ABSTRACT
Cumulus cells and mural granulosa cells (MGCs) have functionally distinct roles in antral follicles, and comparison of their transcriptomes at a global and systems level can propel future studies on mechanisms underlying their functional diversity. These cells were isolated from small and large antral follicles before and after stimulation of immature mice with gonadotropins, respectively. Both cell types underwent dramatic transcriptomic changes, and differences between them increased with follicular growth. Although cumulus cells of both stages of follicular development are competent to undergo expansion in vitro, they were otherwise remarkably dissimilar with transcriptomic changes quantitatively equivalent to those of MGCs. Gene ontology analysis revealed that cumulus cells of small follicles were enriched in transcripts generally associated with catalytic components of metabolic processes, while those from large follicles were involved in regulation of metabolism, cell differentiation, and adhesion. Contrast of cumulus cells versus MGCs revealed that cumulus cells were enriched in transcripts associated with metabolism and cell proliferation while MGCs were enriched for transcripts involved in cell signaling and differentiation. In vitro and in vivo models were used to test the hypothesis that higher levels of transcripts in cumulus cells versus MGCs is the result of stimulation by oocyte-derived paracrine factors (ODPFs). Surprisingly ∼48% of transcripts higher in cumulus cells than MGCs were not stimulated by ODPFs. Those stimulated by ODPFs were mainly associated with cell division, mRNA processing, or the catalytic pathways of metabolism, while those not stimulated by ODPFs were associated with regulatory processes such as signaling, transcription, phosphorylation, or the regulation of metabolism.
Keywords: cumulus cells, mouse, mural granulosa cells, oocyte, oocyte-derived paracrine factors, ovarian follicle, transcriptome
INTRODUCTION
The cellular architecture of the ovarian follicle of most mammalian species becomes clearly diversified at the preantral to antral follicle transition. The granulosa cells of preantral follicles sort into two more differentiated populations at the time of follicular antrum formation: mural granulosa cells (MGCs), which line the follicular wall, and cumulus granulosa cells, which are associated with the oocyte. Generally, cumulus cells are thought to support oocyte development, while MGCs carry out endocrine function(s) indicative of functional as well as cellular architectural diversity. However, these are not strict divisions of labor because interactions between MGCs and cumulus cells can play crucial roles in oocyte maturation and ovulation [1–3]. The two populations have very different fates after the preovulatory surge of luteinizing hormone (LH). The MGCs remain within the ovary and participate in the formation of the corpus luteum. In contrast, the cumulus cells undergo a dramatic morphological change, producing and becoming embedded in a mucinous matrix in a process often referred to as cumulus expansion; they accompany the metaphase II oocyte to the oviduct during ovulation [4–6]. Before the LH surge, the cumulus cells communicate with the oocyte via gap junctions, which allow the exchange of low molecular weight molecules and facilitate both metabolic cooperation between cumulus cells and oocytes and regulation of meiosis [7–11]. The cumulus cells in the first layer around the oocyte are referred to as the corona radiata; and both the corona cells and the cumulus cells in outer ranks communicate with the oocyte via membrane extensions that reach around the corona cells to contact the oocyte, thus forming a pseudostratified communicating epithelium [12]. Oocyte-derived paracrine factors (ODPFs), sometimes in cooperation with other ligands, play crucial roles in the development and function of the cumulus cells [13, 14]. Oocytes are required for the formation of cumulus cells during the preantral to antral follicle transition [15]. This transition is generally correlated with the acquisition of oocyte competence to resume the first meiotic division [16]. These early cumulus cells are competent to undergo expansion in vitro [17] in a manner that appears similar to that of cumulus cells within large Graafian follicles in situ. These specific roles reveal considerable cellular and developmental complexity, but, in fact, very little is known about the global nature of the transcriptomes that support this cellular and functional diversification, which is the goal of this study.
The levels of some transcripts that are different in the cumulus cell and MGC populations in large Graafian follicles even before the LH surge and the induction of cumulus expansion, provide information on how the transcriptome reflects architectural diversity. For example, transcripts encoding enzymes participating in glycolysis and cholesterol production are expressed more highly by cumulus cells than by MGCs. Other transcripts enriched in cumulus cells versus MGCs of Graafian follicles include those encoding an amino acid transporter SLC38A3, the androgen receptor AR, the ligand AMH, and the natriuretic receptor 2 (NPR2) [3, 13, 18]. Expression of these transcripts is seen by in situ hybridization as a gradient with highest expression by cells closest to the oocyte and decreasing with increased distance outward from the oocyte [3, 13, 19]. Some periantral granulosa cells express these transcripts at levels higher than those seen in mural cells closer to the basal lamina. This suggests that periantral MGCs receive ODPF signals at levels sufficient to affect gene expression but that these levels are rapidly diluted with increasing distance from the oocyte. Microsurgical removal of oocytes (oocytectomy, OOX) from isolated cumulus-oocyte complexes (COCs) results in reduced expression of these transcripts, but these levels are maintained by ODPFs, particularly BMP15, GDF9, FGF8, or combinations of these [13, 19, 20]. This demonstrates that ODPFs promote the expression of genes characteristic of the cumulus cell phenotype. On the other hand, follicle-stimulating hormone (FSH) occurs in a diffusion gradient in the follicle opposite to that of ODPFs and promotes the expression of genes characteristic of the MGC phenotype, such as Lhcgr encoding the LH-receptor and Cyp11a1 encoding the P450 cholesterol side chain cleavage enzyme [13, 21]. Actions of FSH are augmented when MGCs contact components of the follicular basal lamina [22, 23]. ODPFs often abrogate the action of FSH and promote the cumulus cell phenotype instead. For example, ODPFs suppress the expression of Lhcgr mRNA by granulosa cells despite stimulation with FSH and culture on basal lamina [24]. Cells in intermediate zones between the gradients of FSH and ODPFs exhibit intermediate phenotypes depending upon their relative proximity to either the basal lamina or the oocyte.
Cumulus expansion in vivo occurs just before ovulation when follicles are stimulated by LH and produce EGF-like growth factors (EGFLGFs), which are first generated by MGCs in response to LH, and then by the cumulus cells via autocrine reinforcement [1, 2]. Cumulus expansion in response to stimulation of the EGF receptor requires the presence of ODPFs [25]. Moreover, expansion requires the expression of at least four factors (HAS2, PTGS2, PTX3, and TNFAIP6) because loss of expression of the genes encoding any of these factors dramatically compromises expansion [5, 26–29]. In addition to these expansion-related factors, the levels of many transcripts in cumulus cells change as a consequence of triggering cumulus expansion by gonadotropins in vivo [30–33]. However, the transcriptomes of cumulus cells and MGCs during the transition of small to large antral follicles (hereafter, SAFs and LAFs, respectively), before the initiation of cumulus expansion and ovulation, have not been described.
Clearly, more global and systems views of the transcriptional complexity underlying the architectural diversification can provide rationale and impetus to future studies of follicular cellular and functional development before the LH surge. Thus, the first objective of this study was to obtain a more global perspective than provided by analyses of single transcripts or pathways by utilizing microarrays to characterize the transcriptomic diversity of cumulus cells and MGCs. Analyses of these data are made by performing pairwise transcriptomic comparisons, with each comparison enhancing our view of the transcriptome of specific cell states and types. The value of this relatively unbiased, but global approach to the transcriptome was demonstrated by a previous study that capitalized on microarray data to discover a key for maintaining oocyte meiotic arrest. From this microarray approach, we found that natriuretic peptide NPPC is a ligand produced by MGCs and this ligand binds to its cognate receptor, NPR2, which is most highly expressed by cumulus cells. NPR2 is a guanylyl cyclase whose product, cGMP, is then transferred from the cumulus cells to oocytes via gap junctions to maintain oocyte meiotic arrest [3, 11]. Now in the current study, we have compared the transcriptomes of cumulus and MGCs in both early antral follicles and in large follicles after eCG-stimulation of immature mice in vivo. Although the cumulus cells isolated from both stages of follicular development are competent to undergo expansion in vitro [17], and are morphologically indistinguishable, we found that cumulus cells undergo transcriptomic changes that are as dramatic as those occurring in MGCs during the SAF to LAF transition.
The second objective of this study was to test the hypothesis that ODPFs are responsible for the higher expression of transcripts in cumulus cells than MGCs. Two approaches were used to test this: 1) comparison of the transcripts expressed at higher levels in freshly isolated cumulus cells with those found previously to be stimulated in cumulus cells by ODPFs in vitro [34] and 2) comparison of the higher cumulus cell transcripts with those found previously to be lower in cumulus cells of mutants deficient in the ODPFs GDF9 and BMP15 versus wild-type (WT) cumulus cells in vivo [35].
MATERIALS AND METHODS
COCs were isolated from the SAFs and LAFs of 22-day-old B6SJLF1 mice that were raised in the research colonies of the authors at The Jackson Laboratory. The Administrative Panel on Laboratory Animal Care at The Jackson Laboratory approved animal protocols, and all the experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. In order to obtain sufficient and uniform populations of cumulus cells and MGCs for microarray analyses, development of SAFs to LAFs was stimulated by intraperitoneal injection of 5 international units eCG 44 h before obtaining the COCs. Thus, SAFs were from unstimulated ovaries of 22-day-old mice, while LAFs were from 22-day-old mice stimulated 44 h previously with eCG. It is uncertain whether the stimulation with exogenous hormones produces the same effects on granulosa cell transcriptomes as would occur with natural stimulation by endogenous hormones.
Only COCs completely enclosed by tightly packed cumulus oophorus were selected for analysis. Two hundred oocyte-cumulus cell complexes were collected using micropipets with the aid of a stereo microscope and washed free of all other cells by serial transfer through three dishes of culture medium. This removes all possibility of contamination by individual MGCs. The cumulus cells were then stripped from the oocytes by drawing them into a fine glass pipet with a diameter slightly smaller than the diameter of the oocytes, which were discarded, and the cumulus cells prepared for RNA extraction. Clumps of MGCs, which are easily distinguishable from oocyte-cumulus cell complexes using a stereo microscope, with a total mass approximately equal to that of the cumulus cells, were collected with micropipets, washed, and prepared for RNA extraction. Given this care, the separation of cell types was absolutely clean. As verification of this, no expression of Lhcgr, a marker of MGCs [13], was found in the cumulus cell preparations, and no expression of Slc38a3, a marker of cumulus cells from LAFs [13], was found in the MGC preparations as shown in Supplemental Figure S1 (all the supplemental data is available online at www.biolreprod.org). The cumulus cells and MGCs were collected from three groups of five pooled mice. Thus, there were three biological replicates of each sample. The samples were amplified and applied to Affymetrics 430V2 arrays by the Jackson Laboratory Gene Expression Service as described previously [19, 34].
Average signal intensities for each probeset within the arrays were calculated by the RMA function provided within the Affymetrix package for R using a custom (Ensembl Transcript) CDF file [36]. The RMA method incorporates convolution background correction, sketch-quantile normalization, and summarization based on a multiarray model fit robustly using the median polish algorithm. For this experiment, three pairwise comparisons were used to statistically resolve transcriptomic differences between treatment levels using the R/maanova analysis package [37, 38]: 1) cumulus cells of SAFs versus LAFs, 2) cumulus cells versus MGCs of SAFs, and 3) cumulus cells versus MGCs of LAFs. Specifically, differentially expressed transcripts were detected using F, a modified F-statistic incorporating shrinkage estimate of variance components from within the R/maanova package [37, 38]. Statistical significance levels of the pairwise comparisons were calculated by permutation analysis (1000 permutations) and adjusted for multiple testing using the false discovery rate (FDR), Q-value, method [39]. Differentially expressed genes are declared at an FDR Q-value threshold of 0.05. Therefore, the FDR was limited by this selection alone to 5%. Furthermore, an additional stringency was imposed by requiring a 1.25-fold difference between contrasted groups for inclusion of transcripts in the cohort considered to be significantly enriched. The effective FDR was, therefore, less than 5%. Moreover, only those transcripts encoded by genes annotated in Mouse Genome Database (MGD) as having known biological functions (http://www.informatics.jax.org/function.shtml) are presented. Transcripts levels whose FDR Q-value was >0.05 and fold difference was <1.25 were considered to be not different. VLAD, a VisuaL Annotation Display tool (v. 1.5.1) (http://proto.informatics.jax.org/prototypes/vlad/) was used to identify biological function gene ontology (GO) terms defined by sets of enriched transcripts. GO terms, established by the GO Consortium (http://www.geneontology.org/), provide a standardized way to group genes or proteins according to their biological or molecular functions. Transcripts allocated to a specific GO term by electronic annotation alone were excluded. Therefore, assignment of a transcript to a GO term was based on curation of experimental data. Supplemental Figure S2 provides an illustration to aid in interpreting VLAD graphics.
For practical reasons, presentation of transcripts in contrasted groups was focused in two ways. First, only the transcripts associated with GO terms and annotated with known biological or molecular functions are presented; the entire lists of transcripts have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, dataset GSE55845). Additional transcriptomic analyses showing the effects of ODPFs on cumulus cell transcripts and on the effect of Gdf9+/− Bmp15−/− double mutation were deposited previously (GSE47967) [34] and (GSE7225) [19], respectively. Second, the top 50 transcripts exhibiting the greatest fold differences within the contrast are presented. Also for practical reasons, many references to general biological/molecular functions of the numerous specific gene products referred to here are not cited but can be found by searching the Mouse Genome Informatics database (http://www.informatics.jax.org/) or other resources.
To assess the ability of cumulus cells to elevate expression of cumulus expansion related transcripts Has2, Ptx3, Ptgs2, and Tnfaip6, in response to EGF, COCs were isolated from small and large follicles of ovaries of 22-day-old mice and cultured for 6 h with or without 10 ng/ml EGF. Transcript levels were assessed by quantitative RT-PCR (qRT-PCR). All procedures and reagents were exactly as described previously [15].
RESULTS
General Considerations of the Samples and Analyses of the Microarray Data
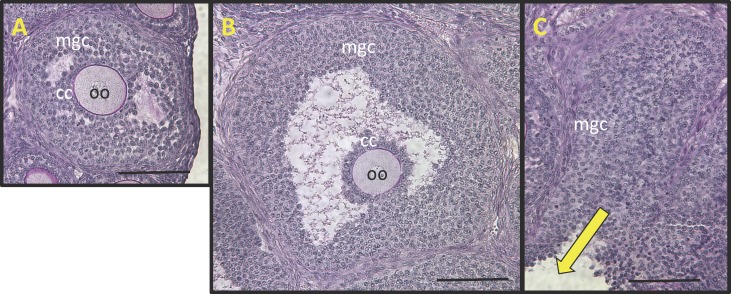
Cumulus and MGCs were obtained from either SAFs (200–350 μm diameter) of unstimulated ovaries or from LAFs (450–550 μm diameter) of eCG-stimulated ovaries (Fig. 1). The cumulus and MGCs of both groups appear morphologically indistinguishable by histological examination (Fig. 1, A and B). In addition to morphological resemblance, the COCs isolated from both groups were competent to undergo expansion in vitro ([17] and results presented here). During the collection process, rupturing follicles with needles expels COCs and clumps of MGCs. Examination of ovarian remnants after puncture of follicles showed that some MGCs were not extruded upon puncture, particularly those most closely associated with the basal lamina (Fig. 1C), suggesting that the MGCs used for analyses are probably a population biased toward the periantral MGCs.
FIG. 1.
Photomicrographs of histological sections of (A) a small antral follicle (SAF) typical of those in the ovaries of 22-day-old B6SJLF1 mice not stimulated with eCG and (B) a large antral follicle (LAF) typical of those in the ovaries 44 h postinjection with 5 international units eCG showing mural granulosa cells (MGC), cumulus cells (CC), and oocyte (oo). C) The follicular remnants after puncturing an LAF with a needle to express (yellow arrow) MGC and the oocyte cumulus cell complex. Note that many MGCs most immediately adjacent to the follicular basal lamina remain unexpressed and therefore not included in the microarray analyses. Bar = 100 μm.
Our previous studies [17] showed that COCs from SAFs underwent expansion in vitro, and, moreover, these experiments were conducted using FSH to stimulate expansion, that is, prior to demonstration of the prominent role of EGFLGFs in promoting cumulus expansion. Therefore, to provide more information on competence of the cumulus cells used for the transcriptomic studies described here, we tested the ability of these COCs to undergo expansion in response to EGF stimulation in vitro. As shown in Supplemental Figure S3, EGF dramatically stimulated the expression of the expansion-related transcripts Has2, Ptx3, Ptgs2, and Tnfaip6 in COCs from both the SAFs from unstimulated ovaries and the LAFs from eCG-stimulated follicles. Nevertheless, in spite of the fact that COCs from both groups appeared maximally expanded (not shown), levels of these transcripts in the cumulus cells of SAFs were 50%–75% of those from LAFs.
In our previous studies, we reported differences in transcript levels between cumulus cells and MGCs using in situ hybridization and/or qRT-PCR or RNase protection assays [3, 13, 19, 20, 40–42]. It was shown that the following transcripts are expressed more highly in cumulus cells than in MGCs: Slc38a3, Npr2, Mvk, Fdps, Pfkp, Ldha, Nog, Smad7, Tpi1, and Eno1. In contrast, the following transcripts are expressed more highly in MGCs than in cumulus cells: Grem1, Twsg1, Tob1, Nrip1, Cyp19a1, Cyp11a1, Lhcgr, Star, and Nppc. All of these transcripts fell within the cutoffs defining the groups that were higher in cumulus or MGCs in the microarray analyses presented here. As indicated in Materials and Methods, the cutoff to discriminate between higher or not higher in cumulus cells or MGCs in this study was set at fold difference > 1.25 and Q < 0.05. Thus, although arbitrary, the set discriminators for being different or not different produced outcomes that were highly correlated with results produced by the various other quantitative approaches. However, Amh, Ar, Sqle, and Cyp51 were excluded from the higher in cumulus cells versus MGCs group despite evidence using alternative, and probably more reliable, methods showing that they are expressed at higher levels in cumulus cells than MGCs. The microarray-based fold increase and Q values for Amh were 1.25-fold and Q = 0.09, for Ar they were 1.12-fold and Q = 0.08, and for Cyp51 they were 1.26-fold and Q = 0.1. Thus, these transcripts fell just outside the boundary that would have included them in the higher in cumulus cell cohort. The microarray data were validated for seven additional transcripts by qRT-PCR (Supplemental Fig. S1). Several issues could account for blurring of the boundary that would put transcripts inside or outside the inclusion groups. These include microarray probe factors, statistical confounding, biological factors (such as bias in the MGC populations that were analyzed toward the periantral MGCs, which express some transcripts typical of cumulus cells at higher levels than the MGCs located nearer to the basal lamina), or other artifacts that could be created by sample collection. Nevertheless, the results here, considered in their global perspective, are probably conservative, although any future studies based on the data presented here should include experimental verification of expression profiles for transcripts of interest.
Contrast: Cumulus Cells Isolated from SAFs Versus Cumulus Cells Isolated from LAFs
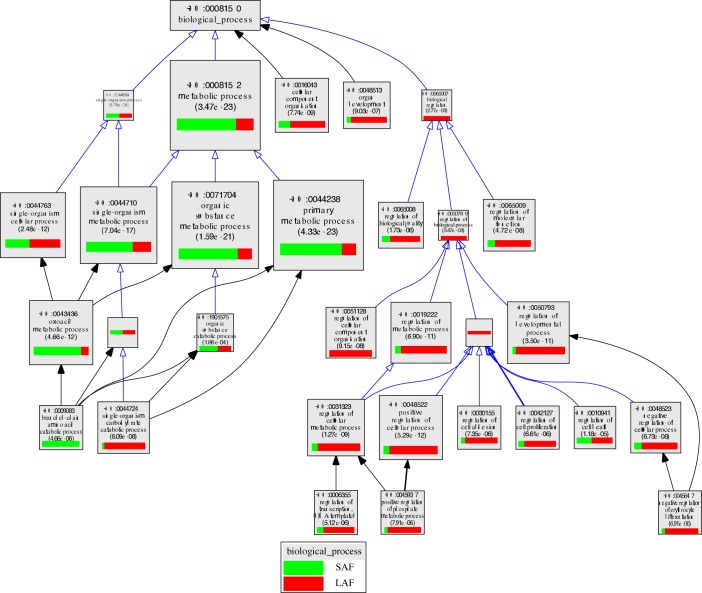
In the contrast between cumulus cells from SAFs versus LAFs, 535 transcripts were expressed at higher levels in cumulus cells from SAFs while 580 were expressed more highly in cumulus cells from LAFs (Fig. 2). Transcripts enriched in the cumulus cells from SAFs encode proteins that participate in metabolic processes, including those with catalytic activities (e.g., GO:0044281, small molecule metabolic process), such as those having transferase (GO:001670) or oxidoreductase activities (GO:0016491). In contrast, those enriched in cumulus cells from LAFs encode proteins involved with the regulation of metabolic and developmental processes, including those with protein-binding functions, such as those having protein- or small molecule-binding activities (e.g., GO:0048522, positive regulation of cellular processes). The relative enrichment of transcripts in the top GO terms are shown in Figure 3 and/or Supplemental Table S1, which presents the complete list of transcripts expressed differently in SAFs and LAFs for each GO term.
FIG. 2.
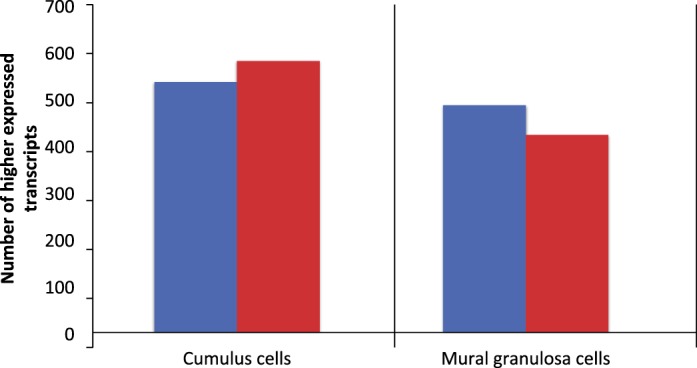
Number of transcripts expressed at higher levels in SAFs of unstimulated ovaries (blue bars) versus those of LAFs 44 h post-eCG-stimulation (red bars) in cumulus cells or MGCs. Only transcripts that encode proteins with annotated biological functions are included.
FIG. 3.
VLAD depiction of GO terms in the contrast of cumulus cells from SAFs versus LAFs. The top 25 GO terms, selected by VLAD on the basis of local maximum P value, are shown. A local maximum term is one whose significance (−log [p]) is greater than its immediate neighbors (children or parents). The size of the rectangle is proportional to the P value; the larger the rectangle the lower, and more significant, is the P value. The green portion of the rectangle shows the relative contribution of cumulus cell transcripts from SAFs to that GO term while the red portion shows the relative contribution of transcripts from LAFs. Supplemental Table S1 presents a complete list of transcripts expressed differently in SAFs and LAFs for each GO term.
Transcripts with greatest fold differences.
A list of the 50 top transcripts expressed more highly in cumulus cells from SAFs versus LAFs is shown on Table 1 and Supplemental Table S2. Serpinf1, encoding an antiangiogenic factor, heads the list followed by the Wt1 transcription factor and Nrip2, which encodes a nuclear corepressor of hormone action. The transcription factor Runx1, encoding a transcription factor, and Agtr2, encoding an angiotensin receptor, are also much more highly expressed by cumulus cells of SAFs.
TABLE 1.
Top 50 fold difference in cumulus cells of SAFs versus LAFs; gene names are presented in Supplemental Table S2.
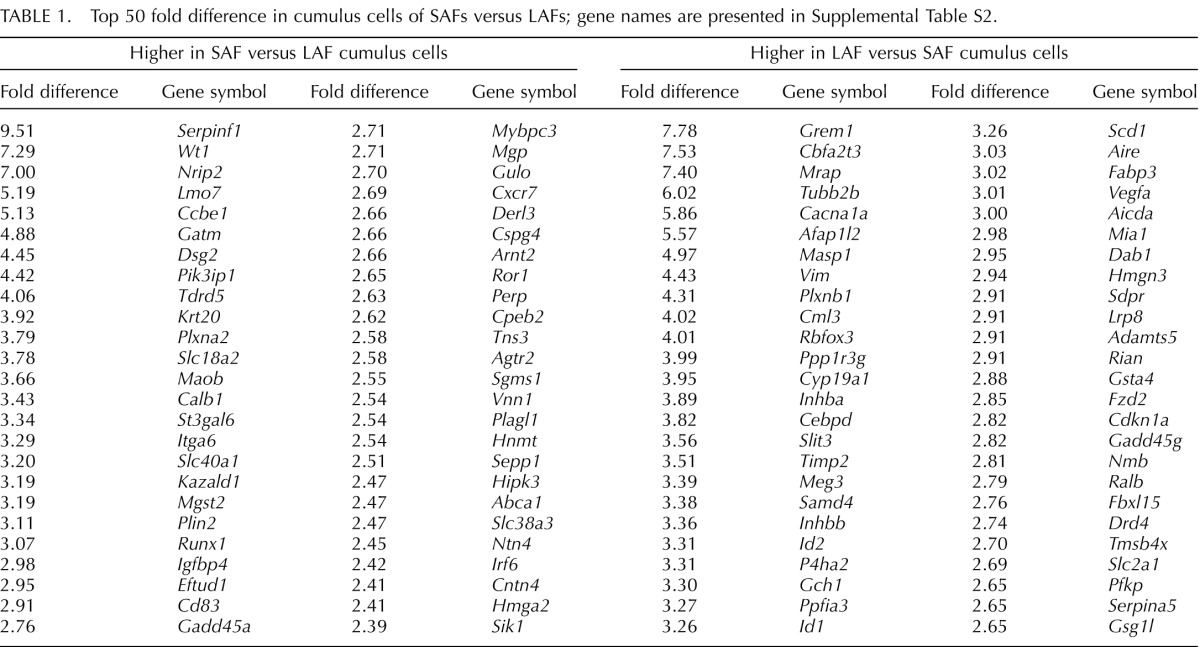
Grem1, Inhba, and Inhbb, encoding ligands in the TGFβ superfamily, were among the transcripts included on the list of the top 50 transcripts most highly expressed in cumulus cells of LAFs compared to SAFs (Table 1 and Supplemental Table S2). Included on this list are Id1 and Id2, which encode DNA-binding inhibitors involved in developmental processes promoted by various bone morphogenetic proteins (BMPs), Vegfa, Fzd2, and Cdkn1a (also known as p21), which encodes a cell cycle-related protein associated with cell differentiation. Oocytes probably regulate the expression of Id1 and Id2 in cumulus cells ([43, 44] and results presented here).
As shown in Figure 3 and/or Supplemental Table S1, transcripts in cumulus cells of SAFs relative to LAFs are enriched in transcripts generally associated with small molecule metabolic pathways (GO:0044281, GO:0019752, GO:0009083). In contrast, cumulus cells of LAFs are enriched in transcripts associated with enzymatic pathways involved in monosaccharide catabolic processes (GO:0046365), including glycolysis (GO:0006096), and cholesterol biosynthetic processes (GO:0006695) in accordance with our previous results [19, 20, 40]. Moreover, the cumulus cells of LAFs were enriched in transcripts associated with developmental processes (GO:0032502), regulation of molecular function (GO:0065009), positive and negative regulation of cellular processes (GO:0048522 and 0048523), and positive regulation of transcription (GO:0045893). Cumulus cells, therefore, become enriched in transcripts encoding components of metabolic pathways important for supporting cellular developmental processes and oocyte metabolism during the SAF to LAF transition.
Contrast: Cumulus Cells Versus MGCs
Cumulus cells expressed 1385 transcripts at levels higher than in MGCs of SAFs, and 2318 were higher in cumulus cells of LAFs (Fig. 4); 944 were commonly higher in cumulus cells versus mural cells in both SAFs and LAFs. Therefore, in SAFs and LAFs together, 2759 different transcripts were expressed at higher levels in cumulus cells than MGCs. MGCs expressed 1217 transcripts at levels higher than in cumulus cells of SAFs and 1814 transcripts were higher in MGCs of LAFs (Fig. 4); 878 transcripts were commonly higher in MGCs in both SAFs and LAFs.
FIG. 4.
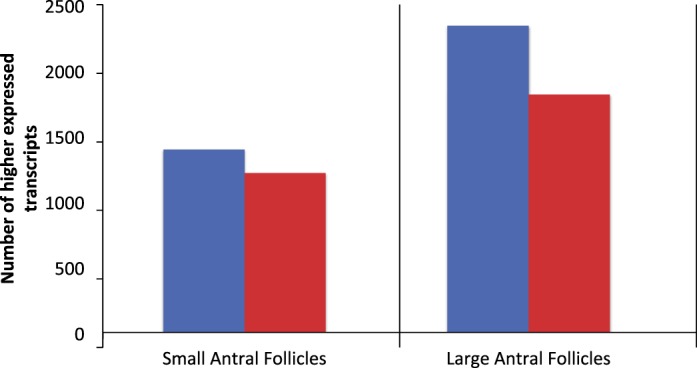
Number of transcripts expressed at levels higher in cumulus cells versus MGCs (blue bars) or higher in MGCs versus cumulus cells (red bars). Samples were taken from the SAFs of 22-day-old mice or the LAFs of 22-day-old mice 44 h poststimulation with eCG. Only transcripts that encode proteins with annotated biological functions are included.
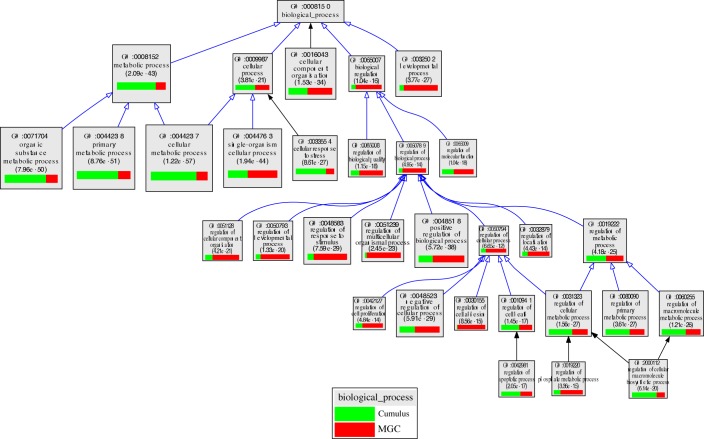
In SAFs, transcripts encoding proteins involved in cell proliferation (e.g., GO:0022402; cell cycle process), cellular response to stress (GO:0033554), and ribosome biogenesis (GO:0042254) were enriched in cumulus cells relative to MGCs. In contrast, transcripts encoding proteins participating in the regulation of cell communication (GO:0010646), regulation of signal transduction (GO:0009966), cell morphogenesis (GO:0000902), regulation of cell adhesion (GO:0030155), the regulation of cellular component movement (GO:0051270), and the negative regulation of cell proliferation (GO:0008285) were enriched in MGCs relative to the cumulus cells in SAFs (Fig. 5 and Supplemental Table S3).
FIG. 5.
VLAD depiction of GO terms in the contrast of cumulus cells versus MGCs from SAFs. The top 25 GO terms, selected by VLAD on the basis of local maximum P value, are shown. The size of the rectangle is proportional to the P value; the larger the rectangle the lower, and more significant, is the P value. The green portion of the rectangle shows the relative contribution of cumulus cell transcripts from SAFs to that GO term while the red portion shows the relative contribution of MGC transcripts from SAFs. Supplemental Table S3 presents a complete list of transcripts expressed differently in SAFs and LAFs for each GO term.
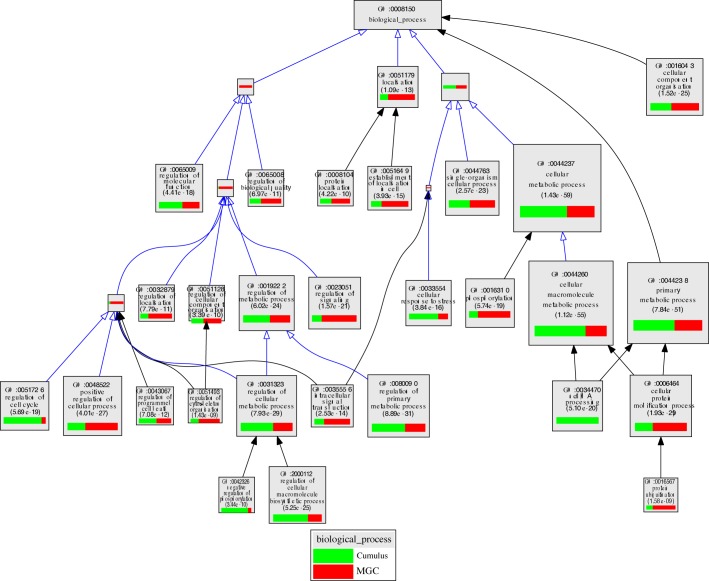
As shown in Figure 6 and/or Supplemental Table S4, cumulus cells of LAFs were enriched, relative to MGCs, in transcripts associated with cell cycle process (GO0022402), noncoding RNA processing (GO:0034470), primary metabolic processes (GO:0044238), which includes cholesterol biosynthetic process (GO:0006695), glycolysis (GO:0006096), and cellular response to stress (GO:0033554). In contrast, MGCs were enriched, relative to cumulus cells, in transcripts encoding proteins involved in the regulation of signaling (GO:0023051), phosphorylation (GO:0016310), protein catabolism (GO:0006511 and GO:0016567), steroid biosynthetic process (GO:0006694), and the regulation of cell communication (GO:0010646).
FIG. 6.
VLAD depiction of GO terms in the contrast of cumulus cells versus MGCs from LAFs. The top 25 GO terms, selected by VLAD on the basis of local maximum P value, are shown. The size of the rectangle is proportional to the P value; the larger the rectangle the lower, and more significant, is the P value. The green portion of the rectangle shows the relative contribution of cumulus cell transcripts from LAFs to that GO term while the red portion shows the relative contribution of transcripts from LAFs. Supplemental Table S4 presents a complete list of transcripts expressed differently in SAFs and LAFs for each GO term.
Transcripts Expressed at Higher Levels in Cumulus Cells Versus MGCs
Transcripts with greatest fold differences.
Table 2 and Supplemental Table S5 show the top 50 lists of transcripts expressed at higher levels in cumulus cells versus MGCs. Among the top 50 transcripts more highly expressed in cumulus cells versus MGCs, 23 are in common in SAF and LAF contrasts; those in the top 10 are particularly notable. They include two members of the FOS proto-oncogene family, Fos and Fosb, and two members of the JUN proto-oncogene family Jun and Junb. Members of these two families form heterodimers to produce the AP1 transcription factor complex, which has been associated with granulosa cell differentiation [45]. Also in these top 10 groups are Klf4, which has been implicated in stem cell-like capacity [46], and Cyr61, which encodes a protein that binds to various integrin receptors and to heparan sulfate proteoglycans involved in cell adhesion and signaling processes. Transcripts commonly expressed in the top 50 also include Rhob, a member of the Rho GTP-binding family, and Arhgdig, which affects the functioning of RHOB and, potentially, amplifies the actions of small G protein-signaling pathways. Also included in this group are Shb, which encodes an adapter protein that interacts with FGFR1 and is important for oocyte and follicular development [47]; Smad7, which encodes a member of TGFβ family and antagonizes the actions of the TGFβ type 1 receptor; Dusp1, which encodes a phosphatase that interacts with MAPK14, MAPK1, and MAPK8 (also known as p38, ERK2, and JNK1, respectively) that are well established to participate in a wide variety of signaling pathways in granulosa cells [48–56]; Ank1, which encodes a protein integral to the plasma membrane involved cell motility and the maintenance of specialized membrane domains; Ednrb, which encodes a G protein-coupled receptor of endothelin that is thought to have important roles in follicular development [57], including the induction of oocyte maturation [58]; Pfkp, whose levels are known to be regulated by ODPFs and encodes an enzyme important for providing products of glycolysis to oocytes [20, 40]; Slc26a3, which encodes a member of the sulfate anion transporter family and mediates chloride and bicarbonate ion exchange, and Cnga1, which encodes a cyclic nucleotide gated ion channel; and Heyl, which encodes a transcription factor that mediates the action of NOTCH family receptors [59] involved in cell fate decisions and thought to participate in the regulation of granulosa cell proliferation [60, 61].
TABLE 2.
Top 50 fold difference higher in cumulus cells versus MGCs; gene names are presented in Supplemental Table S5.
Transcripts common to both SAFa and LAFs (N = 23).
Vegfa is among the top 50 transcripts uniquely expressed more highly in cumulus cells versus MGCs of LAFs (Table 2 and Supplemental Table S5). This transcript encodes a member of the platelet-derived growth factor family of ligands thought to play a role in folliculogenesis [62, 63]. Suggestively, Pdgfrb is also included in the top 50 transcripts in this contrast, although no direct association of VEGFA and PDGFRB has yet been demonstrated. Bmp6 transcripts are also higher in cumulus cells versus MGCs of LAFs as are Trp53 transcripts encoding the tumor suppressor TRP53 (also known as p53).
Transcripts Expressed at Higher Levels in MGCs Versus Cumulus Cells
Transcripts with greatest fold differences.
Some transcripts were expressed at higher levels in MGCs than in cumulus cells in both SAFs and LAFs. Among these on the top 50 lists, 11 were common to both size follicles (Table 3 and Supplemental Table S6) and encode steroidogenic hormone enzymes CYP11A1 (also known as P450 side chain cleavage enzyme) and CYP19A1 (also known as aromatase); the ligand CTGF; transcription factors and coregulators CITED2, NRIP1, and PBX1; apoptosis regulators GSN, STRADB, and TIA1; cell adhesion PCDH8; and receptor/ion channel GABRB2. NRIP1, also known as RIP140, is a nuclear receptor cofactor expressed by MGCs that regulates expression of amphiregulin and is necessary for normal cumulus expansion and ovulation [64]; its expression by cumulus cells is suppressed by oocytes [65].
TABLE 3.
Top 50 fold difference higher in MGCs versus cumulus cells; gene names are presented in Supplemental Table S6.
Transcripts common to both SAFs and LAFs (N = 11).
Transcripts on the top 50 list of transcripts that are uniquely expressed higher in MGCs versus cumulus cells in SAFs (Table 3 and Supplemental Table S6) included those encoding receptors GHR and PLXNB1; ligands BMP2, INHBA, and NTN4; and cytoskeletal- and extracellular-related components ACTA2, COL3A1, FBN2, KRT20, MTAP2, SPNB2, and VIM.
The top 50 list of transcripts uniquely expressed higher in MGCs than cumulus cells in LAFs (Table 3 and Supplemental Table S6) include those encoding receptors LHCGR, PRLR, PTGFR, AHR, and EPHA5; ligands KITL and NPPC; cell adhesion/extracellular matrix-related molecules CD34, VCAM1, CCBE1, MATN2, LAMA4, and VCAN; and enzymes ADH1, CTSH, CYP17A1, ALDH1A1, and PRKAR2B. The ephrin receptor protein EPHA5 has been localized in MGCs by immunocytochemistry, regulated by FSH, and thought to participate in cell adhesion processes [66]. The transcript with the greatest differential between MGCs and cumulus cells in LAFs was Comp. It was previously suggested that Comp expression could be used as a marker of terminal follicular development because it was identified as the transcript most highly regulated by gonadotropin treatment in MGCs and during follicular development in vitro [67]. However, deletion of the Comp gene had no apparent effect on fertility, though quantitative data on fertility were not presented [68].
Role of ODPFs in the Differential Expression of Transcripts in Cumulus Cells and MGCs
These data reveal considerable differences in transcript levels in cumulus cells versus MGCs. Several factors could influence this differential expression: exposure to ODPFs, direct contact with oocytes and/or communication via gap junctions, interactions with basal lamina, different hormone or growth factor concentrations, or other factors influencing cellular microenvironments or physical conditions. Because of our prior findings on the importance of ODPFs, we used two approaches to test the role of ODPFs in promoting higher levels of transcripts in cumulus cells versus MGCs. The first experimental paradigm, an in vitro approach, compared the transcripts expressed at higher levels in cumulus cells freshly isolated from SAFs and LAFs with those elevated in cumulus cells by ODPFs in vitro as reported previously [34]. The second approach, an in vivo one, compared the higher cumulus cell transcripts with those expressed at lower levels in cumulus cells of Gdf9+/− Bmp15−/− double mutant (hereafter, DM) cumulus cells compared to WT cumulus cells in vivo as reported previously [19].
For the in vitro analysis of the role of ODPFs on differential transcript expression, oocytes were microsurgically removed from COCs isolated from eCG-stimulated ovaries and cultured for 20 h without or with coculture with two oocytes/μl of medium. After removal of the oocytes, the cumulus cells are referred to as OOX cumulus cells. We reported effects of ODPFs on the transcriptome of OOX cumulus cells previously [34], and the Jackson Laboratory Gene Expression Service carried out all the sample preparation and microarray protocols. The same boundaries for significance established for the present study, Q < 0.05 and fold change > 1.25, were applied to all data sets. Transcripts whose steady state levels were elevated by ODPFs in OOX cumulus cells were compared to the transcripts differentially expressed by freshly isolated cumulus cells and MGCs. Those transcripts expressed significantly more highly in freshly isolated cumulus cells versus MGCs and those whose expression was stimulated in OOX cumulus cells by oocytes in vitro are probably expressed at higher levels in cumulus cells because of exposure to ODPFs in situ.
All the transcripts with annotation of biological function expressed at higher levels in cumulus cells than MGCs, from both SAFs and LAFs, were compared with those increased by oocytes in OOX cumulus cells. In total, 1099 transcripts were higher in freshly isolated cumulus cells and were also stimulated by ODPFs in OOX-cumulus cells. Unexpectedly, 1660 transcripts that were higher in the freshly isolated cumulus cells compared with MGCs were not significantly stimulated by ODPFs in vitro.
To further test the surprising result that many cumulus transcripts expressed at higher levels than MGCs are not stimulated by ODPFs, we used an in vivo model. In a previous study [19], we assessed the transcriptomes of cumulus cells in Gdf9+/− Bmp15−/− DM ovaries, wherein the oocytes are deficient in the production of ODPFs GDF9 and BMP15 compared to WT cumulus cells. Cumulus expansion is defective in the COCs of these mutant mice, and the defective expansion was not remedied by coculture with WT oocytes, indicating that differentiation of the cumulus cells during follicular development requires these ODPFs [35]. Here we compared the transcripts with significantly lower expression by the DM cumulus cells versus WT cumulus cells with the cohort of transcripts found in the present study to be expressed at levels significantly higher in cumulus cells versus MGCs. Lower expression is the consequence of deficiencies in ODPFs GDF9 and BMP15 throughout follicular development. Thus, this experimental paradigm tests the effects of chronic ODPF deficiency on the cumulus cell transcriptome in vivo in contrast to the model testing the acute effects of ODPFs on normal OOX cumulus cells in vitro. As in the in vitro experiment, the cumulus cell transcripts expressed higher than in MGCs from both SAFs and LAFs were pooled for comparison to the transcripts reduced in the DM cumulus cells. A total of 843 transcripts expressed lower in DM cumulus cells were also expressed higher in normal cumulus cells versus MGCs. Moreover, 1916 were expressed at higher levels in the normal cumulus cells versus MGCs but were unchanged in the DM cumulus cells versus WT. Of the 843 transcripts that were lower in DM cumulus cells, 487 were the same as those stimulated by ODPFs in OOX cumulus cells in vitro. Thus, these 487 transcripts can be considered consensus transcripts whose expression in cumulus cells is stimulated by ODPFs (Supplemental Table S7). Importantly, of the 1916 transcripts that were lower in the DM relative to WT cumulus cells, 1301 were in common with the cohort of transcripts not stimulated by ODPFs in vitro (Supplemental Table S7). Thus, these can be considered consensus transcripts that are expressed more highly in normal cumulus cells, but the higher levels of expression is not due to stimulation by ODPFs.
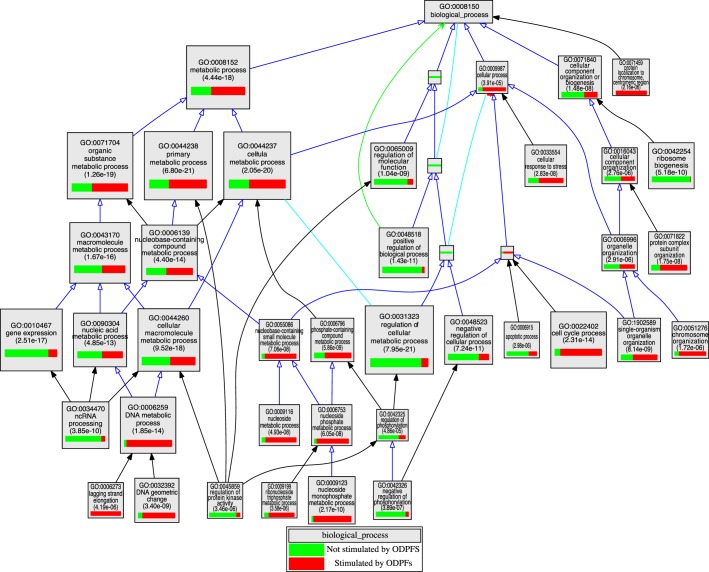
The GO terms associated with consensus ODPF-stimulated versus not stimulated are compared in Figure 7 and Supplemental Table S8. By definition, these transcripts are all more highly expressed in fresh cumulus cells than in MGCs. Analysis of the GO terms associated with the consensus transcripts for ODPF-stimulated or ODPF not stimulated processes was similar to that found for transcripts defined by either the in vitro or in vivo protocols, which are not shown. Transcripts that were higher in freshly isolated cumulus cells and consensus ODPF-stimulated cumulus cells included those enriched in biological function GO terms associated with catalytic processes in small metabolic processes, including glycolysis (GO:0006096, e.g., Pfkp) and cholesterol biosynthesis (GO:0008208, e.g., Mvk) as described previously [19, 20, 40]. Transcripts encoding molecules involved in other metabolic pathways such as GO:0009123 (nucleoside monophosphate metabolic process) were also promoted by consensus ODFPs. Previous studies have demonstrated that oocytes promote the proliferation of granulosa cells [69–71]. Transcripts associated with cell proliferation GO terms such as GO:0022402 (cell cycle process) and GO:0006259 (DNA metabolic process) are therefore not surprisingly found in the consensus ODPF-regulated group. The transcripts encoding receptors EDNRA, EDNRB, EGFR, and ESR1 as well as ligands IGF1, VEGFA, and VEGFB are also among those that are more highly expressed by both cumulus cells in situ and stimulated by ODPFs in OOX cumulus cells.
FIG. 7.
VLAD depiction of GO terms in the contrast of consensus transcripts higher in cumulus cells than MGCs stimulated by ODPFs or not. The top 25 GO terms, selected by VLAD on the basis of local maximum P value, are shown. The size of the rectangle is proportional to the P value; the larger the rectangle the lower, and more significant, is the P value. The green portion of the rectangle shows the relative contribution of cumulus cell transcripts not stimulated by ODPFs to that GO term while the red portion shows the relative contribution of transcripts stimulated by ODPFs. Supplemental Table S8 presents complete list of transcripts expressed differently in SAFs and LAFs for each GO term.
Notable among the consensus transcripts expressed at levels higher in cumulus cells than MGCs but not promoted by ODPFs were the proto-oncogenes Fosb, Jun, and Junb. In addition to these components of the AP1 transcription complex, Ap1b1 and Ap1g1, encoding AP1 complex adaptor proteins, are in the group of consensus transcripts expressed higher in cumulus cells than MGCs but not regulated by ODPFs. Transcripts also apparently not regulated by ODPFs were apoptosis-regulating factors Bad, the transcription factor involved in follicular development Foxo3, all of the early growth response genes (Egr1/2/3), the ligands Efna1, Gh, Ghrl, Shh, Tgfa, Pdgfa, and Wnt4, and transcripts encoding receptors Acvr2b, Adra2c, Fgfr1, Igf1r, Ntrk3, and Pdgfra. In general, GO terms enriched with transcripts not controlled by ODPFs include those that encode regulators of biological processes while those controlled by ODPFs encode proteins involved in cell division and catalytic metabolic pathways (Fig. 7 and Supplemental Table S8).
DISCUSSION
This study has delineated, at a global level, major transcriptomic dynamics that underlie the structural and functional architecture of the ovarian follicle. The analysis has revealed not only transcriptomic complexity, but also unanticipated regulatory differences that control cumulus cell diversification, development, and function. Scrutiny of gene lists and GO terms has revealed functions in cumulus cells and MGCs not previously fully appreciated. For example, although cumulus cells of both stages of follicular development are competent to undergo expansion in vitro, they were otherwise remarkably dissimilar with transcriptomic changes quantitatively equivalent to those of MGCs. GO analysis revealed that cumulus cells of small follicles were enriched in transcripts generally associated with catalytic components of metabolic processes, while those from large follicles were involved in regulation of metabolism, cell differentiation, and adhesion. Contrast of cumulus cells versus MGCs revealed that cumulus cells were enriched in transcripts associated with metabolism and cell proliferation while MGCs were enriched for transcripts involved in cell signaling and differentiation. Together, these findings validate that global systems approaches, such as that taken here, can provide a richness of information that complements other more focused (biased) approaches.
Oocytes are required for the progression of preantral (secondary) follicles to the early antral (tertiary) follicle stage [15]. This pivotal transition in folliculogenesis propels the structural and functional divergence of the cumulus cell and MGC lineages. Although it is well established that gonadotropins drive the development of SAFs to LAFs and promote the expression of genes key to steroidogenesis and the ability to respond to LH by MGCs, an analysis of the developmental changes in gene expression in the cumulus and MGC somatic compartments before the LH-surge (or hCG-treatment) has been lacking. Such information is essential to define the transcriptomic foundation of the cellular and functional architecture of the follicle and to determine how differential gene expression relates to the fates of both the somatic and oocyte compartments.
The cumulus cells of SAFs are competent to undergo expansion in vitro in a manner that appears similar to that of cumulus cells retrieved from LAFs isolated from follicles before the preovulatory LH surge ([17] and results presented here [Supplemental Fig. S3]). Despite this similarity, this study has shown that transcripts of cumulus cells from SAFs are actually quite different than those in cumulus cells from LAFs. In fact, the transcriptome changes in cumulus cells during the SAF to LAF development is qualitatively and quantitatively as significant as the transcriptomic transformation of the MGC population during the same follicular developmental span. Previous studies demonstrated that cumulus cells provide small nutritional and regulatory molecules to the oocyte via the gap junctions that couple the metabolism of these cells. In fact, oocytes, via ODPFs, promote pathways in cumulus cells, such as the glycolytic and cholesterol-generating pathways, in which the oocytes themselves are deficient [19, 40]. Moreover, previous studies have shown that ODPFs promote cumulus cell division [69–71]. A general emerging theme is that the cumulus cell transcriptome is enriched in the transcripts of workhorse molecules that drive both metabolism and cell division. As shown here, as the cumulus cells develop, they also become enriched in transcripts that regulate metabolic and developmental pathways. Curiously, however, most of the regulatory transcripts appear controlled by factors other than ODPFs, although the catalytic pathways themselves are often driven by ODPFs. Moreover, it is not yet clear how, or if, these transcriptomic changes in the cumulus cells relate to the qualitative changes in oocytes during the SAF to LAF developmental transition, when they increase their embryonic developmental competence [72].
Follicular microenvironments are influential in defining the functional diversity of the granulosa cell compartments. As shown both here and in other studies that focused on the expression patterns of individual transcripts or pathways [73], cumulus cells and MGCs differ in their patterns of gene expression and these patterns change during the SAF to LAF transition. What determines these lineage diversifications? ODPFs diffusing from the oocyte are a major influence in establishing the cumulus cell lineage [13]. It has been suggested that factors in apposition to the influence of ODPFs, such as FSH, diffuse from outside of the follicle to establish the reversed gradients in levels of several transcripts, for example, for Lhcgr versus Pfkp. Lhcgr is expressed at highest levels near the follicular-basal lamina [22, 74] while Pfkp is expressed at highest levels near the oocyte [20]. ODPFs suppressed expression of Lhcgr by granulosa cells that was stimulated by both FSH and contact with basal lamina [24], suggesting that ODPFs are the most influential factors in lineage determination. In fact, evidence suggests that ODPFs orchestrate the rate of follicular development [75], probably by promoting essential metabolic pathways and cell division.
We hypothesized that the higher levels of expression of most transcripts expressed higher in cumulus cells are promoted by ODPFs. There were 2759 total transcripts expressed more highly by cumulus cells than MGCs of SAFs and LAFs together. Using data from both the in vitro and in vivo models used to test this hypothesis, a total of 52% (1455) of the transcripts depend upon ODPFs; 39% (843) were acutely stimulated using the in vitro model. Thus, there is a remarkable reliance on ODPFs to promote in cumulus cells higher levels of transcripts that are involved in cell division or catalytic metabolic pathways. Nevertheless, 48% of the transcripts that were higher in cumulus cells were apparently not stimulated by ODPFs. This unanticipated result indicates that factors other than ODPFs are also major determinants of transcriptomic diversity of cumulus cells and MGCs.
Discovering the mechanisms for apparent ODPF-independent elevated expression of some transcripts in cumulus cells versus MGCs is a future challenge. It is possible that oocytes stimulate expression by the cumulus cells via mechanism requiring direct contact rather than ODPFs. However, no differences were detected in the transcriptomes of cumulus cells cultured either as intact oocyte-cumulus cell complexes or OOX cumulus cells cocultured with denuded oocytes. This suggests that oocytes do not affect the cumulus cell transcriptome via juxtacrine or gap junction-mediated communication, at least under the culture conditions used in that study [34]. In the in vitro experimental approach, the action of ODPFs was determined by coculturing OOX cumulus cells with denuded fully grown oocytes. Thus, the ODPFs contained the natural cocktail of factors released by oocytes in vitro and not arbitrarily selected recombinant ODPFs used individually or in combination. This protocol has been used in several previous studies to demonstrate the ability of ODPFs to promote or suppress several transcripts in cumulus cells [13, 19, 24, 34]. It is possible that expression of some cumulus cell transcripts require that ODPFs act in concert with paracrine factors produced within the follicle or endocrine factors, such as FSH or estrogens.
What, if not ODPFs, results in the higher levels of these transcripts in cumulus cells? Clearly, this should be an objective of future studies, but our working hypothesis is presented in Figure 8. Although intrinsic developmental programs cannot be excluded, the relative level of transcript expression in cumulus cells versus MGCs is probably the net result of stimulation by ODPFs in cumulus cells, suppression in MGCs, or both. FSH suppresses levels of Ar and Slc38a3 mRNAs in isolated COCs wherein ODPFs stimulated expression, thus imparting some MGCs-like phenotypic characteristics to cumulus cells [13]. Therefore, FSH could be a key factor in establishing or maintaining the diversity of granulosa cell lineages and is certainly needed, as well as ODPFs, for driving follicular development. Because FSH enters by diffusion from outside the follicle, and much of it is probably bound to FSH receptors abundant on MGCs [76], the concentration of FSH probably decreases with increasing depth into the follicle, minimizing the exposure of cumulus cells to FSH in vivo. Moreover, the follicular basal lamina, contacted by most MGCs [77], augments some actions of FSH [22, 24]. Thus, the combination of FSH and basal lamina could function to decrease the levels of expression of some transcripts in MGCs relative to cumulus cells, which would account for the transcriptomic diversity observed here. Other factors present in the granulosa cell microenvironments—estrogens and growth factors, for example—probably also participate in the architecture of transcriptomic diversity of cumulus cells and MGCs fundamental to cellular function and coordination during follicular development.
FIG. 8.
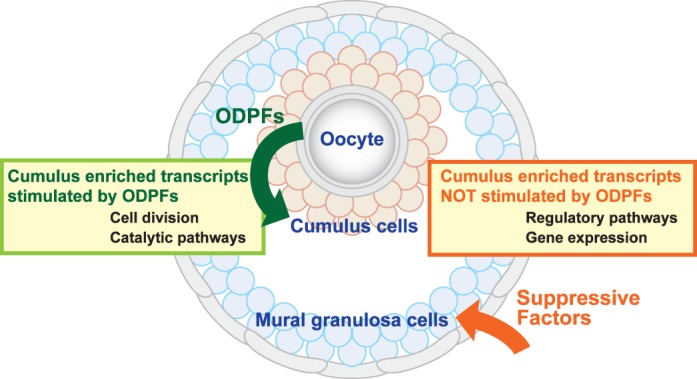
Working model depicting possible mechanisms regulating the expression of two groups of transcripts enriched in cumulus cells relative to MGCs. These groups of transcripts affect the diversity in functions of these cell types. Both groups are expressed at higher levels in cumulus cells than in MGCs. One group of transcripts is elevated in cumulus by oocyte-derived paracrine factors (ODPFs) and the other is not. The group whose expression is stimulated by ODPFs includes, among others, transcripts enriched in the GO terms associated with cell division and catalytic enzymes of metabolic pathways. The transcripts not affected by ODPFs include, among others, transcripts enriched in the GO terms associated with the regulation of these pathways and various aspects of gene expression (see Fig. 7 and Supplemental Table S8). The transcripts unaffected by ODPFs may be expressed at higher levels in cumulus cells than MGCs because expression is suppressed in the MGCs.
ACKNOWLEDGMENT
We are grateful to Tim Stearns of the Jackson Laboratory Computational Sciences Service for statistical analyses of microarray data and for spearheading the Gene Expression Omnibus submission of the data, to Chrystal Snow of The Jackson Laboratory Gene Expression Service for sample preparation and running the microarrays, and to Joel Richardson of the Mouse Genome Informatics Program for heroic assistance in the use of VLAD and for allowing us to have a small part in its evolution. We also thank Dr. Mary Ann Handel for her comments during the preparation of this manuscript
Footnotes
Supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development HD23839, HD 21970 (K.W., K-B.L., K.S., and J.J.E.) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (C.E. and K.S.).
REFERENCES
- Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS. Paracrine and autocrine regulation of epidermal growth factor-like factors in cumulus oocyte complexes and granulosa cells: key roles for prostaglandin synthase 2 and progesterone receptor. Mol Endocrinol. 2006;20:1352–1365. doi: 10.1210/me.2005-0504. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science. 2010;330:366–369. doi: 10.1126/science.1193573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel N, Hillensjo T, Kraicer PF. Maturational effects of gonadotropins on the cumulus-oocyte complex of the rat. Biol Reprod. 1979;20:191–197. doi: 10.1095/biolreprod20.2.191. [DOI] [PubMed] [Google Scholar]
- Chen L, Russell PT, Larsen WJ. Functional significance of cumulus expansion in the mouse: roles for the preovulatory synthesis of hyaluronic acid within the cumulus mass. Mol Reprod Dev. 1993;34:87–93. doi: 10.1002/mrd.1080340114. [DOI] [PubMed] [Google Scholar]
- Salustri A, Yanagishita M, Hascall VC. Synthesis and accumulation of hyaluronic acid and proteoglycans in the mouse cumulus cell-oocyte complex during follicle-stimulating hormone-induced mucification. J Biol Chem. 1989;264:13840–13847. [PubMed] [Google Scholar]
- Anderson E, Albertini DF. Gap junctions between the oocyte and companion follicle cells in the mammalian ovary. J Cell Biol. 1976;71:680–686. doi: 10.1083/jcb.71.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula NB, Epstein ML, Beers WH. Cell-to-cell communication and ovulation. A study of the cumulus-oocyte complex. J Cell Biol. 1978;78:58–75. doi: 10.1083/jcb.78.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM. Roles of cell-to-cell communication in development. Biol Reprod. 1985;32:27–42. doi: 10.1095/biolreprod32.1.27. [DOI] [PubMed] [Google Scholar]
- Albertini DF, Barrett SL. Oocyte-somatic cell communication. Reproduction. 2002:49–54. [PubMed] [Google Scholar]
- Norris RP, Ratzan WJ, Freudzon M, Mehlmann LM, Krall J, Movsesian MA, Wang H, Ke H, Nikolaev VO, Jaffe LA. Cyclic GMP from the surrounding somatic cells regulates cyclic AMP and meiosis in the mouse oocyte. Development. 2009;136:1869–1878. doi: 10.1242/dev.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E, Wilkinson RF, Lee G, Meller S. A correlative microscopical analysis of differentiating ovarian follicles of mammals. J Morph. 1978;156:339–366. doi: 10.1002/jmor.1051560303. [DOI] [PubMed] [Google Scholar]
- Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci. 2007;120:1330–1340. doi: 10.1242/jcs.000968. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su YQ, Sugiura K, Wigglesworth K, Xia G, Eppig JJ. Estradiol promotes and maintains cumulus cell expression of natriuretic peptide receptor 2 (NPR2) and meiotic arrest in mouse oocytes in vitro. Endocrinology. 2011;152:4377–4385. doi: 10.1210/en.2011-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes are required for the preantral granulosa cell to cumulus cell transition in mice. Dev Biol. 2007;305:300–311. doi: 10.1016/j.ydbio.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JJ, Wigglesworth K, Chesnel F. Secretion of cumulus expansion enabling factor by mouse oocytes: relationship to oocyte growth and competence to resume meiosis. Dev Biol. 1993;158:400–409. doi: 10.1006/dbio.1993.1198. [DOI] [PubMed] [Google Scholar]
- Vanderhyden BC, Caron PJ, Buccione R, Eppig JJ. Developmental pattern of the secretion of cumulus-expansion enabling factor by mouse oocytes and the role of oocytes in promoting granulosa cell differentiation. Dev Biol. 1990;140:307–317. doi: 10.1016/0012-1606(90)90081-s. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol Reprod. 2005;73:351–357. doi: 10.1095/biolreprod.105.041798. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Wigglesworth K, O'Brien MJ, Affourtit JP, Pangas SA, Matzuk MM, Eppig JJ. Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development. 2008;135:111–121. doi: 10.1242/dev.009068. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Pendola FL, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol. 2005;279:20–30. doi: 10.1016/j.ydbio.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Richards JS. Perspective: the ovarian follicle—a perspective in 2001. Endocrinology. 2001;142:2184–2193. doi: 10.1210/endo.142.6.8223. [DOI] [PubMed] [Google Scholar]
- Furman A, Rotmensch S, Kohen F, Mashiach S, Amsterdam A. Regulation of rat granulosa cell differentiation by extracellular matrix produced by bovine corneal endothelial cells. Endocrinology. 1986;118:1878–1885. doi: 10.1210/endo-118-5-1878. [DOI] [PubMed] [Google Scholar]
- Huet C, Pisselet C, Mandon-Pepin B, Monget P, Monniaux D. Extracellular matrix regulates ovine granulosa cell survival, proliferation and steroidogenesis: relationships between cell shape and function. J Endocrinol. 2001;169:347–360. doi: 10.1677/joe.0.1690347. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Wigglesworth K, Pendola F, Hirao Y. Murine oocytes suppress expression of luteinizing hormone receptor messenger ribonucleic acid by granulosa cells. Biol Reprod. 1997;56:976–984. doi: 10.1095/biolreprod56.4.976. [DOI] [PubMed] [Google Scholar]
- Peng J, Li Q, Wigglesworth K, Rangarajan A, Kattamuri C, Peterson RT, Eppig JJ, Thompson TB, Matzuk MM. Growth differentiation factor 9: bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc Natl Acad Sci U S A. 2013;110:E776–E785. doi: 10.1073/pnas.1218020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop C, Szanto S, Mukhopadhyay D, Bardos T, Kamath RV, Rugg MS, Day AJ, Salustri A, Hascall VC, Glant TT, Mikecz K. Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development. 2003;130:2253–2261. doi: 10.1242/dev.00422. [DOI] [PubMed] [Google Scholar]
- Ochsner SA, Day AJ, Rugg MS, Breyer RM, Gomer RH, Richards JS. Disrupted function of tumor necrosis factor-alpha-stimulated gene 6 blocks cumulus cell-oocyte complex expansion. Endocrinology. 2003;144:4376–4384. doi: 10.1210/en.2003-0487. [DOI] [PubMed] [Google Scholar]
- Ochsner SA, Russell DL, Day AJ, Breyer RM, Richards JS. Decreased expression of tumor necrosis factor-alpha-stimulated gene 6 in cumulus cells of the cyclooxygenase-2 and EP2 null mice. Endocrinology. 2003;144:1008–1019. doi: 10.1210/en.2002-220435. [DOI] [PubMed] [Google Scholar]
- Varani S, Elvin JA, Yan C, DeMayo J, DeMayo FJ, Horton HF, Byrne MC, Matzuk MM. Knockout of pentraxin 3, a downstream target of growth differentiation factor-9, causes female subfertility. Mol Endocrinol. 2002;16:1154–1167. doi: 10.1210/mend.16.6.0859. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Shimada M, Hernandez-Gonzalez I, Gonzalez-Robanya I, Richards JS. Induced expression of pattern recognition receptors in cumulus oocyte complexes: novel evidence for innate immune-like functions during ovulation. Mol Endocrinol. 2006;20:3228–3239. doi: 10.1210/me.2006-0194. [DOI] [PubMed] [Google Scholar]
- Xu F, Stouffer RL, Muller J, Hennebold JD, Wright JW, Bahar A, Leder G, Peters M, Thorne M, Sims M, Wintermantel T, Lindenthal B. Dynamics of the transcriptome in the primate ovulatory follicle. Mol Hum Reprod. 2011;17:152–165. doi: 10.1093/molehr/gaq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouandaogo ZG, Frydman N, Hesters L, Assou S, Haouzi D, Dechaud H, Frydman R, Hamamah S. Differences in transcriptomic profiles of human cumulus cells isolated from oocytes at GV, MI and MII stages after in vivo and in vitro oocyte maturation. Hum Reprod. 2012;27:2438–2447. doi: 10.1093/humrep/des172. [DOI] [PubMed] [Google Scholar]
- Emori C, Wigglesworth K, Fujii W, Naito K, Eppig JJ, Sugiura K. Cooperative effects of 17beta-estradiol and oocyte-derived paracrine factors on the transcriptome of mouse cumulus cells. Endocrinology. 2013;154:4859–4871. doi: 10.1210/en.2013-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data Nucleic Acids Res 2005. 33 e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kerr MK, Cui XQ, Churchill GA. MAANOVA; A Software Package for the Analysis of Spotted cDNA Microarray Experiments in the Analysis of Gene Expression Data. An Overview of Methods and Software: Methods and Software New York: Springer; 2003. [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirana R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O'Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development. 2007;134:2593–2603. doi: 10.1242/dev.006882. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Li Q, Wigglesworth K, Matzuk MM, Eppig JJ. Fibroblast growth factors and epidermal growth factor cooperate with oocyte-derived members of the TGFbeta superfamily to regulate Spry2 mRNA levels in mouse cumulus cells. Biol Reprod. 2009;81:833–841. doi: 10.1095/biolreprod.109.078485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Eppig JJ. Does bone morphogenetic protein 6 (BMP6) affect female fertility in the mouse? Biol Reprod. 2010;83:997–1004. doi: 10.1095/biolreprod.110.086777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Etherington SL, Young JM, McNeilly AS, Duncan WC. Inhibitor of differentiation (Id) genes are expressed in the steroidogenic cells of the ovine ovary and are differentially regulated by members of the transforming growth factor-beta family. Endocrinology. 2010;151:1247–1256. doi: 10.1210/en.2009-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbraak EJ. van ‘t Veld EM, Groot Koerkamp M, Roelen BA, van Haeften T, Stoorvogel W, Zijlstra C. Identification of genes targeted by FSH and oocytes in porcine granulosa cells. Theriogenology. 2011;75:362–376. doi: 10.1016/j.theriogenology.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Sharma SC, Richards JS. Regulation of AP1 (Jun/Fos) factor expression and activation in ovarian granulosa cells. Relation of JunD and Fra2 to terminal differentiation. J Biol Chem. 2000;275:33718–33728. doi: 10.1074/jbc.M003555200. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Calounova G, Livera G, Zhang XQ, Liu K, Gosden RG, Welsh M. The Src homology 2 domain-containing adapter protein B (SHB) regulates mouse oocyte maturation PLoS One 2010. 5 e11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels ET, Cottom J, Jones JC, Hunzicker-Dunn M. Follicle stimulating hormone (FSH) activates the p38 mitogen-activated protein kinase pathway, inducing small heat shock protein phosphorylation and cell rounding in immature rat ovarian granulosa cells. Endocrinology. 1998;139:3353–3356. doi: 10.1210/endo.139.7.6188. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wigglesworth K, Pendola FL, O'Brien MJ, Eppig JJ. Mitogen-activated protein kinase activity in cumulus cells is essential for gonadotropin-induced oocyte meiotic resumption and cumulus expansion in the mouse. Endocrinology. 2002;143:2221–2232. doi: 10.1210/endo.143.6.8845. [DOI] [PubMed] [Google Scholar]
- Su YQ, Denegre JM, Wigglesworth K, Pendola FL, O'Brien MJ, Eppig JJ. Oocyte-dependent activation of mitogen-activated protein kinase (ERK1/2) in cumulus cells is required for the maturation of the mouse oocyte-cumulus cell complex. Dev Biol. 2003;263:126–138. doi: 10.1016/s0012-1606(03)00437-8. [DOI] [PubMed] [Google Scholar]
- Sela-Abramovich S, Chorev E, Galiani D, Dekel N. Mitogen-activated protein kinase mediates luteinizing hormone-induced breakdown of communication and oocyte maturation in rat ovarian follicles. Endocrinology. 2005;146:1236–1244. doi: 10.1210/en.2004-1006. [DOI] [PubMed] [Google Scholar]
- Hunzicker-Dunn M, Maizels ET. FSH signaling pathways in immature granulosa cells that regulate target gene expression: branching out from protein kinase A. Cell Signal. 2006;18:1351–1359. doi: 10.1016/j.cellsig.2006.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz FJ, O'Brien MJ, Wigglesworth K, Eppig JJ. The preantral granulosa cell to cumulus cell transition in the mouse ovary: development of competence to undergo expansion. Dev Biol. 2006;299:91–104. doi: 10.1016/j.ydbio.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Otsuka F, Inagaki K, Otani H, Takeda M, Suzuki J, Goto J, Ogura T, Makino H. Differential regulation of steroidogenesis by bone morphogenetic proteins in granulosa cells: involvement of extracellularly regulated kinase signaling and oocyte actions in follicle-stimulating hormone-induced estrogen production. Endocrinology. 2007;148:337–345. doi: 10.1210/en.2006-0966. [DOI] [PubMed] [Google Scholar]
- Sriraman V, Modi SR, Bodenburg Y, Denner LA, Urban RJ. Identification of ERK and JNK as signaling mediators on protein kinase C activation in cultured granulosa cells. Mol Cell Endocrinol. 2008;294:52–60. doi: 10.1016/j.mce.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Liu Z, Fan HY, Wang Y, Richards JS. Targeted disruption of Mapk14 (p38MAPKalpha) in granulosa cells and cumulus cells causes cell-specific changes in gene expression profiles that rescue COC expansion and maintain fertility. Mol Endocrinol. 2010;24:1794–1804. doi: 10.1210/me.2010-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Kim H, Kang DW, Yanagisawa M, Ko C. Endothelin B receptor is not required but necessary for finite regulation of ovulation. Life Sci. 2012;91:613–617. doi: 10.1016/j.lfs.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Ye Y, Liang CG, Kawamura N, Gelpke MS, Rauch R, Tanaka T, Hsueh AJ. Paracrine regulation of the resumption of oocyte meiosis by endothelin-1. Dev Biol. 2009;327:62–70. doi: 10.1016/j.ydbio.2008.11.033. [DOI] [PubMed] [Google Scholar]
- Lavery DN, Villaronga MA, Walker MM, Patel A, Belandia B, Bevan CL. Repression of androgen receptor activity by HEYL, a third member of the Hairy/Enhancer-of-split-related family of Notch effectors. J Biol Chem. 2011;286:17796–17808. doi: 10.1074/jbc.M110.198655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J, Espinoza T, McGaughey RW, Rawls A, Wilson-Rawls J. Notch pathway genes are expressed in mammalian ovarian follicles. Mech Dev. 2001;109:355–361. doi: 10.1016/s0925-4773(01)00523-8. [DOI] [PubMed] [Google Scholar]
- Zhang CP, Yang JL, Zhang J, Li L, Huang L, Ji SY, Hu ZY, Gao F, Liu YX. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology. 2011;152:2437–2447. doi: 10.1210/en.2010-1182. [DOI] [PubMed] [Google Scholar]
- Artac RA, McFee RM, Smith RA, Baltes-Breitwisch MM, Clopton DT, Cupp AS. Neutralization of vascular endothelial growth factor antiangiogenic isoforms is more effective than treatment with proangiogenic isoforms in stimulating vascular development and follicle progression in the perinatal rat ovary. Biol Reprod. 2009;81:978–988. doi: 10.1095/biolreprod.109.078097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Seager M, Osman A, Castle-Miller J, Bevan H, Tortonese DJ, Murphy D, Harper SJ, Fraser HM, Donaldson LF, Bates DO. Ovarian VEGF(165)b expression regulates follicular development, corpus luteum function and fertility. Reproduction. 2012;143:501–511. doi: 10.1530/REP-11-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J, Steel JH, Rosell MM, Nikolopoulou E, Lee K, Demayo FJ, White R, Richards JS, Parker MG. The nuclear receptor cofactor receptor-interacting protein 140 is a positive regulator of amphiregulin expression and cumulus cell-oocyte complex expansion in the mouse ovary. Endocrinology. 2010;151:2923–2932. doi: 10.1210/en.2010-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Li Q, Wigglesworth K, Matzuk MM, Eppig JJ. Estrogen promotes the development of mouse cumulus cells in coordination with oocyte-derived GDF9 and BMP15. Mol Endocrinol. 2010;24:2303–2314. doi: 10.1210/me.2010-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buensuceso AV, Deroo BJ. The ephrin signaling pathway regulates morphology and adhesion of mouse granulosa cells in vitro Biol Reprod 2013. 88 25. [DOI] [PubMed] [Google Scholar]
- Skory RM, Bernabe BP, Galdones E, Broadbelt LJ, Shea LD, Woodruff TK. Microarray analysis identifies COMP as the most differentially regulated transcript throughout in vitro follicle growth. Mol Reprod Dev. 2013;80:132–144. doi: 10.1002/mrd.22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson L, Aszodi A, Heinegard D, Hunziker EB, Reinholt FP, Fassler R, Oldberg A. Cartilage oligomeric matrix protein-deficient mice have normal skeletal development. Mol Cell Biol. 2002;22:4366–4371. doi: 10.1128/MCB.22.12.4366-4371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhyden BC, Telfer EE, Eppig JJ. Mouse oocytes promote proliferation of granulosa cells from preantral and antral follicles in vitro. Biol Reprod. 1992;46:1196–1204. doi: 10.1095/biolreprod46.6.1196. [DOI] [PubMed] [Google Scholar]
- Gilchrist RB, Ritter LJ, Armstrong DG. Mouse oocyte mitogenic activity is developmentally coordinated throughout folliculogenesis and meiotic maturation. Dev Biol. 2001;240:289–298. doi: 10.1006/dbio.2001.0451. [DOI] [PubMed] [Google Scholar]
- Hickey TE, Marrocco DL, Amato F, Ritter LJ, Norman RJ, Gilchrist RB, Armstrong DT. Androgens augment the mitogenic effects of oocyte-secreted factors and growth differentiation factor 9 on porcine granulosa cells. Biol Reprod. 2005;73:825–832. doi: 10.1095/biolreprod.104.039362. [DOI] [PubMed] [Google Scholar]
- Pan H, O'Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Eppig JJ. Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Semin Reprod Med. 2009;27:32–42. doi: 10.1055/s-0028-1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Koch Y, Lieberman ME, Lindner HR. Distribution of binding sites for human chorionic gonadotropin in the preovulatory follicle of the rat. J Cell Biol. 1975;67:894–900. doi: 10.1083/jcb.67.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JJ, Wigglesworth K, Pendola FL. The mammalian oocyte orchestrates the rate of ovarian follicular development. Proc Natl Acad Sci U S A. 2002;99:2890–2894. doi: 10.1073/pnas.052658699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JS, Ireland JJ, Rao MC, Bernath GA, Midgley AR, Jr, Reichert LE., Jr. Ovarian follicular development in the rat: hormone receptor regulation by estradiol, follicle stimulating hormone and luteinizing hormone. Endocrinology. 1976;99:1562–1570. doi: 10.1210/endo-99-6-1562. [DOI] [PubMed] [Google Scholar]
- Lipner H, Cross NL. Morphology of the membrana granulosa of the ovarian follicle. Endocrinology. 1968;82:638–641. doi: 10.1210/endo-82-3-638. [DOI] [PubMed] [Google Scholar]