically correct a nonreceptive uterus continues to impede improvements in the efficacy of current assisted reproductive technologies that employ embryo transfer into a receptive uterus [86].
Human cell and mouse studies showed that SRC-2 is essential for rapid progesterone-driven ESC proliferation, a critical cell division phase in the development of the receptive endometrium [85]. This mitotic period serves to rapidly enlarge the ESC population prior to its terminal differentiation into decidual cells [87]. Because rapid expansion of the decidual cell population is key to enabling deep invasion of the conceptus into the maternal compartment [2], SRC-2 is therefore crucial to early formation of the conceptus-endometrial interface. In the context of metabolism, a rapid acceleration of cell division requires a significant increase in both the intake and metabolism of glucose in order to provide: 1) bioenergy in the form of ATP and 2) metabolite intermediates (i.e., amino acids, fatty acids, and nucleotides) to increase biomass to levels that ensure one ESC can generate two daughter cells following mitosis. To meet this bioenergetic and biosynthetic demand, the rate of glycolysis from glucose to pyruvate (the glycolytic flux) is rapidly accelerated to supply sufficient levels of ATP and the requisite glycolytic intermediates to downstream anabolic pathways, which generate the biomass (i.e., organelles and macromolecules) necessary for cell growth prior to mitosis [88]. As long as glucose is abundant, acceleration of the glycolytic flux can rapidly furnish levels of ATP and biosynthetic precursors that far exceed those produced by the slower metabolic process of mitochondrial oxidative phosphorylation [88, 89].
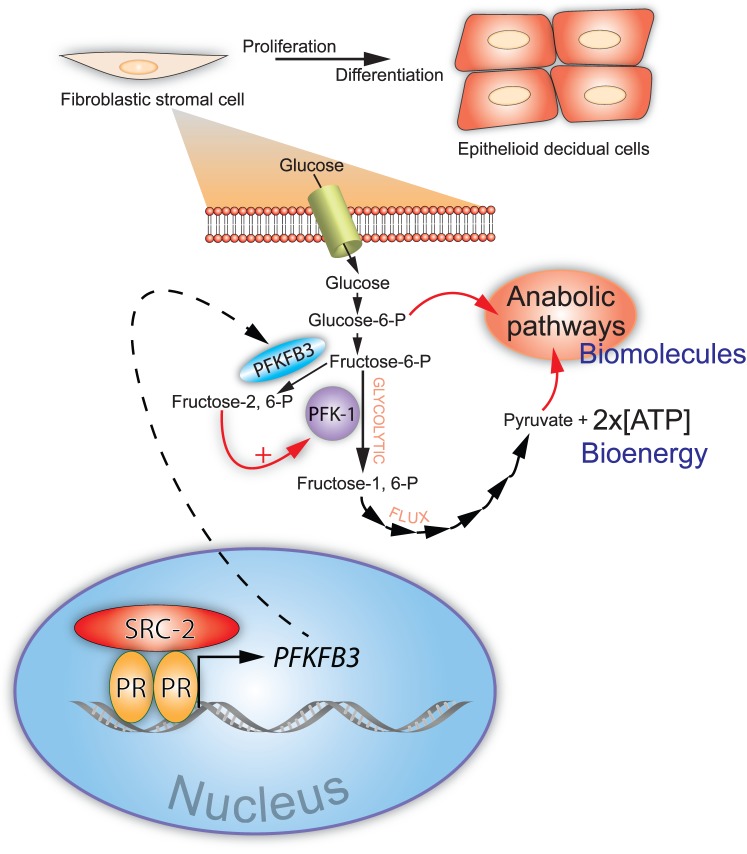
In keeping with its established role as a pleiotropic coregulator of metabolism [90], SRC-2 was shown to be required for progesterone-dependent acceleration of the glycolytic flux in cultured primary human ESCs prior to their decidualization [85]. Specifically, SRC-2 maintains the induction by progesterone of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3), a bifunctional enzyme with essential roles in embryogenesis, postnatal cellular proliferation, and cancer progression [91–93]. Originally detected in the human placenta [94], PFKFB3 was first shown to be induced by progestins in cultured human breast cancer cells [95]. Since then, numerous mitogenic, hypoxic, and inflammatory stimuli have been shown to induce PFKFB3, depending on the cell and signaling context [96–99]; importantly, PFKFB3 also is constitutively expressed in many cancers [100, 101]. Through its kinase domain, PFKFB3 converts fructose-6-phosphate to fructose-2,6-bisphosphate, a potent allosteric activator of phosphofructokinase-1 (PFK-1), which is a critical rate-limiting checkpoint of glycolysis [102, 103] (Fig. 3). With unfettered acceleration of the glycolytic flux through the PFK-1 checkpoint, anabolic pathways, such as the pentose phosphate pathway, can support rapid ESC proliferation and subsequent decidualization (Fig. 3).
FIG. 3.
Acceleration of glycolytic flux by SRC-2 is required for endometrial stromal cell decidualization. Endometrial stromal fibroblasts undergo rapid proliferation prior to differentiation in order to generate sufficient numbers of epithelioid decidual cells. To ensure that a single endometrial stromal cell can generate two daughter cells through rapid cell division, glucose uptake and the rate of glycolysis from glucose to pyruvate (the glycolytic flux) must be significantly increased. The net result of increasing the glycolytic flux is the generation of bioenergy (ATP) and biomolecules to support the formation of two daughter cells from one endometrial stromal cell. The glycolytic flux is accelerated by SRC-2 through coregulation of PR-mediated induction of PFKFB3, a critical positive regulator of the glycolytic flux. Through its regulatory kinase domain, PFKFB3 converts fructose-6-P to fructose-2,6-P, which is a potent allosteric activator of a critical enzymatic checkpoint of glycolysis, PFK-1. Along with a net gain of 2× ATP molecules per glucose molecule catabolized, glycolytic intermediates (i.e., glucose-6-P and pyruvate) provide the necessary precursors for macromolecular and organelle synthesis by downstream anabolic pathways. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, phosphofructokinase-1, steroid receptor coactivator-2, and progesterone receptor are denoted by PFKFB3, PFK-1, SRC-2, and PR, respectively.
Along with advancing our understanding of SRC-2 function in peri-implantation biology, these recent studies pose intriguing questions. 1) Does endometrial SRC-2 regulate other aspects of cellular metabolism that ensure normal ESC decidualization and prevent placental dysfunction? Apart from glucose metabolism, SRC-2 (as with other members of the SRC family) is known to be involved in regulating carbohydrate, lipid, and amino acid metabolism in other target tissues [90]. Because reproductive success is predicated on normal metabolic homeostasis, it will be important to determine whether this coregulator plays a part in other catabolic (and anabolic) pathways required to maintain endometrial functionality. 2) Can aberrant glycolytic flux regulation explain SRC-2′s implicated role in endometrial disorders in patients diagnosed with polycystic ovary syndrome or endometrial cancer? Many cancers rely on the induction of PFKFB3 and the promotion of the glycolytic flux for rapid cellular proliferation [93, 97, 104]. Aberrant expression levels of SRC-2 (along with SRC-3) have been detected in endometrial hyperplasia as well as in the endometrium of patients diagnosed with polycystic ovary syndrome [105], a patient cohort predisposed to endometrial cancer [106, 107]. Whether dysregulation of SRC-2 causes the unscheduled induction of PFKFB3 that drives proliferative disorders of the endometrium awaits further investigation. 3) Does estrogen require PFKFB3 for endometrial epithelial proliferation for normal function and/or dysfunction? As shown for progesterone [95, 108], studies in human breast cancer cells have recently revealed that PFKFB3 is a direct transcriptional target of the ER [109]. Because estrogen is a potent mitogen in the uterine epithelium in normal and abnormal physiologic contexts, establishing a signaling connection between this steroid and accelerated glucose catabolism could provide a new metabolic perspective to this steroid's mechanism of action as well as new opportunities for therapy [93]. 4) Apart from PFKFB3 induction, does SRC-2 coregulate other progesterone signaling pathways that are important for ESC decidualization? In the case of the mouse at least, recent microarray array studies reveal that SRC-2 is critically important for the full induction of the majority of previously known molecular mediators of progesterone-driven ESC decidualization [110]. Whether these findings can be translated to the human ESC remains an open question.
STEROID RECEPTOR COACTIVATOR-3
Endometrial SRC-3: Least Known of the Coregulator Triad
Amplified and overexpressed in ER- and PR-positive human breast cancers [23], SRC-3 (or AIB-1) was first identified as an oncogene [111] and is now considered a key coregulator in the progression and metastasis of many cancer types [111–113]. Along with dwarfism, delayed puberty, stunted mammary gland morphogenesis, and metabolic abnormalities, a striking subfecundity phenotype is exhibited by female mice lacking SRC-3 [114]. Decreased ovulatory capacity, a lower pregnancy frequency, a smaller litter size, and a longer estrous cycle together contribute to the subfertility phenotype of the SRC-3 null female. Follow-up serum hormone analysis indicated that SRC-3 systemically regulates growth hormone signaling pathways as well as the production of ovarian estrogen, explaining in part these diverse reproductive abnormalities [114].
Compared with SRC-1 and SRC-2, markedly less is known concerning the individual functional role of SRC-3 in endometrial biology and dysfunction; this knowledge-gap is significant because expression studies indicate that this coregulator may have important roles in both endometrial contexts. For example, clinical studies by the Lessey group first revealed that SRC-3 expression levels (along with those of SRC-2) are significantly elevated in the endometrium of patients diagnosed with polycystic ovary syndrome [105], suggesting that dysregulation in the expression levels of one or both of these coregulators in this patient cohort may contribute to endometrial dysfunction. Subsequent studies have shown that significant elevation in the expression levels of endometrial SRC-3 is correlated with endometrial hyperplasia and cancer [115–118]. Although providing correlative support, these findings are nonetheless significant because unscheduled upregulation of SRCs is the typical molecular “calling card” for the involvement of this coregulator family in tissue pathogenesis [52]. Therefore, considering the established importance of SRC-3 in aberrant cell behavior and tissue dysfunction [23, 60, 112, 119], studies are urgently required to confirm the predicted role of this coregulator in these uterine functional abnormalities.
In the context of normal uterine function, SRC-3 expression is induced in the primary decidual zone of the murine uterus at Pregnancy Day 5.5 [120], suggesting a role for this coregulator in late decidualization and/or early placentation. Interestingly, absence of SRC-3 (along with SRC-1) in the embryo resulted in a placental morphogenetic impairment that compromises the survival of embryos that lack both coregulators. Using double knockouts for SRC-3 and SRC-1, Chen et al. [63] revealed that both coregulators in the embryo coordinately function to ensure normal labyrinth morphogenesis of the placenta. Accordingly, absence of both coregulators in the developing embryo resulted in placental insufficiency and embryonic lethality at midgestation [63]. However, the individual and combinatorial roles of these coregulators in the maternal compartment during this pregnancy period remain an open question because double knockouts for both coregulators result in embryonic lethality. Given the compelling support for a critical role for SRC-3 in uterine biology, efforts are clearly required to apply innovative mouse genetics to elucidate the role of this uterine coregulator in physiologic and pathophysiologic contexts; development of cell type-specific abrogation or overexpressor mouse models—as used for SRC-2 [84, 85, 121]—may represent the first step toward elucidation in the near future.
PERSPECTIVES
If past progress is prologue, we predict that further innovations in experimental mouse genetics will continue to play an active role in advancing our understanding of the SRC family in the physiologic and pathophysiologic responses of the female reproductive tract in general and of the uterus in particular. For example, the recent CRISPR/Cas9 genome editing revolution [122–124] promises to significantly aid structure/functional analysis of SRC members not only in the murine uterus in vivo but also in clinical translational models, such as primary human endometrial cells in culture or in xenografts. Currently, we have only a rudimentary knowledge as to the extent a given SRC functional domain or PTM contributes to a particular uterine cell response. The easy, rapid, and precise mutational analysis afforded by CRISPR/Cas9 technology is predicted to play a significant role in accelerating our understanding of this understudied field of uterine biology in the not-too-distant future.
Next-generation cre/loxP technology, in which gene function is conditionally ablated in a spatiotemporal manner, will be indispensable for studying SRC function in the later stages of gestation. To date, our knowledge of uterine SRC function is limited to the processes of implantation, decidualization, and early placentation; this limitation is due to technological constraints imposed by conventional engineered mice. Addressing this limitation will be crucial because there is growing support for the involvement of this coregulator family in later stages of pregnancy, including parturition [125, 126]. Furthermore, because SRC family members have been detected in one or more uterine cell types [120], the selective functions of epithelial, stromal, and/or myometrial SRCs to a particular normal or abnormal uterine response will be critical to further resolving the functional role of each uterine SRC at the cellular level. With the availability of cell type-specific cre/loxP strategies [127–131], this type of research is now achievable.
As previously described, unwarranted elevation of SRC expression levels has been correlated with a number of endometrial pathologies [105, 115–118, 132, 133], supporting the proposal that perturbation of cellular SRC levels promotes such disorders. However, mice engineered to model SRC overexpression have not been available to test this proposal. Recently, Szwarc et al. [121] used state-of-the-art cre/loxP genetic strategies to engineer mice that overexpress human SRC-2 specifically in cells that express ER and PR. Although they are at an early stage of investigation, studies reveal that overexpression of SRC-2 in the murine endometrium results in a severe subfertility defect, validating previous predictions drawn from earlier clinical studies [105].
Such innovation notwithstanding, findings in the mouse will always need to be tested for clinical relevance. Primary human endometrial cells in culture have recently proven invaluable for testing the clinical significance of findings made in the mouse concerning uterine SRC function and dysfunction [85]. We believe that partnership between murine in vivo models and human in vitro and ex vivo systems will be essential in addressing the next big questions concerning the contributions of SRCs to endometrial biology and pathobiology. These questions include: 1) Are SRCs part of the link between abnormal metabolism and adverse reproductive outcome? Outside the uterus, members of the SRC family act as pleiotropic modulators of systems metabolism [90]. Because epidemiological and clinical investigations have established a causal connection between impaired metabolic homeostasis (i.e., preconceptional and periconceptional maternal obesity or gestational diabetes) and a broad spectrum of adverse reproductive outcomes (i.e., infertility, increased time to pregnancy, and recurrent early miscarriage), deciphering the role of this coregulator family in this causal connection is an immediate imperative. 2) What are the pivotal NR-mediated and non-NR-mediated signaling pathways that each SRC integrates to ensure normal reproductive function? By virtue of their complex functional domain structure, SRC family members are adept at integrating a remarkable array of input signals (Fig. 1). With the advent of high-throughput genome-wide methodologies, such as RNA-Seq and ChIP-seq, we expect that a comprehensive understanding of the pivotal molecular mechanisms that underpin SRC function in many aspects of female reproductive medicine will be identified. 3) Does the SRC family coordinate with other coregulator families to support normal endometrial function? Although there are nearly 450 coregulators identified to date, only a small number of coregulators (which includes the SRC family) have been shown thus far to be critical for normal endometrial function. However, just as data suggest that SRC family members functionally coordinate and collaborate with each other to maintain normal reproductive function [84, 120, 134], we predict that members of the SRC family most likely cross talk with members of other coregulator classes to achieve the same objective, thus raising the study of reproductive coregulator biology yet again to an unprecedented level of complexity.
Footnotes
Funding support in part by grants from the National Institutes of Health: HD067721 to B.A.L., and CA-077530 and U54-HD28934 to J.P.L.; M.M.S. is supported by Cancer Prevention Research Institute of Texas predoctoral training fellowship grant RP101499.
REFERENCES
- Lessey BA, Young SL. Homeostasis imbalance in the endometrium of women with implantation defects: the role of estrogen and progesterone. Semin Reprod Med. 2014;32:365–375. doi: 10.1055/s-0034-1376355. [DOI] [PubMed] [Google Scholar]
- Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar S, Hantak AM, Bagchi IC, Bagchi MK. Steroid-regulated paracrine mechanisms controlling implantation. Mol Endocrinol. 2014;28:1408–1422. doi: 10.1210/me.2014-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sabbagh M, Lam EW, Brosens JJ. Mechanisms of endometrial progesterone resistance. Mol Cell Endocrinol. 2012;358:208–215. doi: 10.1016/j.mce.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Wetendorf M, DeMayo FJ. The progesterone receptor regulates implantation, decidualization, and glandular development via a complex paracrine signaling network. Mol Cell Endocrinol. 2012;357:108–118. doi: 10.1016/j.mce.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard CJ, Watson PJ, Fairall L, Schwabe JW. An evolving understanding of nuclear receptor coregulator proteins. J Mol Endocrinol. 2013;51:T23–T36. doi: 10.1530/JME-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, Kim BJ, Li C, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Yoon S, Zhao Y, Park SE, Liao L, Xu J, Lydon JP, DeMayo FJ, O'Malley BW, Bagchi MK, Katzenellenbogen BS. Uterine development and fertility are dependent on gene dosage of the nuclear receptor coregulator REA. Endocrinology. 2012;153:3982–3994. doi: 10.1210/en.2012-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Kim TH, Kommagani R, Feng Q, Lanz RB, Jeong JW, DeMayo FJ, Katzenellenbogen BS, Lydon JP, O'Malley BW. Estrogen-regulated prohibitin is required for mouse uterine development and adult function. Endocrinology. 2011;152:1047–1056. doi: 10.1210/en.2010-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe J, Li Q, Mussi P, Liao L, Lydon J, DeMayo F, Xu J. Nuclear receptor coactivator-6 attenuates uterine estrogen sensitivity to permit embryo implantation. Dev Cell. 2012;23:858–865. doi: 10.1016/j.devcel.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Grossman G, Yuan L, Lessey BA, French FS, Young SL, Wilson EM. Hormone control and expression of androgen receptor coregulator MAGE-11 in human endometrium during the window of receptivity to embryo implantation. Mol Hum Reprod. 2008;14:107–116. doi: 10.1093/molehr/gam080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard ME, Simmons CD, Simmen FA, Simmen RC. Kruppel-like factor 9 deficiency in uterine endometrial cells promotes ectopic lesion establishment associated with activated notch and hedgehog signaling in a mouse model of endometriosis. Endocrinology. 2014;155:1532–1546. doi: 10.1210/en.2013-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen AM, Karvonen U, Poukka H, Yan W, Toppari J, Janne OA, Palvimo JJ. A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J Biol Chem. 1999;274:3700–3704. doi: 10.1074/jbc.274.6.3700. [DOI] [PubMed] [Google Scholar]
- Tan J, Hall SH, Hamil KG, Grossman G, Petrusz P, Liao J, Shuai K, French FS. Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol Endocrinol. 2000;14:14–26. doi: 10.1210/mend.14.1.0408. [DOI] [PubMed] [Google Scholar]
- Lee YS, Kim HJ, Lee HJ, Lee JW, Chun SY, Ko SK, Lee K. Activating signal cointegrator 1 is highly expressed in murine testicular Leydig cells and enhances the ligand-dependent transactivation of androgen receptor. Biol Reprod. 2002;67:1580–1587. doi: 10.1095/biolreprod.102.006155. [DOI] [PubMed] [Google Scholar]
- Song KH, Park YY, Park KC, Hong CY, Park JH, Shong M, Lee K, Choi HS. The atypical orphan nuclear receptor DAX-1 interacts with orphan nuclear receptor Nur77 and represses its transactivation. Mol Endocrinol. 2004;18:1929–1940. doi: 10.1210/me.2004-0043. [DOI] [PubMed] [Google Scholar]
- Leach DA, Need EF, Trotta AP, Grubisha MJ, DeFranco DB, Buchanan G. Hic-5 influences genomic and non-genomic actions of the androgen receptor in prostate myofibroblasts. Mol Cell Endocrinol. 2014;384:185–199. doi: 10.1016/j.mce.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Shang Z, Niu Y, Cai Q, Chen J, Tian J, Yeh S, Lai KP, Chang C. Human kallikrein 2 (KLK2) promotes prostate cancer cell growth via function as a modulator to promote the ARA70-enhanced androgen receptor transactivation. Tumour Biol. 2014;35:1881–1890. doi: 10.1007/s13277-013-1253-6. [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH-PAS proteins in cancer. Nat Rev Cancer. 2013;13:827–841. doi: 10.1038/nrc3621. [DOI] [PubMed] [Google Scholar]
- Kim J, Li H, Stallcup M. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- Chen Y-H, Kim J, Stallcup M. GAC63, a GRIP1-dependent nuclear receptor coactivator. Mol Cell Biol. 2005;25:5965–5972. doi: 10.1128/MCB.25.14.5965-5972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litterst CM, Pfitzner E. Transcriptional activation by STAT6 requires the direct interaction with NCoA-1. J Biol Chem. 2001;276:45713–45721. doi: 10.1074/jbc.M108132200. [DOI] [PubMed] [Google Scholar]
- Litterst CM, Pfitzner E. An LXXLL motif in the transactivation domain of STAT6 mediates recruitment of NCoA-1/SRC-1. J Biol Chem. 2002;277:36052–36060. doi: 10.1074/jbc.M203556200. [DOI] [PubMed] [Google Scholar]
- Giraud S, Bienvenu F, Avril S, Gascan H, Heery DM, Coqueret O. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J Biol Chem. 2002;277:8004–8011. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim HJ, Kim JW, Lee JW. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol Endocrinol. 1999;13:1924–1933. doi: 10.1210/mend.13.11.0365. [DOI] [PubMed] [Google Scholar]
- Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275:30801–30805. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science. 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Lodrini M, Munz T, Coudevylle N, Griesinger C, Becker S, Pfitzner E. P160/SRC/NCoA coactivators form complexes via specific interaction of their PAS-B domain with the CID/AD1 domain. Nucleic Acids Res. 2008;36:1847–1860. doi: 10.1093/nar/gkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PY, Hsieh TY, Chou WY, Huang SM. Modulation of glucocorticoid receptor-interacting protein 1 (GRIP1) transactivation and co-activation activities through its C-terminal repression and self-association domains. FEBS J. 2006;273:2172–2183. doi: 10.1111/j.1742-4658.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- Webster NJ, Green S, Jin JR, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- Jenster G, Spencer TE, Burcin MM, Tsai SY, Tsai MJ, O'Malley BW. Steroid receptor induction of gene transcription: a two-step model. Proc Natl Acad Sci U S A. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo ML, Kraus WL. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates. Mol Cell Biol. 2003;23:335–348. doi: 10.1128/MCB.23.1.335-348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, Nelson WE, Probst MR, Rosenfeld MG, Hankinson O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol. 2002;22:4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na SY, Lee SK, Han SJ, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappaB-mediated transactivations. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- Li S, Shang Y. Regulation of SRC family coactivators by post-translational modifications. Cell Signal. 2007;19:1101–1112. doi: 10.1016/j.cellsig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Han SJ, Lonard DM, O'Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Lonard DM, O'Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2013;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu RC, O'Malley B. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–4319. [PubMed] [Google Scholar]
- Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- Strehl S, Nebral K, Konig M, Harbott J, Strobl H, Ratei R, Struski S, Bielorai B, Lessard M, Zimmermann M, Haas OA, Izraeli S. ETV6-NCOA2: a novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin Cancer Res. 2008;14:977–983. doi: 10.1158/1078-0432.CCR-07-4022. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Gorunova L, Bjerkehagen B, Boye K, Heim S. Chromosome aberrations and HEY1-NCOA2 fusion gene in a mesenchymal chondrosarcoma. Oncol Rep. 2014;32:40–44. doi: 10.3892/or.2014.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Miyachi M, Sakamoto K, Ouchi K, Yagyu S, Kikuchi K, Kuwahara Y, Tsuchiya K, Imamura T, Iehara T, Kakazu N, Hojo H, et al. PAX3-NCOA2 fusion gene has a dual role in promoting the proliferation and inhibiting the myogenic differentiation of rhabdomyosarcoma cells Oncogene (in press) Published online ahead of print 11 November 2013. DOI:10.1038/onc.2013.491. [DOI] [PubMed] [Google Scholar]
- Sumegi J, Streblow R, Frayer RW, Dal Cin P, Rosenberg A, Meloni-Ehrig A, Bridge JA. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteyries S, Perot C, Adelaide J, Imbert M, Lagarde A, Pautas C, Olschwang S, Birnbaum D, Chaffanet M, Mozziconacci MJ. NCOA3, a new fusion partner for MOZ/MYST3 in M5 acute myeloid leukemia. Leukemia. 2008;22:663–665. doi: 10.1038/sj.leu.2404930. [DOI] [PubMed] [Google Scholar]
- Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, Mancini MA, Tsai SY, Tsai MJ, O'Malley BW. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37:321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon JP, O'Malley BW. Minireview: steroid receptor coactivator-3: a multifarious coregulator in mammary gland metastasis. Endocrinology. 2011;152:19–25. doi: 10.1210/en.2010-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu Z, Xu J. The cooperative function of nuclear receptor coactivator 1 (NCOA1) and NCOA3 in placental development and embryo survival. Mol Endocrinol. 2010;24:1917–1934. doi: 10.1210/me.2010-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98:511–519. doi: 10.1016/j.fertnstert.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. Peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14:422–469. [Google Scholar]
- Eskenazi B, Warner ML. Epidemiology of endometriosis. Obstet Gynecol Clin North Am. 1997;24:235–258. doi: 10.1016/s0889-8545(05)70302-8. [DOI] [PubMed] [Google Scholar]
- Beliard A, Noel A, Foidart JM. Reduction of apoptosis and proliferation in endometriosis. Fertil Steril. 2004;82:80–85. doi: 10.1016/j.fertnstert.2003.11.048. [DOI] [PubMed] [Google Scholar]
- Nasu K, Yuge A, Tsuno A, Nishida M, Narahara H. Involvement of resistance to apoptosis in the pathogenesis of endometriosis. Histol Histopathol. 2009;24:1181–1192. doi: 10.14670/HH-24.1181. [DOI] [PubMed] [Google Scholar]
- Han SJ, Hawkins SM, Begum K, Jung SY, Kovanci E, Qin J, Lydon JP, DeMayo FJ, O'Malley BW. A new isoform of steroid receptor coactivator-1 is crucial for pathogenic progression of endometriosis. Nat Med. 2012;18:1102–1111. doi: 10.1038/nm.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJ, He YL, Peng DX. Expression of metalloproteinase-9 in ectopic endometrium in women with endometriosis. Di Yi Jun Yi Da Xue Xue Bao. 2002;22:467–469. [PubMed] [Google Scholar]
- Ueda M, Yamashita Y, Takehara M, Terai Y, Kumagai K, Ueki K, Kanda K, Hung YC, Ueki M. Gene expression of adhesion molecules and matrix metalloproteinases in endometriosis. Gynecol Endocrinol. 2002;16:391–402. [PubMed] [Google Scholar]
- Halme J. Release of tumor necrosis factor-alpha by human peritoneal macrophages in vivo and in vitro. Am J Obstet Gynecol. 1989;161:1718–1725. doi: 10.1016/0002-9378(89)90957-5. [DOI] [PubMed] [Google Scholar]
- Eisermann J, Gast MJ, Pineda J, Odem RR, Collins JL. Tumor necrosis factor in peritoneal fluid of women undergoing laparoscopic surgery. Fertil Steril. 1988;50:573–579. doi: 10.1016/s0015-0282(16)60185-1. [DOI] [PubMed] [Google Scholar]
- Fujimoto J, Hirose R, Sakaguchi H, Tamaya T. Expression of oestrogen receptor-alpha and -beta in ovarian endometriomata. Mol Hum Reprod. 1999;5:742–747. doi: 10.1093/molehr/5.8.742. [DOI] [PubMed] [Google Scholar]
- Xue Q, Lin Z, Cheng YH, Huang CC, Marsh E, Yin P, Milad MP, Confino E, Reierstad S, Innes J, Bulun SE. Promoter methylation regulates estrogen receptor 2 in human endometrium and endometriosis. Biol Reprod. 2007;77:681–687. doi: 10.1095/biolreprod.107.061804. [DOI] [PubMed] [Google Scholar]
- Brandenberger AW, Lebovic DI, Tee MK, Ryan IP, Tseng JF, Jaffe RB, Taylor RN. Oestrogen receptor (ER)-alpha and ER-beta isoforms in normal endometrial and endometriosis-derived stromal cells. Mol Hum Reprod. 1999;5:651–655. doi: 10.1093/molehr/5.7.651. [DOI] [PubMed] [Google Scholar]
- Zeitoun K, Takayama K, Michael MD, Bulun SE. Stimulation of aromatase P450 promoter (II) activity in endometriosis and its inhibition in endometrium are regulated by competitive binding of steroidogenic factor-1 and chicken ovalbumin upstream promoter transcription factor to the same cis-acting element. Mol Endocrinol. 1999;13:239–253. doi: 10.1210/mend.13.2.0229. [DOI] [PubMed] [Google Scholar]
- Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, Chluba J, Langsley G, Weitzman JB. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72:810–820. doi: 10.1158/0008-5472.CAN-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov AV, Sounni NE, Remacle AG, Strongin AY. Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J Biol Chem. 2009;284:12727–12734. doi: 10.1074/jbc.M900273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxburgh CS, McMillan DC. Cancer and systemic inflammation: treat the tumour and treat the host. Br J Cancer. 2014;110:1409–1412. doi: 10.1038/bjc.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol. 2002;22:5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Soyal S, Fernandez-Valdivia R, Gehin M, Chambon P, Demayo F, Lydon J, O'Malley B. Steroid receptor coactivator 2 is critical for progesterone-dependent uterine function and mammary morphogenesis in the mouse. Mol Cell Biol. 2006;26:6571–6583. doi: 10.1128/MCB.00654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommagani R, Szwarc MM, Kovanci E, Gibbons WE, Putluri N, Maity S, Creighton CJ, Sreekumar A, DeMayo FJ, Lydon JP, O'Malley BW. Acceleration of the glycolytic flux by steroid receptor coactivator-2 is essential for endometrial decidualization. PLoS Genet. 2013;9:e1003900. doi: 10.1371/journal.pgen.1003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BS, Daneshmand ST, Garner FC, Aguirre M, Hudson C, Thomas S. Evidence of impaired endometrial receptivity after ovarian stimulation for in vitro fertilization: a prospective randomized trial comparing fresh and frozen-thawed embryo transfer in normal responders. Fertil Steril. 2011;96:344–348. doi: 10.1016/j.fertnstert.2011.05.050. [DOI] [PubMed] [Google Scholar]
- Sroga JM, Ma X, Das SK. Developmental regulation of decidual cell polyploidy at the site of implantation. Front Biosci (Schol Ed) 2012;4:1475–1486. doi: 10.2741/s347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CB. Rethinking the regulation of cellular metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:23–29. doi: 10.1101/sqb.2012.76.010496. [DOI] [PubMed] [Google Scholar]
- Stashi E, York B, O'Malley BW. Steroid receptor coactivators: servants and masters for control of systems metabolism. Trends Endocrinol Metab. 2014;25:337–347. doi: 10.1016/j.tem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney J, Telang S, Yalcin A, Clem A, Wallis N, Bucala R. Targeted disruption of inducible 6-phosphofructo-2-kinase results in embryonic lethality. Biochem Biophys Res Commun. 2005;331:139–146. doi: 10.1016/j.bbrc.2005.02.193. [DOI] [PubMed] [Google Scholar]
- Chesney J. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase and tumor cell glycolysis. Curr Opin Clin Nutr Metab Care. 2006;9:535–539. doi: 10.1097/01.mco.0000241661.15514.fb. [DOI] [PubMed] [Google Scholar]
- Clem B, Telang S, Clem A, Yalcin A, Meier J, Simmons A, Rasku MA, Arumugam S, Dean WL, Eaton J, Lane A, Trent JO, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther. 2008;7:110–120. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- Fukasawa M, Tsuchiya T, Takayama E, Shinomiya N, Uyeda K, Sakakibara R, Seki S. Identification and characterization of the hypoxia-responsive element of the human placental 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene. J Biochem. 2004;136:273–277. doi: 10.1093/jb/mvh137. [DOI] [PubMed] [Google Scholar]
- Novellasdemunt L, Obach M, Millan-Arino L, Manzano A, Ventura F, Rosa JL, Jordan A, Navarro-Sabate A, Bartrons R. Progestins activate 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) in breast cancer cells. Biochem J. 2012;442:345–356. doi: 10.1042/BJ20111418. [DOI] [PubMed] [Google Scholar]
- Duran J, Obach M, Navarro-Sabate A, Manzano A, Gomez M, Rosa JL, Ventura F, Perales JC, Bartrons R. Pfkfb3 is transcriptionally upregulated in diabetic mouse liver through proliferative signals. FEBS J. 2009;276:4555–4568. doi: 10.1111/j.1742-4658.2009.07161.x. [DOI] [PubMed] [Google Scholar]
- Novellasdemunt L, Bultot L, Manzano A, Ventura F, Rosa JL, Vertommen D, Rider MH, Navarro-Sabate A, Bartrons R. PFKFB3 activation in cancer cells by the p38/MK2 pathway in response to stress stimuli. Biochem J. 2013;452:531–543. doi: 10.1042/BJ20121886. [DOI] [PubMed] [Google Scholar]
- Minchenko A, Leshchinsky I, Opentanova I, Sang N, Srinivas V, Armstead V, Caro J. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene: its possible role in the Warburg effect. J Biol Chem. 2002;277:6183–6187. doi: 10.1074/jbc.M110978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando M, Uehara I, Kogure K, Asano Y, Nakajima W, Abe Y, Kawauchi K, Tanaka N. Interleukin 6 enhances glycolysis through expression of the glycolytic enzymes hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3. J Nippon Med Sch. 2010;77:97–105. doi: 10.1272/jnms.77.97. [DOI] [PubMed] [Google Scholar]
- Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–5887. [PubMed] [Google Scholar]
- Kessler R, Bleichert F, Warnke JP, Eschrich K. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3) is up-regulated in high-grade astrocytomas. J Neurooncol. 2008;86:257–264. doi: 10.1007/s11060-007-9471-7. [DOI] [PubMed] [Google Scholar]
- Uyeda K, Furuya E, Luby LJ. The effect of natural and synthetic D-fructose 2,6-bisphosphate on the regulatory kinetic properties of liver and muscle phosphofructokinases. J Biol Chem. 1981;256:8394–8399. [PubMed] [Google Scholar]
- Van Schaftingen E, Jett MF, Hue L, Hers HG. Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proc Natl Acad Sci U S A. 1981;78:3483–3486. doi: 10.1073/pnas.78.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A, Clem BF, Simmons A, Lane A, Nelson K, Clem AL, Brock E, Siow D, Wattenberg B, Telang S, Chesney J. Nuclear targeting of 6-phosphofructo-2-kinase (PFKFB3) increases proliferation via cyclin-dependent kinases. J Biol Chem. 2009;284:24223–24232. doi: 10.1074/jbc.M109.016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CW, Wilson EM, Apparao KB, Lininger RA, Meyer WR, Kowalik A, Fritz MA, Lessey BA. Steroid receptor coactivator expression throughout the menstrual cycle in normal and abnormal endometrium. J Clin Endocrinol Metab. 2002;87:2960–2966. doi: 10.1210/jcem.87.6.8572. [DOI] [PubMed] [Google Scholar]
- Coulam CB, Annegers JF, Kranz JS. Chronic anovulation syndrome and associated neoplasia. Obstet Gynecol. 1983;61:403–407. [PubMed] [Google Scholar]
- Pillay OC, Te Fong LF, Crow JC, Benjamin E, Mould T, Atiomo W, Menon PA, Leonard AJ, Hardiman P. The association between polycystic ovaries and endometrial cancer. Hum Reprod. 2006;21:924–929. doi: 10.1093/humrep/dei420. [DOI] [PubMed] [Google Scholar]
- Hamilton JA, Callaghan MJ, Sutherland RL, Watts CK. Identification of PRG1, a novel progestin-responsive gene with sequence homology to 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Mol Endocrinol. 1997;11:490–502. doi: 10.1210/mend.11.4.9909. [DOI] [PubMed] [Google Scholar]
- Imbert-Fernandez Y, Clem BF, O'Neal J, Kerr DA, Spaulding R, Lanceta L, Clem AL, Telang S, Chesney J. Estradiol stimulates glucose metabolism via 6-phosphofructo-2-kinase (PFKFB3) J Biol Chem. 2014;289:9440–9448. doi: 10.1074/jbc.M113.529990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommagani R, Szwarc MM, Kovanci E, Creighton CJ, O'Malley BW, Demayo FJ, Lydon JP. A murine uterine transcriptome, responsive to steroid receptor coactivator-2, reveals transcription factor 23 as essential for decidualization of human endometrial stromal cells. Biol Reprod. 2014;90:75. doi: 10.1095/biolreprod.114.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Arzayus M, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, Sellers W, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, Ittmann M, Tsai SY, Tsai MJ. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O'Malley BW, Xu J. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley BW. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci U S A. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer NN, Richer JK, Spoelstra NS, Torkko KC, Lyle PL, Singh M. Steroid receptor coactivator AIB1 in endometrial carcinoma, hyperplasia and normal endometrium: correlation with clinicopathologic parameters and biomarkers. Mod Pathol. 2006;19:1593–1605. doi: 10.1038/modpathol.3800696. [DOI] [PubMed] [Google Scholar]
- Uchikawa J, Shiozawa T, Shih HC, Miyamoto T, Feng YZ, Kashima H, Oka K, Konishi I. Expression of steroid receptor coactivators and corepressors in human endometrial hyperplasia and carcinoma with relevance to steroid receptors and Ki-67 expression. Cancer. 2003;98:2207–2213. doi: 10.1002/cncr.11760. [DOI] [PubMed] [Google Scholar]
- Kershah SM, Desouki MM, Koterba KL, Rowan BG. Expression of estrogen receptor coregulators in normal and malignant human endometrium. Gynecol Oncol. 2004;92:304–313. doi: 10.1016/j.ygyno.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Fujimoto J, Sun WS, Tamaya T. Clinical implications of steroid receptor coactivator (SRC)-3 in uterine endometrial cancers. J Steroid Biochem Mol Biol. 2007;104:237–240. doi: 10.1016/j.jsbmb.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Liao L, Chen X, Wang S, Parlow A, Xu J. Steroid receptor coactivator 3 maintains circulating insulin-like growth factor I (IGF-I) by controlling IGF-binding protein 3 expression. Mol Cell Biol. 2008;28:2460–2469. doi: 10.1128/MCB.01163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JW, Lee K, Han S, Aronow B, Lydon J, O'Malley B, DeMayo F. The p160 steroid receptor coactivator 2, SRC-2, regulates murine endometrial function and regulates progesterone-independent and -dependent gene expression. Endocrinology. 2007;148:4238–4250. doi: 10.1210/en.2007-0122. [DOI] [PubMed] [Google Scholar]
- Szwarc MM, Kommagani R, Jeong JW, Wu SP, Tsai SY, Tsai MJ, O'Malley BW, DeMayo FJ, Lydon JP. Perturbing the cellular levels of steroid receptor coactivator-2 impairs murine endometrial function. PLoS One. 2014;9:e98664. doi: 10.1371/journal.pone.0098664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMayo FJ, Spencer TCRISPR. Bacon: a sizzling technique. Biol Reprod. 2014;91:79. doi: 10.1095/biolreprod.114.123935. [DOI] [PubMed] [Google Scholar]
- Condon JC, Jeyasuria P, Faust JM, Wilson JW, Mendelson CR. A decline in the levels of progesterone receptor coactivators in the pregnant uterus at term may antagonize progesterone receptor function and contribute to the initiation of parturition. Proc Natl Acad Sci U S A. 2003;100:9518–9523. doi: 10.1073/pnas.1633616100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karteris E, Zervou S, Pang Y, Dong J, Hillhouse EW, Randeva HS, Thomas P. Progesterone signaling in human myometrium through two novel membrane G protein-coupled receptors: potential role in functional progesterone withdrawal at term. Mol Endocrinol. 2006;20:1519–1534. doi: 10.1210/me.2005-0243. [DOI] [PubMed] [Google Scholar]
- Winuthayanon W, Hewitt SC, Orvis GD, Behringer RR, Korach KS. Uterine epithelial estrogen receptor alpha is dispensable for proliferation but essential for complete biological and biochemical responses. Proc Natl Acad Sci U S A. 2010;107:19272–19277. doi: 10.1073/pnas.1013226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Rubel CA, Large MJ, Wetendorf M, Fernandez-Valdivia R, Jeong JW, Spencer TE, Behringer RR, Lydon JP, Demayo FJ. Epithelial progesterone receptor exhibits pleiotropic roles in uterine development and function. FASEB J. 2012;26:1218–1227. doi: 10.1096/fj.11-193334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jia Y, Franken P, Smits R, Ewing PC, Lydon JP, DeMayo FJ, Burger CW. Anton Grootegoed J, Fodde R, Blok LJ. Loss of APC function in mesenchymal cells surrounding the Mullerian duct leads to myometrial defects in adult mice. Mol Cell Endocrinol. 2011;341:48–54. doi: 10.1016/j.mce.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, Hammer RE, Herz J, Kuhn M. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc Natl Acad Sci U S A. 2002;99:7142–7147. doi: 10.1073/pnas.102650499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring B, Shynlova O, Tsui P, Eckardt D, Janssen-Bienhold U, Hofmann F, Feil S, Feil R, Lye SJ, Willecke K. Ablation of connexin43 in uterine smooth muscle cells of the mouse causes delayed parturition. J Cell Sci. 2006;119:1715–1722. doi: 10.1242/jcs.02892. [DOI] [PubMed] [Google Scholar]
- Quezada S, Avellaira C, Johnson MC, Gabler F, Fuentes A, Vega M. Evaluation of steroid receptors, coregulators, and molecules associated with uterine receptivity in secretory endometria from untreated women with polycystic ovary syndrome. Fertil Steril. 2006;85:1017–1026. doi: 10.1016/j.fertnstert.2005.09.053. [DOI] [PubMed] [Google Scholar]
- Villavicencio A, Bacallao K, Avellaira C, Gabler F, Fuentes A, Vega M. Androgen and estrogen receptors and co-regulators levels in endometria from patients with polycystic ovarian syndrome with and without endometrial hyperplasia. Gynecol Oncol. 2006;103:307–314. doi: 10.1016/j.ygyno.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Mark M, Yoshida-Komiya H, Gehin M, Liao L, Tsai MJ, O'Malley B, Chambon P, Xu J. Partially redundant functions of SRC-1 and TIF2 in postnatal survival and male reproduction. Proc Natl Acad Sci U S A. 2004;101:4453–4458. doi: 10.1073/pnas.0400234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chiao P, Zhang X, Zhang X, Lazar MA, Seto E, Young HA, Ye J. Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter. J Biol Chem. 2005;280:21091–21098. doi: 10.1074/jbc.M500754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Heaton JH, Gelehrter TD. Role of steroid receptor coactivators in glucocorticoid and transforming growth factor beta regulation of plasminogen activator inhibitor gene expression. Mol Endocrinol. 2006;20:1025–1034. doi: 10.1210/me.2005-0145. [DOI] [PubMed] [Google Scholar]
- Batsche E, Desroches J, Bilodeau S, Gauthier Y, Drouin J. Rb enhances p160/SRC coactivator-dependent activity of nuclear receptors and hormone responsiveness. J Biol Chem. 2005;280:19746–19756. doi: 10.1074/jbc.M413428200. [DOI] [PubMed] [Google Scholar]
- Baldwin A, Huh KW, Munger K. Human papillomavirus E7 oncoprotein dysregulates steroid receptor coactivator 1 localization and function. J Virol. 2006;80:6669–6677. doi: 10.1128/JVI.02497-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O'Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O'Malley BW. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by I kappa B kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Mawji NR, Bruchovsky N, Sadar MD. Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J Biol Chem. 2002;277:38087–38094. doi: 10.1074/jbc.M203313200. [DOI] [PubMed] [Google Scholar]
- Gianni M, Parrella E, Raska I, Jr., , Gaillard E, Nigro EA, Gaudon C, Garattini E, Rochette-Egly C. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 2006;25:739–751. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]