ABSTRACT
Progesterone receptor membrane component 1 (PGRMC1) mediates the antiapoptotic action of progesterone (P4). PGRMC1 interacts with plasminogen activator inhibitor 1 RNA-binding protein (PAIRBP1), but the functional significance of this interaction is unknown. To examine the function of PGRMC1-PAIRBP1 interaction, PAIRBP1 was depleted from spontaneously immortalized granulosa cells (SIGCs) and the effects on the expression and localization of PGRMC1 as well as P4's ability to bind to SIGCs and prevent apoptosis was assessed. Depleting PAIRBP1 enhanced cellular 3H-P4 binding and did not alter the expression or cellular localization of PGRMC1 but attenuated P4's antiapoptotic action. Transfection of a PGRMC1-green fluorescent protein (GFP) peptide mimic, which binds PAIRBP1 as demonstrated by in situ proximity assay, doubled the rate at which SIGCs undergo apoptosis compared to cells transfected with either the empty GFP expression vector or Pairbp1 small interfering RNA. Moreover, P4 did not prevent these cells from undergoing apoptosis. Similar studies conducted with granulosa cells isolated from immature rats also showed that PGRMC1 interacts with PAIRBP1 and that transfection of PGRMC1-GFP peptide mimic accelerates the rate of granulosa cell apoptosis by 4-fold even in the presence of serum and P4. These studies support the concept that the interaction between PAIRBP1-PGRMC1 is an essential component of the mechanism through which P4 inhibits apoptosis. Surprisingly, PGRMC1-PAIRBP1 interaction is not required for P4 binding or the cellular localization of PGRMC1 but rather appears to couple PGRMC1 to downstream components of the P4-PGRMC1 signal transduction pathway.
Keywords: apoptosis, granulosa cells, mechanisms of hormone action, ovary, progesterone/progesterone receptor
INTRODUCTION
Progesterone (P4), which is present in antral fluid throughout the course of antral follicle development [1], inhibits rat [2–6] and human [7–9] granulosa cell mitosis and apoptosis. Because of its ability to influence both granulosa cell mitosis and apoptosis, P4 appears to function as an intraovarian “governor” controlling the rate at which antral follicles grow and undergo atresia. In addition, P4 regulates the viability and steroidogenic potential of luteal cells [10]. As a result, P4 plays a key role in regulating the reproductive cycle and the female reproductive life span [11, 12].
Interestingly, granulosa cells of small antral follicles do not express the nuclear progesterone receptor (PGR) [13, 14], thereby eliminating PGR as a mediator of P4's actions. Granulosa cells of small antral follicles as well as spontaneously immortalized granulosa cells (SIGCs) express progesterone receptor membrane component 1 (PGRMC1), which localizes to the plasma membrane, cytoplasm, and nucleus [15–17]. Moreover, small interfering RNA (siRNA) depletion of PGRMC1 reduces the ability of SIGCs to bind 3H-P4 and subsequently P4's capacity to inhibit apoptosis [16, 18]. In addition, partially purified PGRMC1-green fluorescent protein (GFP) fusion protein specifically binds 3H-P4 through a single high-affinity P4 binding site (kd 10–40 nM) [16, 18]. These studies indicate that PGRMC1 mediates P4's antiapoptotic action.
Unfortunately, little is known about PGRMC1's mechanism of action. One aspect of PGRMC1's action involves binding to plasminogen activator inhibitor 1 RNA-binding protein (PAIRBP1), also known by several aliases, including SERPINE1 mRNA-binding protein (SERBP1) and CGI-55 [19, 20]. Granulosa cells of small antral follicles and SIGCs express both PAIRBP1 and PGRMC1. Unlike PGRMC1, PAIRBP1 is present only at the plasma membrane and in the cytoplasm of these cells [19, 20]. Finally, the PGRMC1 sequence between amino acids 70 and 130 is required for PGRMC1 to bind PAIRBP1 [18].
It is likely that PAIRBP1 is somehow involved in mediating P4's actions. This concept is further supported by the findings that forced expression of PAIRBP1 increases cellular responsiveness to P4 by 10-fold [19]. In addition, an antibody to PAIRBP1 attenuates P4's antiapoptotic actions in SIGCs [19], granulosa cells [20], and luteal cells [20].
There appear to be at least three possible mechanisms through which a PAIRBP1 interaction with PGRMC1 could facilitate P4's actions. First, PAIRBP1 could regulate the cellular localization of PGRMC1. Second, PAIRBP1 could interact with PGRMC1 to form an optimal “binding pocket” for P4, thereby enhancing 3H-P4 binding. Finally, PAIRBP1 could act to couple PGRMC1 to downstream components of the P4-PGRMC1 signaling pathway. Because these are not mutually exclusive mechanisms, each of these possibilities was tested in the experiments presented in this paper. These experiments used either siRNA to deplete PAIRBP1 or a PGRMC1 peptide mimic to disrupt endogenous PAIRBP1-PGRMC1 interaction.
MATERIALS AND METHODS
Generation of GFP-Fusion Proteins
Total mRNA was isolated from SIGCs and cDNA generated as previously described [19]. The entire Pgrmc1 coding region was then amplified using the following primer pair: forward: 3′-TTCTCGAGATGGCTGCCGAGGATGTG-5′ (with XhoI site) and reverse: 3′-AGAAGCTTGTCACTCTTCCGAGC-5′ (with HindIII site). Pgrmc1 was then cloned into pEGFP N1 vector (Clontech) at XhoI and HindIII restriction sites. The resulting construct, referred to as Pgrmc1-Gfp, was sequenced to ensure that it correctly encoded PGRMC1. To generate the PGRMC1 peptide mimic, Pgrmc1-Gfp was used as a template and the following primers were used to amplify the sequence that encodes amino acids 70–130: forward: 3′-TTCTCGAGATGCGTGACTTCACC-5′; reverse: 3′-AGAAGCTTGTCCAGGCAAAATGTGG-5′. The construct encoding this Pgrmc1-Gfp peptide mimic was verified by restriction enzyme digest. The amino acid sequence of the PGRMC1 peptide mimic is RDFTPAELRRYDGVQDPRILMAINGKVFDVTKGRKFYGPEGPY GVFAGRDASRGLATFCL (http://www.ncbi.nlm.nih.gov/protein/AAH62073).
SIGC and Granulosa Cell Culture
SIGCs were cultured in Dulbecco modified Eagle medium/F12 supplemented with 5% fetal bovine serum (HyClone), 100 U/ml penicillin G, and 100 μg/ml streptomycin as previously described [18]. Rat granulosa cells were isolated from immature 23- to 24-day-old rats using the protocol described by Campbell [21], which was approved by the Animal Care Committee of the University of Connecticut Health Center. Granulosa cells were plated either on cover glass in 35-mm culture dishes for immunocytochemical studies or in 24-well plates at 2 × 105 cells/well for experiments in which apoptosis was assessed. After 3 h the culture medium was replaced to remove any nonattached cells and the cultures continued for an additional 16 h.
Pgrmc1-Gfp Plasmid Transfection and Pairbp1 siRNA Treatment
For transfection of GFP-fusion proteins, 4 × 105 SIGCs were seeded in 35-mm dishes. Twenty-four hours later, 2 μg of Pgrmc1-Gfp plasmid DNA was transfected into SIGCs using Lipofectamine 2000 (Invitrogen) in Opti-MEM medium in 2.5% serum. A similar protocol was used to transfect granulosa cells that were plated at 2 × 105 cells/well in 24-well plates and using Lipofectamine 2000 as described with the exception that the cells were transfected in Opti-MEM supplemented with 2.5% steroid-free serum (HyClone, Thermo Scientific) in the presence or absence of 1 μM P4.
PAIRBP1 was depleted from SIGCs using Pairbp1 siRNA as follows. SIGCs (4 × 105 cells) were plated either on cover glasses that were placed in 35-mm culture dishes for experiments involving immunofluorescence localization studies or in 35-mm culture dishes for studies of apoptosis and mRNA. For 3H-P4 binding studies, SIGCs were plated in 96-well plates. Regardless of the culture plates used, the medium was removed after 24 h of culture and half of the culture dishes were transfected with Pairbp1 siRNA (Cat. No. AM16708 siRNA ID# 261504; Ambion). The remaining dishes were transfected with scramble siRNA (Cat. No. AM4611; Ambion). All the transfections were done in the presence of 2.5% serum using Lipofectamine 2000 according to the manufacturer's instructions. Pairbp1 and scramble siRNAs were used at a final concentration of 60 nM. After transfection the cells were incubated for 48 h in Opti-MEM medium with a final concentration of 2.5% fetal bovine serum and then assessed for either Pairbp1 and Pgrmc1 mRNA levels, 3H-P4 binding, or P4-regulated apoptosis.
Real-Time PCR for Pgrmc1 and Pairbp1
Real-time PCR for Pgrmc1 mRNA measurements was performed with the primers published by Peluso et al. [18] using the Script one-site RT-PCR kit with SYBR green (BioRad Laboratories). The relative level of Pgrmc1 mRNA was determined with Bio-Rad CFX96 software using the ΔCT method. Values were expressed as a percentage of the scramble control treatment.
Quantitative measurements of Pairbp1 mRNA were done using primers to rat Pairbp1 that were designed using Real Time PCR design software from Biosearch Technologies. Primers used were forward: 5′-GTGGCGCTTAA GAAAGAAGG-3′ and reverse: 5′-CGCCTTTCTGGTCTCCTATC-3′. The sequence for the Pairbp1 probe was 5′ FAM-TGATCAGGTCTTCTTCCAACTCGCC–BHQ-1 3′. Real-time PCR was performed on a CFX96 Real Time system with SsoFast Probe Supermix (BioRad Laboratories). The relative level of Pairbp1 mRNA was determined using the ΔCT method. Values were expressed as a percentage of the scramble control treatment.
Western Blots
SIGCs were lysed in RIPA buffer (50 mm Tris, 150 mm sodium chloride, 1.0 mm EDTA, 1% Nonidet P-40, and 0.25% sodium-deoxycholate; pH 7.0) that was supplemented with complete protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail 1 (Calbiochem) and then centrifuged at 14 000 relative centrifugal force at 4°C for 10 min. Protein was determined using the BCA protein assay (Bio-Rad).
The levels of PAIRBP1 and PGRMC1 were determined by Western blot analysis using a mouse monoclonal to PAIRBP1 (Cat. No. ab57285, dilution 1:1000; Abcam) or rabbit polyclonal antibody to PGRMC1 (Cat. No. HPA002877, dilution 1:750; Sigma), respectively. PGRMC1-GFP fusion proteins were detected using a rabbit polyclonal GFP antibody (Cat. No. 2555, dilution 1:1000; Cell Signaling Technology). For Western analysis, lysates were run on a 10% acrylamide gel, transferred to nitrocellulose, and then incubated with 5% nonfat dry milk overnight at 4°C. The nitrocellulose blots were incubated with the primary antibodies overnight at 4°C and processed for Western blot analysis using horseradish peroxidase-labeled secondary antibodies and ECL Western Blotting Analysis System (Amersham Biosciences).
Immunofluorescence and Co-localization Studies
Localization of PAIRBP1, PGRMC1, and PGRMC1-GFP fusion proteins was assessed by immunofluorescence. Cells were plated on cover glass, treated according to the experimental design, and then fixed in 4% paraformaldehyde. After fixation, samples were permeabilized with 0.1% Triton-X 100 in PBS for 7 min, blocked with 5% normal goat serum in PBS for 1 h at room temperature, and then incubated overnight at 4°C with the primary antibody or a combination of two primary antibodies when colocalization studies were conducted. Primary antibodies used in this study were mouse monoclonal anti-PAIRBP1 (Cat. No. ab57285, 1:200 dilution; Abcam), rabbit polyclonal anti-PGRMC1 (Cat. No. HPA002877, 1:250 dilution; Sigma), and rabbit monoclonal anti-GFP (Cat. No. G10362, 1:200 dilution; Life Technologies-Molecular Probes).
PAIRBP1, PGRMC1, and GFP primary antibodies were detected with Alexa Fluor 488-labeled anti-mouse, Alexa Fluor 546-labeled anti-rabbit, and Alexa Fluor 350-labeled anti-rabbit antibodies (1:800 dilution; Life Technologies-Molecular Probes), respectively. All primary and secondary antibodies were diluted in 0.1% BSA in PBS. After immunostaining, the slides were stained with 4,6-diamidino-2-phenylindole (DAPI) to visualize the DNA, mounted with ProLong antifade reagent (Life Technologies-Invitrogen), and observed using a Zeiss Axio Observer inverted microscope equipped with a Lumen 200 Fluorescence Illumination Systems (Prior Scientific). The images were captured with a QImaging Retiga EXi CCD digital camera (QImaging).
The presence of the intrinsic GFP fluorescence (green) was confirmed by the immunocytochemical detection of GFP using the anti-GFP antibody and Alexa Fluor 350-labeled anti-rabbit antibody (blue fluorescence). In this case DNA staining with DAPI was omitted.
Assessment of PAIRBP1-PGRMC1 Interactions
PAIRBP1 interaction with PGRMC1 was detected by an in situ proximity ligation assay (PLA). The PLA was performed according to the manufacturer's instructions (OLINK Bioscience), using either PAIRBP1 and PGRMC1 primary antibody pairs or PAIRBP1 and GFP primary antibody pairs, if PAIRP1-PGRMC1-GFP interaction was assessed. Briefly, the PLA utilized secondary antibody pairs that were labeled with complementary oligonucleotide DNA sequences. An interaction was detected when the two probes were in close proximity and could hybridize. The double-stranded DNA was then amplified and detected by a fluorescent probe, which was visualized as an individual fluorescent (red) spot (http://www.olink.com/). In the present studies, negative controls were performed by omitting one of the two primary antibodies.
To quantify the effect of P4 on the interaction between PAIRBP1 and endogenous PGRMC1, SIGCs were cultured in serum, serum-free medium, or serum-free medium supplemented with 1 μM P4 for 5 h. For each treatment group, images were captured in five different areas using identical exposure times and gain settings. The mean fluorescence intensity per cell was determined with the iVision-Mac Image Acquisition and Analysis Software (BioVision Technologies) using the DAPI staining to identify the cells. This entire experiment was repeated three times. The relative fluorescence units per cell associated with each treatment was calculated and used as an indicator of the degree of PAIRBP1-PGRMC1 interaction.
A similar approach was used to monitor the ability of PGRMC1-GFP peptide mimic to interact with PAIRBP1. The only modification was that a rabbit GFP antibody replaced the PGRMC1 antibody.
Assessment of Apoptosis
In order to assess the effect of Pairbp1 siRNA treatment on P4's ability to prevent apoptosis, SIGCs were transfected with Pairbp1 siRNA, cultured in Opti-MEM supplemented with 2.5% serum for 48 h, and then placed in serum-free medium in the presence or absence of 1 μM P4. After 5 h, the cells were stained with DAPI (0.3 μM) for 10 min at 37°C in the dark to reveal apoptotic nuclei. Phase and fluorescent images of the same field were captured and the percentage of apoptotic (DAPI-stained) cells determined [18]. Note that these cells were not fixed, which allowed DAPI to enter and stain the nuclei of only dying cells. That the dying cells were apoptotic was confirmed by the presence of condensed and fragmented nuclei. Moreover, identification of apoptotic cells by DAPI staining corresponds to TUNEL-stained cells [16].
To determine whether the PGRMC1-GFP peptide mimic was capable of inhibiting P4's anti-apoptotic action, SIGCs were plated on 35-mm dishes. After 24 h, the cells were transfected with 0.5 μg/dish of either an expression vector that encodes PGRMC1-GFP peptide mimic or an empty GFP vector. After an additional 24 h, the serum-supplemented medium was removed and the cells placed in serum-free medium supplemented with P4 (1 μM). After 5 h, the cells were stained with DAPI. Random areas within each cell culture were sequentially observed under a fluorescein isothiocyanate (FITC) filter set to identify GFP-transfected cells and a DAPI filter set to detect cells undergoing apoptosis. Images of each area were captured under identical optical conditions and stored in a computer. By comparing the images from the same area, the transfection status (FITC-green fluorescence) and viability (apoptosis; DAPI-blue fluorescence) of each cell could be determined. Approximately 100 transfected cells per culture dish were evaluated for apoptosis. The percentage of transfected apoptotic cells per treatment dish was calculated.
Similar studies were conducted with granulosa cells with the exception that transfected cells were assessed for apoptosis after 24 h of culture with medium supplemented with steroid-free serum with or without P4.
3H-P4 Binding to Intact SIGCs
SIGCs were plated in 96-well culture plates (4 × 104/well) and transfected with either scramble or Pairbp1 siRNA. After 48 h of culture, saturation-binding studies were conducted in which the effect of increasing concentrations of 3H-P4 on the amount of 3H-P4 specifically bound to each SIGC was determined. Briefly, SIGCs were incubated at 4°C in 25 μl TEMGD (Tris-EDTA-molybdate-glycerol-dithiothreitol) buffer with digitonin for 30 min and then increasing amounts of 3H-P4 in the presence or absence of 50 μM P4 at 4°C for 1 h. The cells were then scraped from the culture dish and filtered through Whatman glass microfiber filters. After five washes in cold PBS, the filters were counted in a scintillation counter. Specific 3H-P4 binding was determined by subtracting the amount (counts per minute) of 3H-P4 bound to SIGCs in the presence of 50 μM P4 from the amount of 3H-P4 bound to SIGCs in the absence of P4. Specific 3H-P4 binding was expressed as the number of binding sites/cell by converting counts per minute to number of binding sites using the equation provided by GraphPad (http://www.graphpad.com/quickcalcs/radcalcform.cfm). The binding experiments were repeated four times and values expressed as means ± one standard error.
Statistical Analysis
For the apoptosis studies, each treatment group in each experiment was conducted in either triplicate or quadruplicate and the entire experiment repeated three times. Individual values from each experiment were pooled and used to generate means ± standard errors for each treatment. Student t tests were used to determine if the means were different between two groups. When the means of three or more groups were compared, a one-way ANOVA followed by a Dunnett multiple comparisons test was used. The 3H-P4 binding characteristics were determined using the saturation binding equation for specific binding to one site as provided in the Prism 5.0 software package (GraphPad Software). An ANOVA was used to assess the effect of PAIRBP1 siRNA on the number of P4 binding sites. All statistical analysis was done using GraphPad Prism 5.0 software. Regardless of the statistical test, P values ≤ 0.05 were considered to be significant.
RESULTS
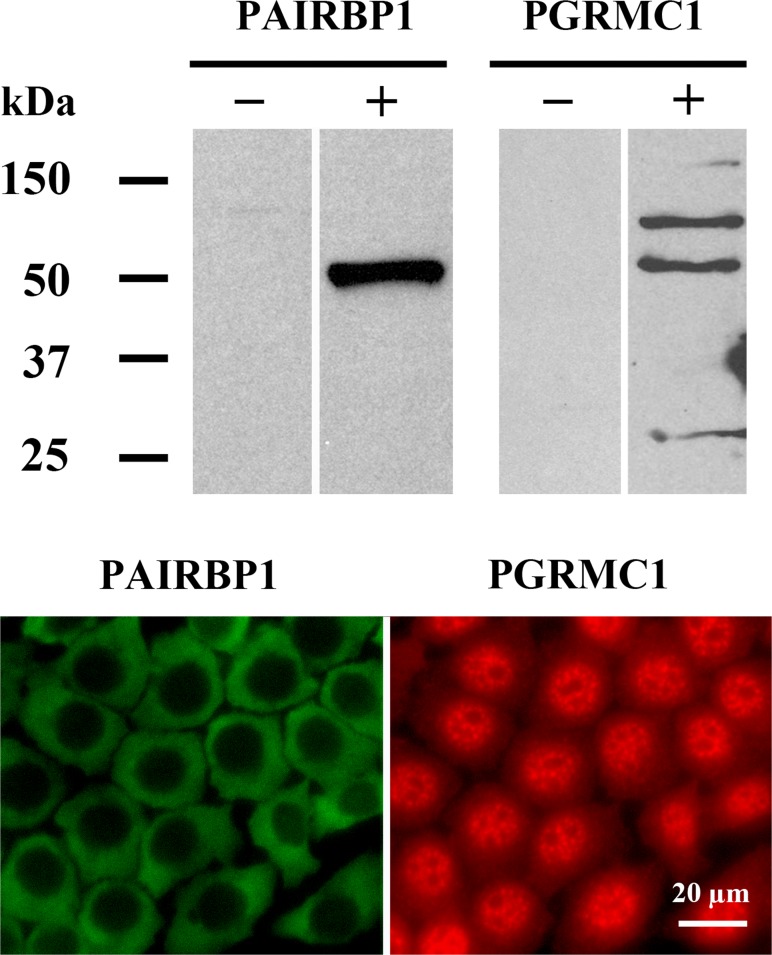
Previous Western blots conducted using an antibody produced by Aves Labs detected PAIRBP1 as a ≈55 kDa band in lysates prepared from rat granulosa cell and SIGCs [19]. Western blots using the commercially available antibody to PAIRBP1 from Abcam also detected PAIRBP1 as a ≈55-kDa band (Fig. 1). PGRMC1 was observed in SIGC lysates as multiple major bands at ≈27, ≈56, and ≈75 kDa (Fig. 1), which is consistent with published Western blots [17, 18]. Immunocytochemical analysis revealed that PAIRBP1 was present only in the cytoplasm, whereas PGRMC1 was present in both the cytoplasm and the nucleus (Fig. 1).
FIG. 1.
Expression of PAIRBP1 and PGRMC1 in SIGCs cultured in serum-supplemented medium as assessed by Western blot (upper panel) and immunocytochemistry (lower panel). PAIRBP1 was detected as a single band of ≈55 kDa and localized to the cytoplasm. In contrast, PGRMC1 was detected as multiple bands and distributed throughout the cell. Negative controls for the Western blots were conducted by omitting the primary antibodies and are indicated with a minus sign. Negative controls were also conducted for the immunocytochemistry and did not show any staining (not shown).
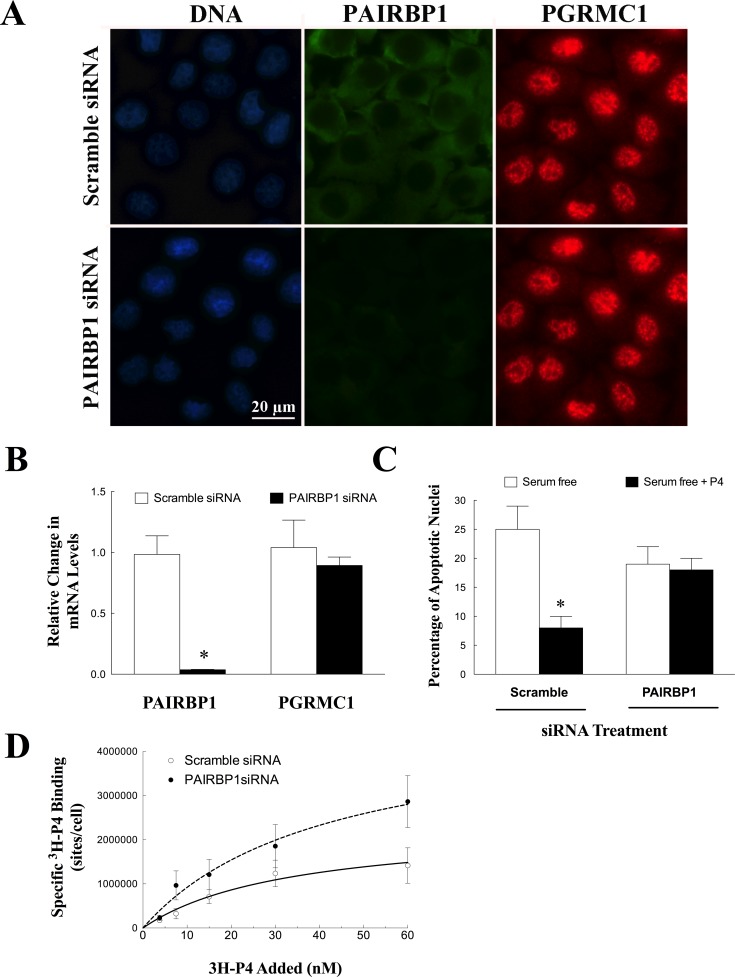
Treatment with Pairbp1 siRNA depleted PAIRBP1 protein (Fig. 2A) and mRNA (Fig. 2B) levels without altering the localization or amount of Pgrmc1 mRNA (Fig. 2, A and B). Moreover, depletion of PAIRBP1 attenuated P4's ability to prevent apoptosis (Fig. 2C). 3H-P4 specifically bound to SIGCs with a Kd of ≈26 nM with a Bmax of ≈2.2 × 106 binding sites/cell (Fig. 2D). After treatment with Pairbp1 siRNA, 3H-P4 bound to SIGCs with a similar Kd (≈25 nM), but the number of P4 binding sites doubled (Bmax ≈ 4.7 × 106 binding sites/cell; P = 0.001) (Fig. 2D).
FIG. 2.
The effect of Pairbp1 siRNA treatment on the expression and localization of PAIRBP1 and PGRMC1 in SIGCs. Compared to scramble (control) siRNA treatment, Pairbp1 siRNA selectively depleted PAIRBP1 protein as assessed by immunocytochemistry (A) and mRNA as measured by real-time PCR (B) without altering Pgrmc1 mRNA or localization in SIGCs maintained in serum-supplemented medium. In C the effect of Pairbp1 siRNA on the ability of progesterone (P4) to suppress SIGC apoptosis induced by serum withdrawal is shown. In B and C, values are expressed as a mean ± one standard error, with * indicating a value that is significantly different from scramble control (P < 0.05). The capacity of SIGCs treated with either scramble or Pairbp1 (D) siRNA and maintained in serum-supplemented medium to specifically bind 3H-P4 is shown. Values represent the mean ± one standard error for each dose of 3H-P4. Data was fitted using a single-binding-site model and represented by either a solid (scramble control) or dashed (PAIRBP1 siRNA) line.
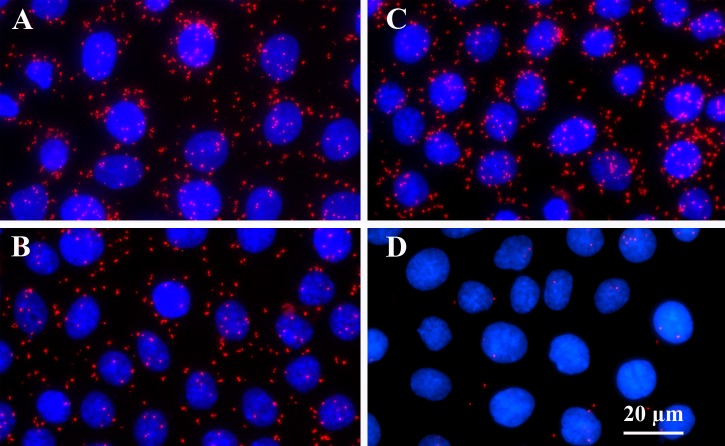
Although the siRNA studies demonstrate that PAIRBP1 plays an important role in P4's antiapoptotic action, they do not provide any insight into the mechanism of action. PAIRBP1's role in P4's action likely relates to its ability to interact with PGRMC1 [20]. Through the use of PLA, the present study demonstrated that endogenous PAIRBP1 interacts with endogenous PGRMC1 (Fig. 3). In serum-supplemented medium, the amount of PAIRBP1-PGRMC1 interaction as measured by the fluorescent signal generated by the PLA was 131 ± 6 relative fluorescent units (RFU)/cell (Fig. 3A). After 5 h in serum-free culture, the PAIRBP1-PGRMC1 interaction was reduced by ≈30% (Fig. 3B; 90 ± 5 RFU/cell, P < 0.05). However, P4 treatment maintained the PAIRBP1-PGRMC1 interaction at 131 ± 8 RFU/cell (Fig. 3C). In the absence of either PAIRBP1 or PGRMC1 antibody, the PLA signal was negligible (Fig. 3D).
FIG. 3.
PAIRBP1-PGRMC1 interaction in SIGCs maintained in serum (A) or cultured for 5 h in serum-free medium (B) or serum-free medium plus P4 (C). Negative control is shown in D. PAIRBP1-PGRMC1 interaction was determined by in situ PLA and revealed by a red dot.
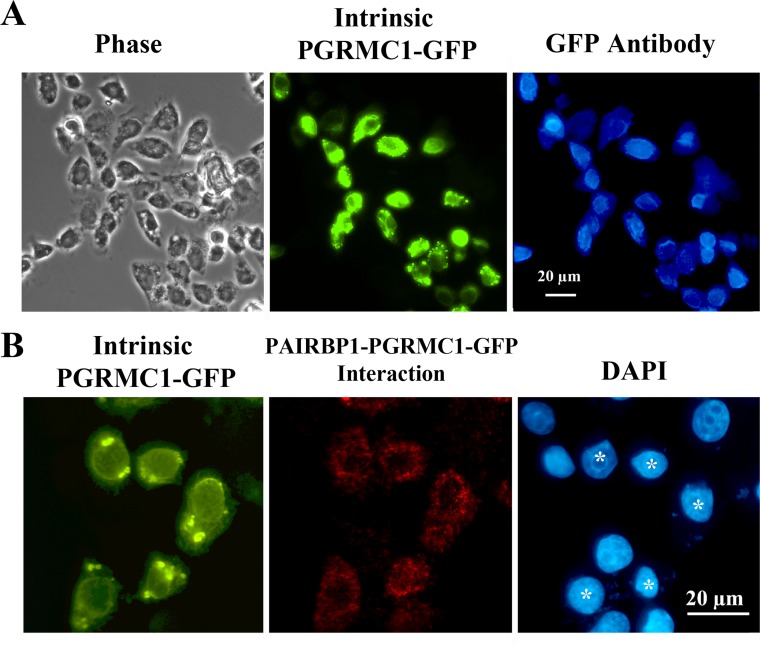
In order to use PLA to confirm our previous finding that endogenous PAIRBP1 interacts with PGRMC1-GFP [18, 20], it was first necessary to demonstrate that PGRMC1-GFP not only could be detected by its intrinsic GFP fluorescence but also could be immunodetected by a GFP antibody. This was demonstrated as shown in Figure 4A. Having established this, the GFP antibody was used in conjunction with the PAIRBP1 antibody in a PLA to reveal the interaction between PGRMC1-GFP and PAIRBP1 (Fig. 4B).
FIG. 4.
PAIRBP1-PGRMC1-GFP interaction in SIGCs maintained in serum as assessed by in situ PLA. In order to detect an interaction with PGRMC1-GFP, PGRMC1-GFP must be detectable both by its intrinsic GFP fluorescence and by an antibody to GFP. This is illustrated in A, in which PGRMC1-GFP-transfected SIGCs were fixed and stained for GFP using a GFP antibody and an Alexa Fluor 350-labeled anti-rabbit antibody. The same field of cells was observed under phase or fluorescent optics with filters to detect intrinsic GFP (green fluorescence) or immunodetectable GFP (blue fluorescence). Note that immunodetectable GFP is observed only in cells with intrinsic GFP fluorescence. In B in situ PLA detected PAIRBP1-PGRMC1-GFP interaction (red) only in SIGCs that were transfected with PGRMC1-GFP (green). Note that the transfected cells are marked by * in DAPI-stained preparation.
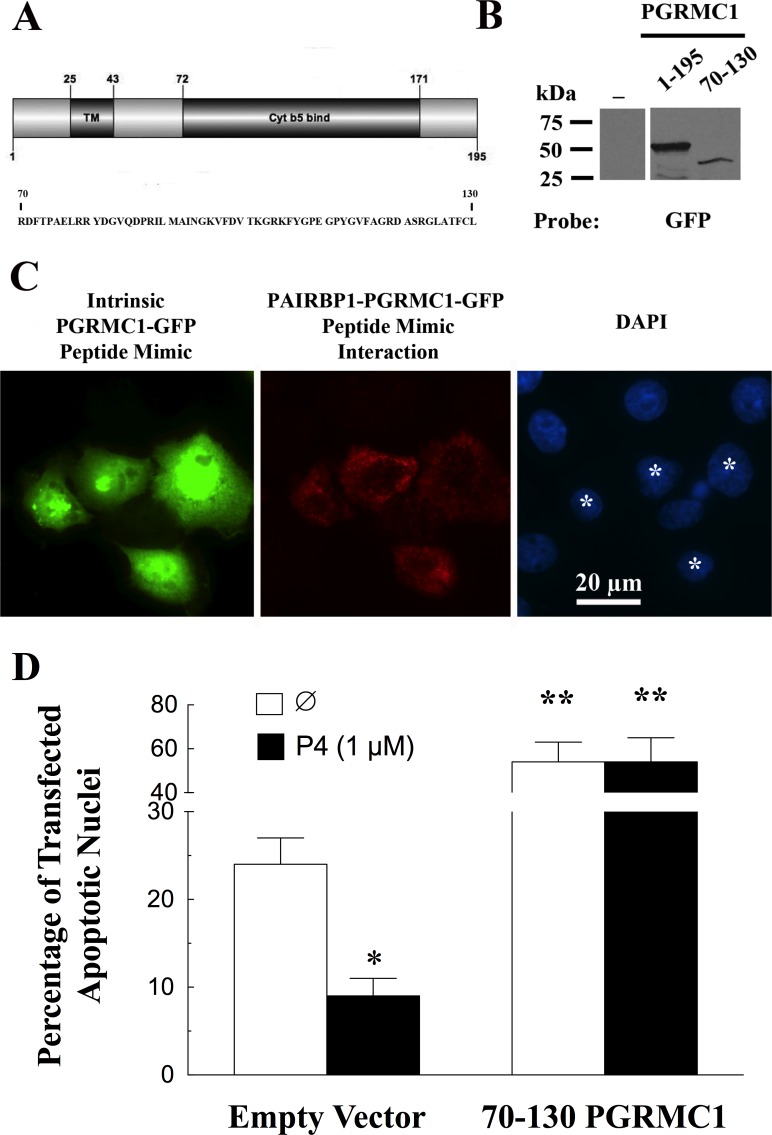
Previous studies have demonstrated that the PGRMC1 sequence between amino acids 70 and 130 was required for PGRMC1-GFP to bind PAIRBP1 [18]. This segment of PGRMC1 encodes about half of the cytochrome b5 binding domain (Fig. 5A). The observation that this 60-amino-acid sequence is the site of PGRMC1-PAIRBP1 interaction suggests that it could be used to disrupt PAIRBP1-PGRMC1 interaction. With the ability to use PLA to detect PGRMC1-GFP fusion protein interactions, a GFP expression construct was made that encoded the 70–130 amino acids of PGRMC1. This 70–130 PGRMC1-GFP fusion protein (i.e., a PGRMC1 peptide mimic) was transfected into SIGCs and its expression confirmed by both Western blot (Fig. 5B) and intrinsic GFP fluorescence, which was observed throughout the transfected cells (Fig. 5C). Moreover, the PGRMC1 peptide mimic interacted with PAIRBP1 within the cytoplasm, as demonstrated by PLA (Fig. 5C). SIGCs were viable 24 h after being transfected with either the PGRMC1 peptide mimic (Fig. 5C) or the empty GFP vector (image not shown). Interestingly, about 25% of the cells transfected with the empty GFP vector underwent apoptosis within 5 h of being placed in serum-free medium, and P4 significantly reduced the percentage of these GFP expressing cells that underwent apoptosis. In contrast, when placed in serum-free medium, >50% of cells expressing the PGRMC1 peptide mimic underwent apoptosis, and P4 was no longer able to suppress apoptosis (Fig. 5D).
FIG. 5.
The ability of the PGRMC1 peptide mimic (70-130-PGRMC1-GFP) to interact with endogenous PAIRBP1. In A the sequence of the PGRMC1 peptide mimic is shown, indicating that it comprises approximately half of the cytochrome b5 binding domain. B) Western blot indicating that the PGRMC1 peptide mimic (70–130) is expressed in SIGCs and detected as a single band that has a kDa, which is appropriately less than the full-length PGRMC1-GFP fusion protein (1–195). As seen in C the in situ PLA (red) detected the interaction between PAIRBP1 and the PGRMC1-GFP peptide mimic in those cells transfected with PGRMC1-GFP peptide mimic (green). The transfected cells are marked by * in DAPI-stained preparation. The effect of the PGRMC1 peptide mimic (70-130-PGRMC1-GFP) on the ability of P4 to inhibit apoptosis is shown in D. In this experiment cells were transfected with either the empty GFP vector or the PGRMC1-GFP peptide mimic and maintained in serum-supplemented medium for 24 h. After 24 h, the SIGCs were cultured for 5 h in serum-free medium in the presence or absence of P4 and then the transfected cells (green) assessed for apoptotic nuclei by DAPI staining (blue). Values are expressed as means ± one standard error, with * indicating a value that is less (P < 0.05) and ** indicating values greater than (P < 0.05) nontreated (∅) cells transfected with empty vector.
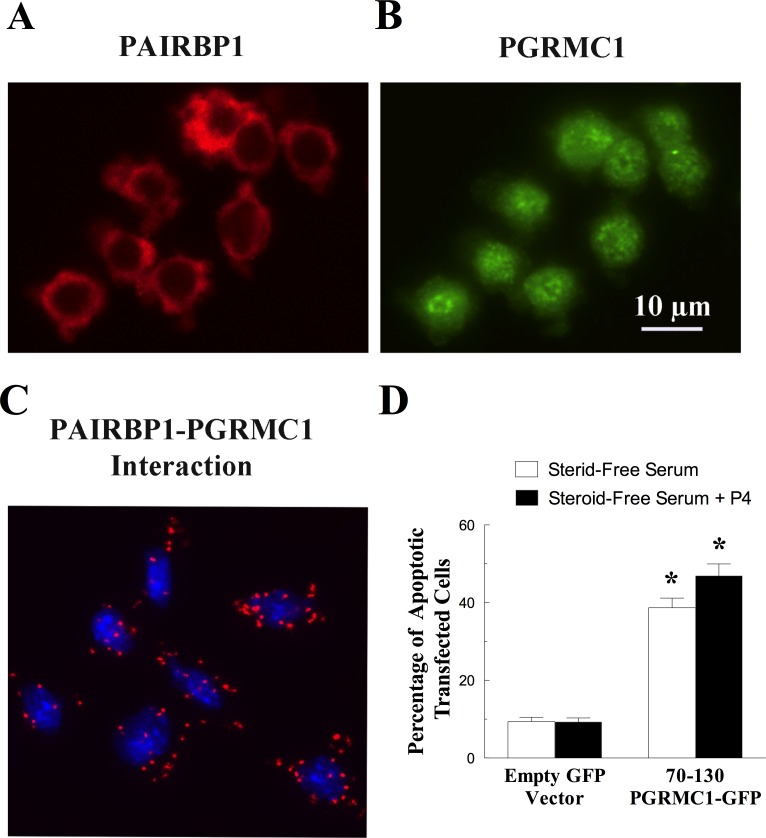
Immunocytochemical studies on granulosa cells isolated from immature rat ovaries revealed that PAIRBP1 was localized to the cytoplasm (Fig. 6A), whereas PGRMC1 was present throughout the cell (Fig. 6B), confirming the findings in SIGCs. As with SIGCs, these two proteins interact in granulosa cells, as shown by PLA (Fig. 6C). Unlike its effect in SIGCs, the PGRMC1 peptide mimic induced about 40% of the granulosa cells to undergo apoptosis even in the presence of steroid-free serum and P4 (Fig. 6D).
FIG. 6.
The localization and interaction of PAIRBP1 and PGRMC1 in granulosa cells maintained in serum-supplemented medium. The localizations of PAIRBP1 shown in red (A) and PGRMC1 shown in green (B) were determined by immunocytochemistry, and the interaction between PAIRBP1 and PGRMC1 was revealed by PLA (red dots, C). In the PLA image, the nuclei were stained with DAPI (blue). The graph in D shows the effect of transfecting the PGRMC1 peptide mimic (70-130-PGRMC1-GFP) on the rate of apoptosis in granulosa cells cultured in the steroid-free serum with or without P4. In this graph, the values are expressed as means ± one standard error, with * indicating a value that is greater than (P < 0.05) granulosa cells transfected with empty vector.
DISCUSSION
Our previous studies implicated PAIRBP1 as an important component of the mechanism through which P4 inhibits granulosa cell [20], luteal cell [20], and SIGC apoptosis [19, 20]. These initial studies revealed that overexpression of PAIRBP1 increases the sensitivity of the cells to the antiapoptotic action of P4 by 10-fold [19]. Conversely, pretreatment with a PAIRBP1 antibody attenuated P4's biological actions [19]. The present study expands these previous studies by demonstrating that Pairbp1 siRNA selectively depletes Pairbp1 mRNA and protein and completely attenuates P4's antiapoptotic action. This siRNA-based study provides the first conclusive evidence of PAIRBP1's involvement in P4's signaling pathway.
Although an important element in P4's action, PAIRBP1 is an unlikely candidate to directly bind P4 and mediate its action, because it possesses only a hyaluronan binding site and several phosphorylation sites (see http://www.phosphosite.org/proteinAction.do?id=7064&show AllSites=false). Interestingly, PAIRBP1 interacts with PGRMC1 [17, 18], which has been shown to transduce P4's antiapoptotic action [17, 18]. As such, PAIRBP1 could facilitate P4's actions by serving as either a scaffolding protein [17] or a coactivator/regulator of PGRMC1.
The present studies demonstrate that PAIRBP1 does not regulate PGRMC1's expression or localization, because Pairbp1 siRNA treatment virtually eliminates Pairbp1 mRNA and protein levels without altering PGRMC1 expression. Moreover, depleting PAIRBP1 does not significantly alter the cellular localization of PGRMC1 as judged by immunocytochemistry, although more detailed morphological and biochemical studies are required to definitively support this conclusion.
Even though immunocytochemical studies did not reveal major changes in the cellular distribution of PGRMC1, depletion of PAIRBP1 could alter PGRMC1's ability to bind 3H-P4. In fact, we initially proposed that PAIRBP1 interaction with PGRMC1 is required to form a functional binding site for P4 [17]. Based on our siRNA studies, this is clearly not the case. In fact, the present saturation binding studies reveal that depleting PAIRBP1 does not reduce P4 binding but rather greatly increases the capacity of the cells to bind P4. This suggests that the interaction between PAIRBP1 and PGRMC1 functions to limit PGRMC1's ability to bind P4.
How then does depleting PAIRBP1 attenuate P4's antiapoptotic action in the presence of PGRMC1? The most likely explanation is that PAIRBP1-PGRMC1 interaction functions to couple PGRMC1 to other components of the P4's antiapoptotic signal transduction pathway. The observation that P4 enhances the interaction of PAIRBP1 and PGRMC1 as judged by PLA further supports this concept.
If direct interaction between PAIRBP1 and PGRMC1 is an essential component of the P4-PGRMC1 signal cascade, then disrupting the interaction between these two proteins would ablate P4's action. Our previous studies have shown that the amino acid sequence RDFTPAELRRYDGVQDPRILMAINGKVFDVTKGRKFYGPEGPYGVFAGRDASRGLATFCL, which is located between PGRMC1 amino acid numbers 70 and 130, is required for PGRMC1 to bind to PAIRBP1 [18]. Although the entire 60-amino-acid sequence may not be required, we hypothesized that this sequence could be used as a peptide mimic that would interfere with PAIRBP1-PGRMC1 interaction. This 60-amino-acid PGRMC1 mimic was cloned into the GFP expression vector, transfected into cells, and shown by PLA assay to directly interact with endogenous PAIRBP1. Thus the PGRMC1 peptide mimic likely reduces the interaction between endogenous PAIRBP1 and PGRMC1. Interestingly, the presence of this PGRMC1 peptide mimic does not alter SIGC viability as long as the SIGCs are maintained in serum-supplemented medium. Because the serum was not heat inactivated, the mostly likely reason for the viability of the cells transfected with PGRMC1-GFP peptide mimic is the presence of growth factors in the serum that activate survival pathways that do not involve PAIRBP1-PGRMC1 interaction. However, when placed in a stressful environment such as in the absence of serum, the cells undergo apoptosis at a rate more than two to three times greater than GFP-transfected control cells. Moreover, P4 cannot reduce the stress-induced increase in the rate of apoptosis. This finding supports the concept that promoting or maintaining PAIRBP1-PGRMC1 interaction is an important aspect of P4's antiapoptotic action.
This concept is further strengthened by the observations made on cultured rat granulosa cells. Granulosa cells transfected with the empty GFP vector remain viable when cultured for 24 h in serum in the presence or absence of P4. In contrast, transfection of the 70–130 PGRMC1 peptide mimic induces ≈40% of the transfected cells to undergo apoptosis even in serum-supplemented medium with P4. This contrast between SIGCs and primary granulosa cells suggests that the primary granulosa cells are more dependent on PAIRBP1-PGRMC1 interaction than SIGCs. The reason for their increased dependency on PAIRBP1-PGRMC1 interaction for survival is unclear but may be related to the fact that SIGCs are immortalized and constitutively overexpress p53 compared to primary granulosa cells [22].
Although this 60-amino-acid sequence is present in PGRMC1 of all mammalian species, the PGRMC1 family member, PGRMC2, also has a 60-amino-acid segment, which is 80% identical to that of PGRMC1 [23]. Recently we have also shown that PGRMC2 interacts with PAIRBP1 (data not shown), so it is probable that the PGRMC1 peptide mimic also disrupts PAIRBP1's interaction with PGRMC2. Regardless, the limited presence of this 60-amino-acid sequence supports the concept that the PGRMC1 peptide mimic only interferes with the ability of PGRMC1 and possibly PGRMC2 to interact PAIRBP1.
The cellular response to the PGRMC1 peptide mimic also raises two important questions. The first question relates to the role that PAIRPB1 plays in modulating the function of PGRMC1. Recently we have shown that PGRMC1 localizes to the nucleus and in response to P4 functions to suppress the transcriptional activation of Tcf/Lef transcription factors [24]. The ability of P4 to suppress Tcf/Lef activity is dependent in part on the sumoylation status of PGRMC1 [24]. Sumoylation involves the covalent linking of small ubiquitin-like modifier (SUMO) proteins to specific target proteins [25–27]. The linkage of SUMO proteins to a specific protein is mediated by several enzymes, including E3 SUMO ligases and SUMO-activating enzyme subunit 2 [26]. Importantly, PAIRBP1, which is referred to as CGI-55, has been shown to bind these enzymes [28]. As such P4 could act to enhance PAIRBP1-PGRMC1 interaction, thereby stimulating PAIRBP1 to act as a scaffolding protein and “deliver” the sumoylating enzymes to PGRMC1. As a result, PGRMC1 would undergo sumoylation, and, once in the nucleus, modulate the expression of genes that promote cell survival [29, 30]. Thus, ability of PAIRBP1 to couple PGRMC1 to downstream components of its signal pathway could be a role that PAIRBP1 plays in the P4-PGRMC1 signal transduction pathway. This possibility is actively being investigated.
The second question relates to why the PGRMC1 peptide mimic accelerates the rate at which SIGCs undergo apoptosis when placed in serum-free culture condition. If the PGRMC1 peptide mimic only disrupts PAIRBP1's interaction with either PGRMC1 or PGRMC2, then the rate of apoptosis should be similar to that observed with Pairbp1 siRNA treatment. However, the rate of apoptosis in the presence of the PGRMC1 peptide mimic is at least 2-fold greater than that observed in the Pairbp1 siRNA-treated cells. It is important to appreciate that the 60-amino-acid sequence of the PGRMC1 mimic corresponds to the first 60 amino acids of the cytochrome 5b domain of PGRMC1. The cytochrome 5b domain of PGRMC1 has been shown to bind heme [31, 32] and presumably hemoproteins. Moreover, nuclear magnetic resonance structural analysis of Arabidopsis PGRMC1 homolog, At2g24940.1, revealed the presence of a crevice that is formed by helix 2, helix 3, and helix 4, with the loop between helix 3 and 4 being the likely site to which heme and hemoproteins bind (for review see Cahill [23]). Because helixes 1–3 are within the 70- to 130-amino-acid sequence of PGRMC1 [23], the PGRMC1 peptide mimic likely interferes with the ability of PGRMC1 and possibly PGRMC2 to interact with hemoproteins. Hemoproteins include respiration cytochromes, gas sensors, P450 enzymes, catalases, peroxidases, nitric oxide synthases, and guanyl cyclases [33]. Because hemoproteins regulate numerous cellular functions, including cell viability [33], it is possible that the PGRMC1 peptide mimic also disrupts the ability of hemoproteins to interact with PGRMC1 or PGRMC2. Thus, the putative ability of the PGRMC1 peptide mimic to attenuate the interaction between hemoproteins and PGRMC family member could explain why it induces a higher rate of apoptosis than the Pairbp1 siRNA treatment.
Taken together, the present studies reveal a unique and novel role for PAIRBP1 in the regulation of PGRMC1's action. In addition, we have identified a PGRMC1 peptide mimic that appears to function as a competitive inhibitor of PAIRBP1-PGRMC1 interaction. This PGRMC1 peptide mimic greatly enhances the rate at which cells undergo stress-induced apoptosis, suggesting that this peptide mimic affects more than just PAIRBP1-PGRMC1 interaction. Because the amino acid sequence of the peptide mimic is unique to PGRMC1 and 2, it has the potential to specifically inhibit P4's actions that are mediated through PGRMC family members while not influencing P4 signals that are transduced through either PGR or progestin and adipoQ receptors (PAQR). As such, this PGRMC1 peptide mimic could have therapeutic potential in the treatment of ovarian dysfunction as well as ovarian [34, 35], placental [34], uterine [36, 37], breast [38–40], and lung [41] cancer, because PGRMC1 is highly expressed in these cancers.
ACKNOWLEDGMENT
The authors would like to thank Drs. James Pru of Washington State University and Alberto Luciano of the University of Milan for their critical review of this manuscript and Dr. Robert Burghardt of Texas A&M University for providing the spontaneously immortalized granulosa cells.
Footnotes
This work was supported by NIH grant R01 HD 052740 awarded to J.J.P.
REFERENCES
- Fujii T, Hoover DJ, Channing CP. Changes in inhibin activity, and progesterone, oestrogen and androstenedione concentrations, in rat follicular fluid throughout the oestrous cycle. J Reprod Fertil. 1983;69:307–314. doi: 10.1530/jrf.0.0690307. [DOI] [PubMed] [Google Scholar]
- Luciano AM, Pappalardo A, Ray C, Peluso JJ. Epidermal growth factor inhibits large granulosa cell apoptosis by stimulating progesterone synthesis and regulating the distribution of intracellular free calcium. Biol Reprod. 1994;51:646–654. doi: 10.1095/biolreprod51.4.646. [DOI] [PubMed] [Google Scholar]
- Luciano AM, Peluso JJ. Effect of in vivo gonadotropin treatment on the ability of progesterone, estrogen, and cyclic adenosine 5′-monophosphate to inhibit insulin-dependent granulosa cell mitosis in vitro. Biol Reprod. 1995;53:664–669. doi: 10.1095/biolreprod53.3.664. [DOI] [PubMed] [Google Scholar]
- Peluso JJ. Placing progesterone in the apoptotic pathway. Trends Endocrinol Metab. 1997;8:267–271. doi: 10.1016/s1043-2760(97)00078-7. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A. Progesterone mediates its anti-mitogenic and anti-apoptotic actions in rat granulosa cells through a progesterone-binding protein with gamma aminobutyric acidA receptor-like features. Biol Reprod. 1998;58:1131–1137. doi: 10.1095/biolreprod58.5.1131. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A. Progesterone maintains large rat granulosa cell viability indirectly by stimulating small granulosa cells to synthesize basic fibroblast growth factor. Biol Reprod. 1999;60:290–296. doi: 10.1095/biolreprod60.2.290. [DOI] [PubMed] [Google Scholar]
- Chaffkin LM, Luciano AA, Peluso JJ. Progesterone as an autocrine/paracrine regulator of human granulosa cell proliferation. J Clin Endocrinol Metab. 1992;75:1404–1408. doi: 10.1210/jcem.75.6.1464640. [DOI] [PubMed] [Google Scholar]
- Chaffkin LM, Luciano AA, Peluso JJ. The role of progesterone in regulating human granulosa cell proliferation and differentiation in vitro. J Clin Endocrinol Metab. 1993;76:696–700. doi: 10.1210/jcem.76.3.8445028. [DOI] [PubMed] [Google Scholar]
- Makrigiannakis A, Coukos G, Christofidou-Solomidou M, Montas S, Coutifaris C. Progesterone is an autocrine/paracrine regulator of human granulosa cell survival in vitro. Ann N Y Acad Sci. 2000;900:16–25. doi: 10.1111/j.1749-6632.2000.tb06212.x. [DOI] [PubMed] [Google Scholar]
- Stouffer RL. Progesterone as a mediator of gonadotrophin action in the corpus luteum: beyond steroidogenesis. Hum Reprod Update. 2003;9:99–117. doi: 10.1093/humupd/dmg016. [DOI] [PubMed] [Google Scholar]
- Anzalone CR, Hong LS, Lu JK, LaPolt PS. Influences of age and ovarian follicular reserve on estrous cycle patterns, ovulation, and hormone secretion in the Long-Evans rat. Biol Reprod. 2001;64:1056–1062. doi: 10.1095/biolreprod64.4.1056. [DOI] [PubMed] [Google Scholar]
- Levi AJ, Raynault MF, Bergh PA, Drews MR, Miller BT, Scott RT Jr. Reproductive outcome in patients with diminished ovarian reserve. Fertil Steril. 2001;76:666–669. doi: 10.1016/s0015-0282(01)02017-9. [DOI] [PubMed] [Google Scholar]
- Park OK, Mayo KE. Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol Endocrinol. 1991;5:967–978. doi: 10.1210/mend-5-7-967. [DOI] [PubMed] [Google Scholar]
- Park-Sarge OK, Mayo KE. Regulation of the progesterone receptor gene by gonadotropins and cyclic adenosine 3′,5′-monophosphate in rat granulosa cells. Endocrinology. 1994;134:709–718. doi: 10.1210/endo.134.2.8299566. [DOI] [PubMed] [Google Scholar]
- Engmann L, Losel R, Wehling M, Peluso JJ. Progesterone regulation of human granulosa/luteal cell viability by an RU486-independent mechanism. J Clin Endocrinol Metab. 2006;91:4962–4968. doi: 10.1210/jc.2006-1128. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Liu X, Gawkowska A, Johnston-MacAnanny E. Progesterone activates a progesterone receptor membrane component 1-dependent mechanism that promotes human granulosa/luteal cell survival but not progesterone secretion. J Clin Endocrinol Metab. 2009;94:2644–2649. doi: 10.1210/jc.2009-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Losel R, Wehling M. Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone's antiapoptotic action. Endocrinology. 2006;147:3133–3140. doi: 10.1210/en.2006-0114. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Romak J, Liu X. Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology. 2008;149:534–543. doi: 10.1210/en.2007-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Fernandez G, Wu CA. Involvement of an unnamed protein, RDA288, in the mechanism through which progesterone mediates its antiapoptotic action in spontaneously immortalized granulosa cells. Endocrinology. 2004;145:3014–3022. doi: 10.1210/en.2004-0067. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Losel R, Wehling M. Expression and function of PAIRBP1 within gonadotropin-primed immature rat ovaries: PAIRBP1 regulation of granulosa and luteal cell viability. Biol Reprod. 2005;73(2):261–270. doi: 10.1095/biolreprod.105.041061. [DOI] [PubMed] [Google Scholar]
- Campbell KL. Ovarian granulosa cells isolated with EGTA and hypertonic sucrose: cellular integrity and function. Biol Reprod. 1979;21:773–786. doi: 10.1095/biolreprod21.4.773. [DOI] [PubMed] [Google Scholar]
- Stein LS, Stoica G, Tilley R, Burghardt RC. Rat ovarian granulosa cell culture: a model system for the study of cell-cell communication during multistep transformation. Cancer Res. 1991;51:696–706. [PubMed] [Google Scholar]
- Cahill MA. Progesterone receptor membrane component 1: an integrative review. J Steroid Biochem Mol Biol. 2007;105:16–36. doi: 10.1016/j.jsbmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Lodde V, Liu X. Progesterone regulation of progesterone receptor membrane component 1 (PGRMC1) sumoylation and transcriptional activity in spontaneously immortalized granulosa cells. Endocrinology. 2012;153:3929–3939. doi: 10.1210/en.2011-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ET. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 2011;585:2891–2896. doi: 10.1016/j.febslet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- Lemos TA, Kobarg J. CGI-55 interacts with nuclear proteins and co-localizes to p80-coilin positive-coiled bodies in the nucleus. Cell Biochem Biophys. 2006;44:463–474. doi: 10.1385/CBB:44:3:463. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Decerbo J, Lodde V. Evidence for a genomic mechanism of action for progesterone receptor membrane component-1. Steroids. 2012;77(10):1007–1012. doi: 10.1016/j.steroids.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso JJ, Liu X, Gawkowska A, Lodde V, Wu CA. Progesterone inhibits apoptosis in part by PGRMC1-regulated gene expression. Mol Cell Endocrinol. 2010;320:153–161. doi: 10.1016/j.mce.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K, Thompson AM, Goldbeck RA, Shi X, Whitman S, Oh E, Zhiwu Z, Vulpe C, Holman TR. Spectroscopic and biochemical characterization of heme binding to yeast Dap1p and mouse PGRMC1p. Biochemistry. 2005;44:16729–16736. doi: 10.1021/bi0511585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Strushkevich NV, Harnastai IN, Iwamoto H, Gilep AA, Takemori H, Usanov SA, Nonaka Y, Hori H, Vinson GP, Okamoto M. Molecular identification of adrenal inner zone antigen as a heme-binding protein. FEBS J. 2005;272:5832–5843. doi: 10.1111/j.1742-4658.2005.04977.x. [DOI] [PubMed] [Google Scholar]
- Tsiftsoglou AS, Tsamadou AI, Papadopoulou LC. Heme as key regulator of major mammalian cellular functions: molecular, cellular, and pharmacological aspects. Pharmacol Ther. 2006;111:327–345. doi: 10.1016/j.pharmthera.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Peluso JJ. Progesterone signaling mediated through progesterone receptor membrane component-1 in ovarian cells with special emphasis on ovarian cancer. Steroids. 2011;76:903–909. doi: 10.1016/j.steroids.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso JJ, Liu X, Saunders MM, Claffey KP, Phoenix K. Regulation of ovarian cancer cell viability and sensitivity to cisplatin by progesterone receptor membrane component-1. J Clin Endocrinol Metab. 2008;93:1592–1599. doi: 10.1210/jc.2007-2771. [DOI] [PubMed] [Google Scholar]
- Zachariades E, Foster H, Goumenou A, Thomas P, Rand-Weaver M, Karteris E. Expression of membrane and nuclear progesterone receptors in two human placental choriocarcinoma cell lines (JEG-3 and BeWo): effects of syncytialization. Int J Mol Med. 2011;27:767–774. doi: 10.3892/ijmm.2011.657. [DOI] [PubMed] [Google Scholar]
- Panda H, Chuang TD, Luo X, Chegini N. Endometrial miR-181a and miR-98 Expression is altered during transition from normal into cancerous state and target PGR, PGRMC1, CYP19A1, DDX3X, and TIMP3. J Clin Endocrinol Metab. 2012;97(7):E1316–E1326. doi: 10.1210/jc.2012-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed IS, Rohe HJ, Twist KE, Craven RJ. Pgrmc1 (progesterone receptor membrane component 1) associates with epidermal growth factor receptor and regulates erlotinib sensitivity. J Biol Chem. 2010;285:24775–24782. doi: 10.1074/jbc.M110.134585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed IS, Rohe HJ, Twist KE, Mattingly MN, Craven RJ. Progesterone receptor membrane component 1 (Pgrmc1): a heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J Pharmacol Exp Ther. 2010;333:564–573. doi: 10.1124/jpet.109.164210. [DOI] [PubMed] [Google Scholar]
- Craven RJ. PGRMC1: a new biomarker for the estrogen receptor in breast cancer. Breast Cancer Res. 2008;10:113. doi: 10.1186/bcr2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir SU, Ahmed IS, Arnold S, Craven RJ. Elevated progesterone receptor membrane component 1/sigma-2 receptor levels in lung tumors and plasma from lung cancer patients. Int J Cancer. 2012;131:E1–E9. doi: 10.1002/ijc.26432. [DOI] [PubMed] [Google Scholar]