ABSTRACT
The antral compartment in the ovary consists of two populations of oocytes that differ by their ability to resume meiosis and to develop to the blastocyst stage. For reasons still not entirely clear, antral oocytes termed surrounded nucleolus (SN; 70% of the population of antral oocytes) develop to the blastocyst stage, whereas those called not-surrounded nucleolus (NSN) arrest at two cells. We profiled transcriptomic, proteomic, and morphological characteristics of antral oocytes and observed that NSN oocyte arrest is associated with lack of cytoplasmic lattices coincident with reduced expression of MATER and ribosomal proteins. Cytoplasmic lattices have been shown to store maternally derived mRNA and ribosomes in mammalian oocytes and embryos, and MATER has been shown to be required for cytoplasmic lattice formation. Thus, we isolated antral oocytes from a Matertm/tm mouse and we observed that 84% of oocytes are of the NSN type. Our results provide the first molecular evidence to account for inability of NSN-derived embryos to progress beyond the two-cell stage; these results may be relevant to naturally occurring preimplantation embryo demise in mammals.
Keywords: cytoplasmic lattices, Mater, oocyte maturation, oocyte to embryo transition, oogenesis, preimplantation embryo development
INTRODUCTION
Mammalian oogenesis encompasses complex mechanisms required for the maturation of primordial follicles into preovulatory follicles followed by the release of antral oocytes. Many factors are involved in oocyte maturation and among them the action of hormones (e.g., follicle-stimulating hormone and luteinizing hormone) and the exchange of substances between the oocyte and the surrounding somatic cells play critical roles. Moreover, the oocyte of the ovulating follicle secretes growth factors that are important for the expansion of cumulus cells, regulation of extracellular stability, and, last of all, ovulation.
The antral compartment in the ovary consists of two populations of oocytes that differ mainly by their ability to resume meiosis and to develop to the blastocyst stage following fertilization. Antral oocytes distinguishable as having a surrounded nucleolus (SN; 70% of the population of antral oocytes) are able to develop to the blastocyst stage, whereas those with a not-surrounded nucleolus (NSN) arrest at the two-cell stage [1]. Our recent studies showed more extensive localization of Oct4 in SN than in NSN oocytes, together with higher levels of expression of some oogenesis-specific genes, such as Gdf9, Bmp15, and Nobox, at the antral stage [2, 3]. Based on these data, we considered whether physiological features of SN oocytes may be related to the expression of key genes likely to function in critical developmental windows that predict outcomes for antral oocytes. Despite extensive studies of antral oocytes, the antral compartment, and the signals/molecules exchanged between somatic and germ cells, it is still not clear why antral SN and NSN oocytes are developmentally different and what factors, molecules, or mechanisms endow the SN type with competence for embryo development. Recently, however, it has been suggested that cytoplasmic lattices (CPLs) reflect storage for maternally derived mRNA and ribosomes in mammalian germinal vesicle stage (GV) oocytes and preimplantation embryos [4]. In the mouse, the Mater gene (maternal antigen that embryos require) has been shown to be required for CPL formation, and the development of oocytes lacking Mater is arrested at the two-cell stage [5].
To highlight this intriguing and still unclear situation, we determined the transcriptomic, proteomic, and morphological profiles of differentiated GV oocytes; furthermore, we examined the CPL network in both NSN and SN oocytes.
MATERIALS AND METHODS
Reagents
Female mice were purchased from Charles River; M2 medium was purchased from Millipore. Reagents for microarray analysis were purchased from Agilent Technologies. An RNeasy Mini Kit was purchased from Qiagen and all other reagents were purchased from Sigma unless otherwise noted.
Animals
Adult B6C3F1 4- to 5-wk-old female mice were maintained in a room with controlled temperature and humidity with phases of 12L:12D and fed ad libitum. All investigations were conducted in accordance with the guiding principles of European (no. 86/609/CEE) and Italian (nos. 116/92, 8/94) laws protecting animals used for scientific research.
For the collection of antral oocytes, females (wild-type and Matertm/tm) were injected intraperitoneally with 3.75 IU of equine chronic gonadotropin and 48 h later oocytes were isolated from the ovaries.
Isolation of Oocytes
Antral oocytes from both wild-type and Matertm/tm mouse (39 wk old; kind gift of Prof. Dean, NIDDK) were collected by puncturing the surface of the ovaries with a sterile needle and were then washed in M2 medium and stained with the fluorochrome Hoechst 33342 to distinguish chromatin organization in the NSN and SN types [2]. Different assays were performed in parallel using different batches of oocytes from wild-type mice. In particular, isolated oocytes in groups of 50 were then collected in 0.5 μl of M2 and stored in liquid nitrogen until further utilization for microarray analysis, and isolated groups of 300 SN and 300 NSN were processed for proteomics analysis. Two groups of 10 oocytes/type (10 SN and 10 NSN oocytes) were fixed for transmission electron microscopy.
Microarray Analysis
RNA labeling and hybridization on microarray was done independently for SN and NSN oocytes with two replications. Two sets of 50 antral oocytes were collected in M2 medium and then were labeled with Cy3-CTP. Fluorescently labeled microarray targets were prepared using a Low RNA Input Fluorescent Linear Amplification Kit (Agilent) with two-round amplification. Cy5-CTP-labeled reference target was produced from a mixture of Stratagene Universal Mouse Reference RNA and MC1 cell RNA. Targets were purified using an RNeasy Mini Kit and then quantified on a NanoDrop scanning spectrophotometer (NanoDrop Technologies). Target cRNA was hybridized to the National Institute on Aging (NIA) Mouse 44K Microarray v3.0 (whole-genome 60-mer oligo arrays, design ID 015087; Agilent) according to the manufacturer's protocol (Two-Color Microarray-Based Gene Expression Analysis Protocol, product #G4140-90050, version 5.0.1). Slides were scanned with an Agilent DNA Microarray Scanner (model G2505-64120) at 100% and 10% photomultiplier tubes in both channels, with a scan resolution of 5 μm. All hybridizations compared one Cy3-CTP-labeled experimental target to the single Cy5-CTP-labeled reference target that was used for normalization. All DNA microarray data are available at the public depository (Gene Expression Omnibus [GEO], National Center for Biotechnology Information [NCBI], http://www.ncbi.nlm.nih.gov/geo/, GEO accession number GSE34671) and at the NIA Array Analysis software (http://lgsun.grc.nia.nih.gov/ANOVA/).
Microarray data were uploaded in the NIA Array Analysis software and the background threshold was determined according to the plot of error function versus the expression level. For each gene on the array, mean log signal intensity was calculated on the basis of the signal intensities obtained from the two replicate hybridizations. Only genes with log mean intensity less than 2 were filtered out to remove noisy data near background levels. A hierarchical clustering was performed using the same NIA Array Analysis tool. The NIA Mouse Gene Index, mm8 [6], was used to find the genes overrepresented and underrepresented between the antral oocytes.
Proteomic Analysis
Three hundred SN and 300 NSN trypsin-digested oocytes were processed by means of two-dimensional micro-liquid chromatography coupled to an ion trap mass spectrometer (also referred to as multidimensional protein identification technology [MudPIT]) and analyzed as previously described [7]. The experimental mass spectra produced by MudPIT analyses were correlated to the in silico peptide sequences of the rat protein database retrieved from NCBI in 2010 (http://www.ncbi.nlm.nih.gov/); data processing, including that for posttranslational modifications, such as acetylation, deimination, and methylation, was performed by means of Bioworks 3.3.1, based on the SEQUEST algorithm [8]. The validity of peptide/spectrum matches was assessed using SEQUEST-defined parameter thresholds. Specifically, matching between spectra was only retained if they had a minimum Xcorr of 1.5 for +1, 2.0 for +2, and 2.5 for +3 charge state. In addition, to obtain very stringent results, the threshold of peptide probability was set at ≤10−3. Protein lists were compared and the differential expression evaluation was performed by Multidimensional Algorithm Protein Map (MAProMa) software as reported previously [9]. Protein interaction network was performed by means of Cytoscape software (Institute for System Biology), an open source software project for integrating biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework [10].
Preparation of Oocytes for Transmission Electron Microscopy
Oocytes in the collecting solution were centrifuged at 5000 rpm for 20 min and pellets were processed for transmission electron microscopy. Fixation was performed by immersion through gentle replacement of the supernatant with 2.5% glutaraldehyde (EM grade) and 4% paraformaldehyde, with 0.1% tannic acid and 0.01 M MgCl2, in 0.1 M sodium cacodylate buffer (pH 7.3) solution for 2 h at room temperature, followed by 4 h at 4°C. Oocytes were postfixed for 1 h in osmium tetroxide 1.33% in 0.1 M s-collidine buffer, stained en bloc with 2% uranyl acetate, and then dehydrated in a graded ethanol series. Finally, the specimens were embedded in epoxy resin Epon 812. Semithin (0.2 μm) and ultrathin (40–60 nm) sections were obtained using an ultramicrotome Reichert Ultracut S provided with a diamond knife. The semithin sections were stained with toluidine blue and ultrathin sections, after collection on 200-mesh grids, were counterstained with lead citrate. Observations and electron micrographs were made using a Zeiss EM 10 transmission electron microscope operating at 80 kV with an objective aperture of 30 or 60 μm; images were recorded on Kodak 4489 Electron Image film and finally digitized on an Epson Perfection V750 Pro scanner at 1600 dpi.
Morphometric Computerized Analysis
Lipid droplet content in the oocytes was evaluated in single oocytes on five sections from a series: two peripheral, two tangential to the nuclear envelope, and one equatorial. For each section relating to a single oocyte, the steps preparing the quantitative evaluation with NIH ImageJ software were 1) equalization of the oocyte image for a much better discrimination of image details and features in order to obtain the most accurate selection of the regions of interest (ROIs; lipid droplets); 2) thresholding the continuous tone image in order to obtain a binary image in which the ROI corresponded exactly to the lipid droplets; and 3) measurement of the ROIs in the five sections/oocyte, saving the results collected from each single oocyte in a file to treat data for statistical evaluations and comparisons between SN and NSN oocytes.
RESULTS
Microarray Analysis Shows Overexpression of 19 Genes in NSN Antral Oocytes
We observed that transcriptomic profiles are largely similar between SN and NSN oocytes. Among 44 000 probes (representing the majority of known genes), 43 522 probes were not expressed differently, 459 were more highly expressed in SN than in NSN oocytes, and 19 were more highly expressed in NSN than in SN oocytes (Fig. 1A and Supplemental Tables S1 and S2; all Supplemental Data are available online at www.biolreprod.org). Genes overexpressed in SN or NSN oocytes were then clustered into classes according to presumed biological functions (Fig. 1B). Expression of 27 ribosomal proteins was greater in SN oocytes relative to NSN oocytes (Fig. 1C), suggesting that long-term storage of ribosomal protein mRNA differs between antral SN and NSN oocytes. Consequently, reduced expression of these 27 ribosomal proteins in NSN oocytes suggests that these oocytes likely mature with a cytoplasm not fully equipped to sustain future embryo development, as the lack of storage of maternally derived ribosomes seems to suggest [11]. Because oocyte mRNA synthesis stops at the germinal vesicle breakdown stage and new mRNA production does not resume until the two-cell stage, ribosomal protein synthesis must be carried out on a maternal mRNA template [12]. The deficient ribosome usage and subsequent cessation of protein activity could be the prelude to the two-cell developmental arrest of the NSN-derived embryo.
FIG. 1.
Microarray analysis on 50 oocytes/type shows an overexpression of ribosomal proteins in SN oocytes. A) Scatter plot of 44K features on the NIA 60-mer oligo microarray comparing gene expression in antral oocytes (SN, green dots; NSN, red dots). Microarray data were analyzed by ANOVA-false discovery rate (FDR) statistics (averaged log intensity, FDR ≤ 0.05 and P ≤ 0.01). B) Bars representing transcripts detected in antral oocytes by ANOVA statistics and grouped according to the main Gene Ontology (GO) biological process categories they belong to. C) Heat map showing hierarchical clustering of significant ribosomal genes with one-fold expression change between SN and NSN oocytes. Expression levels are represented by the color; green indicates lower expression and red indicates higher expression.
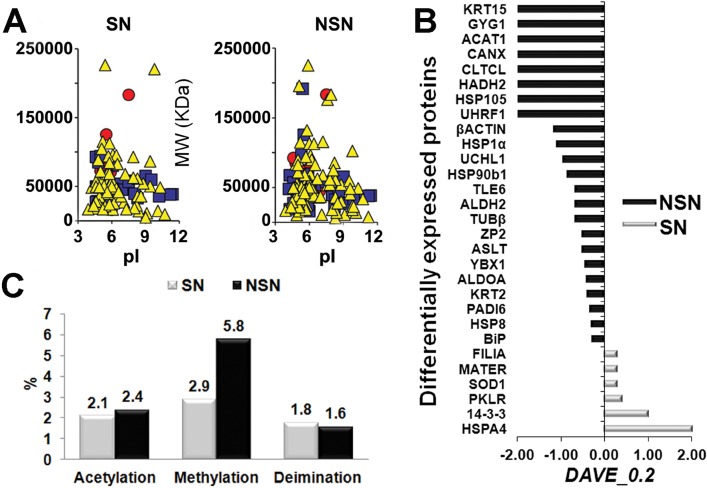
Proteomic Analysis Reveals Subtle Differences Between SN and NSN Antral Oocytes
We next investigated the proteomic profiles of 600 antral oocytes (300 SN and 300 NSN) by MudPIT (Fig. 2A). Because both SN and NSN oocytes reach the metaphase II (MII) stage and are competent for fertilization and the first cellular divisions, we analyzed the data using a very stringent threshold to identify the most likely critical proteins linked to subtle differences that distinguish SN and NSN oocytes. We observed six proteins that were up-regulated in SN relative to NSN and 23 that were up-regulated in NSN relative to SN oocytes.
FIG. 2.
Proteomic profile on 300 oocytes/type reveals the up-regulation of several proteins in the NSN oocytes together with a higher number of peptide modifications. A) Two-dimensional map of SN and NSN oocytes obtained from the related identified protein and plotted by MAProMa software. Specifically, proteins are plotted according to their theoretical isoelectric point (pI) and molecular weight (MW KDa), and the color/shape code for each identified protein is related to its identification confidence by SEQUEST score value (yellow/triangle <15, blue/square from >15 to <35, and red/circle >35). B) Differentially abundant proteins characterized through DAVE and DCI algorithms from MAProMa software (black/negative and grey/positive values correspond to up-regulated proteins in NSN and SN cells, respectively). C) Posttranslational modifications of peptides (acetylation, methylation, and deimination) in SN versus NSN oocytes.
Our proteomic results revealed up- and down-regulation of several proteins crucial for cellular processes (Figs. 2B and 3 and Supplemental Tables S3 and S4); in particular, when compared to SN oocytes, NSN oocytes showed a significant down-regulation of the proteins MATER and FILIA, and up-regulation of peptidylarginine deiminase type 6 (PADI6).
FIG. 3.
Map of the interactome networks correlating the main biological processes/functions. Identified proteins were plotted on the networks by means of Cytoscape application. Node colors correspond to specific proteins up-regulated in SN (red) or NSN (blue) oocytes: pink are proteins equally distributed in the oocytes; white are not-identified proteins. Blue, green, yellow, and red connecting lines represent protein-protein, genetic, and metabolic pathways and protein-DNA interactions, respectively. The dashed circle indicates the magnification of the proteins belonging to the subcortical maternal complex and to the proteins directly or indirectly connected to it.
We also analyzed the protein data to search for significant peptide modifications, and found that NSN oocytes had a higher number of methylated and acetylated peptides than SN oocytes (Fig. 2C and Supplemental Table S5). In the light of the following data, it is tempting to speculate that the greater modifications of the NSN proteome might be another cause for the failure of meiosis to resume: 1) keratins, titin, and thymosin-beta 10 are among the most highly methylated and acetylated proteins; keratin is one of the most well-known PAD substrates for deimination, and it is involved in the reorganization of cytoskeletal sheets in preparation for oocyte maturation [13]; 2) in muscle fibers, titin-based tension is calcium responsive, and those calcium currents are responsible for oocyte resumption of meiosis [14]; 3) thymosin-beta 10 is a G-actin binding protein whose function is still unknown, although the gene has been shown to influence cell proliferation towards apoptosis [15].
MATER Expression Is Significantly Lower in NSN Oocytes
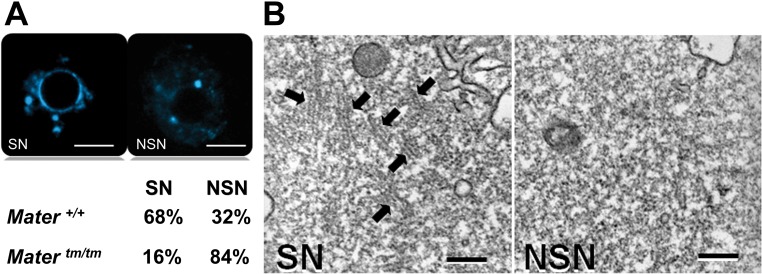
Because we showed down-regulation of MATER and underexpression of ribosomal proteins in NSN oocytes, we speculated that, unlike Mater+/+ GV oocytes, the Matertm/tm antral population should be composed mostly of NSN oocytes. To test this hypothesis, we analyzed Matertm/tm antral oocytes, showing that 84% of these GV oocytes were of the NSN type (Fig. 4A). Matertm/tm mice express residual amounts of MATER protein [16]; this can explain why the remaining 16% of Matertm/tm antral oocytes were of the SN type.
FIG. 4.
MATER and the CPLs are essential for meiotic acquisition. A) Comparison between the percentages of SN and NSN oocytes in Mater+/+ and Matertm/tm females (3 mo old). SN and NSN nucleoli are stained with Hoechst 33342. Bars = 10 μm. B) Transmission electron microscopy images of SN and NSN oocytes. Black arrows point to CPLs. Bars = 200 nm.
We also performed transmission electron microscopy analysis on SN and NSN oocytes, revealing that the NSN type contained none or very few CPLs as compared to SN oocytes (Fig. 4B). For this reason, the total absence (or great reduction) of CPLs can be considered a NSN-specific molecular morphology feature. In addition, considering that Matertm/tm eggs lack subcortical maternal complex [17] and that MATER is down-regulated in NSN oocytes in wild-type and mutated animals, the data, taken together, confirm that structural defects of the subcortical maternal complex (due to the down-regulation/absence of some of its components) are characteristic of NSN oocytes (Fig. 5A).
FIG. 5.
CPLs and lipid droplet content are good candidate markers for oocyte developmental competence. A) Model of GV SN and NSN oocytes showing the abundance or absence of CPLs (pink lines) and ribosomes (dark blue dots) freely dispersed in the cytoplasm. A magnification of a portion of the subcortical maternal complex (SCMC) is schematically represented (dotted box) as a model showing the interactions between the components (red, MATER; yellow, FILIA; green, PADI6; blue, TLE6; black, OOEP) and their overexpression (filled shape) or underexpression (empty shape) in the antral SN and NSN oocytes. In this model, the functional SN maternal complex (due to MATER and FILIA overexpression) is compared to a nonfunctional NSN maternal complex characterized by MATER and FILIA underexpression. For this reason, the SCMC disassembly of NSN oocytes is probably due to the inability of all four proteins to bind to MATER. B) Number of lipid droplets content in SN and NSN oocytes after morphometric computerized image analysis performed with ImageJ software. Data is displayed as means ± SEM. *P < 0.05 versus SN oocytes (Student t-test). C, D) Semithin sections of SN and NSN oocytes stained with toluidine blue. Black arrows point to lipid droplets. Bars = 10 μm. C', D') Binary images from C and D after ImageJ thresholding segmentation for the evaluation of oocyte lipid droplet content.
PADI6 is another constituent of the subcortical maternal complex and colocalizes with MATER throughout the cytoplasm in association with CPLs [13, 17, 18]. It has been suggested that the absence of PADI6 leads to a lack of CPL formation because of a citrullination deficiency [13]. However, our data suggest that a deficiency of citrullination does not affect the formation of CPL, but rather that the underexpression of MATER and the absence of subcortical maternal complex components may be the main cause. The abundance of PADI6 and citrullination in NSN is, in fact, able to affect protein-protein interactions, altering the three-dimensional folding and/or the activity of the substrate proteins [19].
Morphometric Analysis of Lipid Droplet Content: Another Marker to Differentiate SN and NSN Oocytes
We next sought to determine if additional markers of biological function might distinguish SN and NSN oocytes; for this purpose, we analyzed the lipid droplet content of the two types of antral oocytes, because it was recently shown that a decrease of CPLs in Mater null oocytes corresponds to an increase in the volume of these lipid droplets [5]. Except for their role in cellular energy storage and their association with apoptosis [20] and cancer [21], the contribution of lipid droplets during oocyte maturation is still poorly known. Some authors recently found a correlation between MATER and the antiapoptotic pathway [21, 22], leading to the conclusion that the accumulation of lipid droplets in Mater null oocytes could be a sign related to apoptotic activity. Through the morphometric analysis, we demonstrated that NSN oocytes contain significantly more lipid droplets than did SN oocytes (Fig. 5A–D').
DISCUSSION
In this paper, we explore whether mouse antral SN and NSN GV oocytes could be clearly distinguished via transcriptomics, proteomics, and morphological analyses. Although several studies have already performed genome-wide microarray analyses on preimplantation embryos [23, 24], oocytes as a function of aging [25], and general oogenesis [26, 27], no studies have reported analysis restricted to isolated antral SN and NSN oocytes. Besides, recent papers [28–30] have also reported proteomic analysis of undifferentiated GV, MII, and zygotes; however, nobody has yet described comparisons between SN and NSN oocytes.
For these reasons, our study provides, for the first time, the conceptual framework to understand naturally occurring embryo preimplantation losses through a link between molecular morphology and cell biology.
At first, we hypothesize that inability of NSN oocytes to complete embryonic development may be due to cytoplasmic defects and/or to reduced activity or expression of key maternal proteins. We observe that developmental failure of NSN antral oocytes is coincident with lack of CPLs and reduced expression of MATER protein and ribosomal proteins. In contrast, SN oocyte cytoplasm contains a significantly higher number of CPLs and associated ribosomes. MATER protein is the main constituent of the subcortical maternal complex [17] and is required for the formation of CPLs [5]. It is also involved in ribosome biogenesis and RNA transcription and translation [18]; thus, MATER is critical for acquisition of meiotic competence. In fact, Matertm/tm oocytes do not form (or form to a lesser degree) CPLs and are not able to progress beyond the two-cell stage [5]. To demonstrate definitively that NSN oocytes are CPL deficient and have lower levels of MATER, we compare antral oocytes from a Matertm/tm mouse to wild type and observe that the vast majority of oocytes are of the NSN type. This tells us that Matertm/tm embryo arrest depends only on the heritage the embryos have originally acquired being born as NSN oocytes.
The down-regulation of MATER in NSN oocytes correlated with the underexpression of ribosomal proteins and the absence of CPLs (all factors involved in ribosome biogenesis and translation machinery) together with the down-regulation of several other important maternal proteins (i.e., all the proteins belonging to the heat shock factor family) gives us interesting information regarding antral oocyte physiology. For this reason, we believe that it is extremely important to look at the chromatin configuration of oocytes of mutated animals [4, 18, 31]; besides the information on Matertm/tm oocytes we provide here, a paper has recently been published reporting, in agreement with our way of thinking, that Dppa3−/− oocytes are 66% of the NSN type [32]. A question arises: is the phenotype of oocytes derived from mutated maternal genes really due to gene “shutdown/manipulation” or is it due to the morphological characteristics (SN or NSN) of the oocyte itself?
MATER, together with FILIA, FLOPED, PADI6, and TLE6 proteins, forms the subcortical maternal complex located in the subcortical region of the oocytes and early embryos and is essential for preimplantation embryo development beyond the two-cell stage [17]. MATER, the major constituent of the complex, interacts directly with FLOPED and TLE6, whereas FILIA only binds to MATER [17]. Based on our proteomic data, we speculate that MATER down-regulation in NSN oocytes causes complex disassembly because of the inability of the other proteins to bind to MATER. However, further experiments are necessary to confirm this hypothesis.
We also analyze the lipid droplet contents of antral oocytes as further evidence of the role played by CPLs in the resumption of meiosis and of lipid droplets as being good candidate markers for oocyte developmental competence. In fact, NSN oocyte cytoplasm contains more lipid droplets than SN oocytes. Our recent results obtained with Fourier-transformed infrared technology support this hypothesis, showing that there are significant spectral differences in the lipid absorption region between SN and NSN oocytes, with the highest content of long-chain saturated fatty acids in the NSN type [33].
We believe that these data are important also for human reproduction, especially considering that few live births are derived from in vitro maturation and fertilization of human oocytes isolated from small and antral follicles, hence the importance of selecting good oocytes (i.e., SN type) able to complete preimplantation embryo development. For this reason, we think that future studies should focus on the ultimate biological meaning of NSN oocytes, trying to explain the reasons for their existence, which leads to a natural decrease (i.e., not counterselected) in embryonic developmental yield. Exploitation of these data could lead to a hypothetical 30% increase of preimplantation embryo yields, rendering artificial reproduction both in humans and in zootechnics more efficient for the benefit of human health and animal production. Furthermore, the data will contribute to the bioethical debate on embryo status and personhood, helping to ensure that decisions (philosophical and bioethical) about embryos are founded on updated scientific information. In brief, the data give some basic knowledge about the miniature, wonderfully organized laboratory of molecular biology that is the oocyte.
Supplementary Material
ACKNOWLEDGMENT
We are deeply grateful to J. Dean for providing Matertm/tm mice; R. Cockerham and C. Vasco for discussions; Y. Piao, Y. Qian, and A. Sharov for technical assistance with the microarray experiments; D. Di Silvestre, F. Basilico, and L. Benazzi for experimental assistance with the proteomic studies; M. Avella for experimental assistance with isolation of the Matertm/tm oocytes; and A. Farina for technical assistance with the preparation of the samples for transmission electron microscopy.
Footnotes
Supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Aging (grant 1 ZIA AG000662 to M.S.H.K.). M.M. and C.A.R. acknowledge the financial support of the Ministero della Salute, Ricerca Finalizzata/giovani ricercatori anno 2009 of the Fondazione IRCCS Ospedale San Matteo, Pavia (I). All DNA microarray data are available at the public depository (Gene Expression Omnibus [GEO], National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/geo/, GEO accession number GSE34671) and at the National Institute on Aging Array Analysis software (http://lgsun.grc.nia.nih.gov/ANOVA/).
REFERENCES
- Zuccotti M, Piccinelli A. Giorgi Rossi P, Garagna S, Redi CA. Chromatin organisation during mouse oocyte growth. Mol Reprod Dev. 1995;41:479–485. doi: 10.1002/mrd.1080410410. [DOI] [PubMed] [Google Scholar]
- Monti M, Garagna S, Redi CA, Zuccotti M. Gonadotropins affect Oct4 gene expression during mouse oocyte growth. Mol Reprod Dev. 2006;3:685–691. doi: 10.1002/mrd.20471. [DOI] [PubMed] [Google Scholar]
- Monti M, Redi CA. Oogenesis specific genes (Nobox, Oct4, Bmp15, Gdf9, Oogenesin1 and Oogenesin2) are differentially expressed during natural and gonadotropin induced mouse follicular development. Mol Reprod Dev. 2009;76:994–1003. doi: 10.1002/mrd.21059. [DOI] [PubMed] [Google Scholar]
- Yurttas P, Vitale PM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Conrood S. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kan R, Anguish A, Nelson LM, Conrood SA. Potential role for MATER in cytoplasmic lattice formation in murine oocytes. PLoS One. 2010;5:1–8. doi: 10.1371/journal.pone.0012587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MSH. A web-based tool for principal component and significant analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Comunian C, Rusconi F, Di Palma A, Brunetti P, Catalucci D, Mauri PL. A comparative MudPIT analysis identifies different expression profiles in heart compartments. Proteomics. 2011;11:2320–2328. doi: 10.1002/pmic.201000479. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Mauri P, Scarpa A, Nascimbeni AC, Benazzi L, Parmagnani E, Mafficini A, Della Peruta M, Bassi C, Miyazaki K, Sorio C. Identification of proteins released by pancreatic cancer cells by multidimensional protein identification technology: a strategy for identification of novel cancer markers. FASEB J. 2005;19:1125–1127. doi: 10.1096/fj.04-3000fje. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin D, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;11:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Nakajima R, Nagata M, Aoki F. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod. 2008;23:1377–1384. doi: 10.1093/humrep/den096. [DOI] [PubMed] [Google Scholar]
- Taylor KD, Piko L. Expression of ribosomal protein genes in the mouse oocytes and early embryos. Mol Reprod Dev. 1992;31:182–188. doi: 10.1002/mrd.1080310304. [DOI] [PubMed] [Google Scholar]
- Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P, Robben TJ, Conrood S, Gossen JA. Peptidylarginine deiminase (PAD) is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273:25–31. doi: 10.1016/j.mce.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Labeit D, Watanabe K, Witt C, Fujita H, Wu Y, Lahmers S, Funck T, Labeit S, Granzier H. Calcium-dependent molecular spring elements in the giant protein titin. Proc Natl Acad Sci U S A. 2003;23:13716–13721. doi: 10.1073/pnas.2235652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AK. Thymosin beta-10 accelerates apoptosis. Cell Mol Biol Res. 1995;41:167–180. [PubMed] [Google Scholar]
- Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development. 2008;135:259–269. doi: 10.1242/dev.011445. [DOI] [PubMed] [Google Scholar]
- Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, De Pol A, Vanevski K, Dorwand H, Sena P, Palumbo C, Bondy CA, Nelson LM. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology. 2004;145:1427–1434. doi: 10.1210/en.2003-1160. [DOI] [PubMed] [Google Scholar]
- Snow AJ, Puri P, Acker-Palmer A, Bouwmeester T, Vijiayaraghavan S, Kline D. Phosphorylation-dependent interaction of tyrosine 3-monoxygenase/tryptophan 5-monoxygenase activation protein (YWHA) with PADI6 following oocyte maturation in mice. Biol Reprod. 2008;79:337–347. doi: 10.1095/biolreprod.108.069328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Maraldi T, Riccio M, Sena P, Marzona L, Nicoli A, La Marca A, Marmiroli S, Bertacchini J, La Sala G, De Pol A. MATER protein as substrate of PKC epsilon in human cumulus cells. Mol Hum Reprod. 2009;15:499–506. doi: 10.1093/molehr/gap048. [DOI] [PubMed] [Google Scholar]
- Gorin MA, Pan Q. Protein kinase C epsilon: an oncogene and emerging tumor biomarker. Mol Cancer. 2009;8:1–8. doi: 10.1186/1476-4598-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation embryo development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global genes expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MSH. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Gen. 2004;13:2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- Kageyama S, Gunji W, Nakasato M, Murakami Y, Nagata M, Aoki F. Analysis of transcription factor expression during oogenesis and preimplantation development in mice. Zygote. 2007;15:117–128. doi: 10.1017/S096719940700411X. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Merico V, Sacchi L, Bellone M, Brink TC, Stefanelli M, Redi CA, Bellazzi R, Adjaye J, Garagna S. Oct-4 regulates the expression of Stella and Foxj2 at the Nanog locus: implications for the developmental competence of mouse oocytes. Hum Reprod. 2009;24:2225–2237. doi: 10.1093/humrep/dep191. [DOI] [PubMed] [Google Scholar]
- Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci U S A. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurttas P, Morency E, Conrood SA. Use of proteomics to identify highly abundant maternal factors that drive the egg-to-embryo transition. Reproduction. 2010;139:809–823. doi: 10.1530/REP-09-0538. [DOI] [PubMed] [Google Scholar]
- Pfeiffer MJ, Siatkowski M, Paudel Y, Balbach ST, Baeumer N, Crosetto N, Drexler HC, Fuellen G, Boiani M. Proteomic analysis of mouse oocytes reveals 28 candidates factors of the “reprogrammome.”. J Prot Res. 2011;10:2140–2153. doi: 10.1021/pr100706k. [DOI] [PubMed] [Google Scholar]
- Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Nakamura T, Nakano T. Essential role of Dppa3 for chromatin condensation in mouse oocytogenesis. Biol Reprod. 2011;40:1–8. doi: 10.1095/biolreprod.111.095018. [DOI] [PubMed] [Google Scholar]
- Ami D, Mereghetti P, Natalello A, Doglia SM, Zanoni M, Redi CA, Monti M. FT-IR spectra signatures of mouse antral oocytes: molecular markers of oocytes maturation and developmental competence. Biochim Biophys Acta. 2011;1813:1220–1222. doi: 10.1016/j.bbamcr.2011.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.