Abstract
Lung adenocarcinomas from never smokers account for approximately 15 to 20% of all lung cancers and these tumors often carry genetic alterations that are responsive to targeted therapy. Here we examined mutation status in 10 oncogenes among 89 lung adenocarcinomas from never smokers. We also screened for oncogene fusion transcripts in 20 of the 89 tumors by RNA-Seq. In total, 62 tumors had mutations in at least one of the 10 oncogenes, including EGFR (49 cases, 55%), K-ras (5 cases, 6%), BRAF (4 cases, 5%), PIK3CA (3 cases, 3%), and ERBB2 (4 cases, 5%). In addition to ALK fusions identified by IHC/FISH in four cases, two previously known fusions involving EZR- ROS1 and KIF5B-RET were identified by RNA-Seq as well as a third novel fusion transcript that was formed between exons 1–9 of SND1 and exons 2 to 3′ end of BRAF. This in-frame fusion was observed in 3/89 tested tumors and 2/64 additional never smoker lung adenocarcinoma samples. Ectopic expression of SND1-BRAF in H1299 cells increased phosphorylation levels of MEK/ERK, cell proliferation, and spheroid formation compared to parental mock-transfected control. Jointly, our results suggest a potential role of the novel BRAF fusion in lung cancer development and therapy.
Lung adenocarcinoma is the most common type of lung cancer worldwide and occurs in both smokers and never smokers (NS). It is estimated that up to 70% of all lung cancers in women living in East Asia are unrelated to smoking, while in North America about 15% of all lung cancers in both men and women occur in NS1,2,3. The different mutation spectra of lung cancer from smokers and nonsmokers have been well described and account for significantly different therapeutic responses in these patients4,5,6. For example, NSCLC adenocarcinomas in female never smokers tend to have a much higher frequency of EGFR gene mutations and EML4-ALK transcript fusions7. These patients benefit from targeted drugs such as gefinitib and crizotinib with significant tumor response and improved progression-free survival in advanced NSCLC over conventional therapy7,8. In contrast, up to 50% of lung adenocarcinomas from smokers carry K-ras gene mutations, and they often respond poorly to tyrosine kinase inhibitor (TKI) treatment9,10.
As a result of these discoveries, treatment strategies for lung adenocarcinoma in advanced stages have evolved significantly from the traditional platinum-based chemotherapy to a gene-based targeted approach for first line therapy when tumors carried targetable mutations7. Screening for oncogene mutations in primary lung tumors is becoming a clinical standard to guide individual treatment options and to identify new targets11. However, most patients treated with targeted kinase inhibitors eventually relapse, making it essential that new targets be identified in order to improve overall survival of the patients.
In this study, we used a mass spectrometry-based multiplex assay (MassArray technology, Sequenom, San Diego, CA) capable of quantitatively identifying single nucleotide changes in the DNA to screen for 187 mutations in 10 oncogenes (EGFR, K-RAS, BRAF, ERBB2, MET, PIK3CA, AKT1/AKT2, KIT, and JAK2)12. We then employed whole transcriptome sequencing (RNA-Seq) to screen for transcript fusions involving an oncogene. This study reports the high frequency of oncogene mutations and the identification of a novel SND1-BRAF fusion transcript in about 3% of lung adenocarcinoma from never smokers.
Results
Oncogene mutation detection in cancer tissues
We used a custom designed panel of 16 multiplex Mass Array assays to focus on oncogenes commonly mutated in lung adenocarcinoma (Supplementary Table S1). In total, 62 of the 89 tested tumors had mutations in at least one of the tested genes. EGFR gene mutations were detected in 49/89 cases (55%), while K-ras gene mutations were observed only in 5/89 cases (6%). Mutations involving BRAF were observed in 4 cases (5%), PIK3CA in 3 cases (3%), and ERBB2 in 4 cases (5%) (Table 1 and Supplementary Table S2). Two tumors had compound mutations in the EGFR gene: Lu-246 with S768I and 773_V774insNPH mutations while sample Lu-243 had S768I and L858R mutations. None of the tumors with K-ras, BRAF, or ERBB2 point mutations overlapped with each other or with those having EGFR mutations. However, all three cases with PIK3CA mutations also carried a mutation in the EGFR gene. No statistically significant correlations were observed between mutation status and the patients' age, gender, or tumor stage.
Table 1. Summary of clinical information and mutation status for 89 tested samples.
| Variable* | No. of Patients | EGFR (%) | BRAF2 (%) | ERBB2 (%) | K-ras (%) | PIK3CA (%) | Other Fusions3 (%) | |
|---|---|---|---|---|---|---|---|---|
| (Adenocarcinoma1) | ||||||||
| Total | 89 | 49 (55.0) | 7 (7.9) | 4 (4.5) | 5 (5.6) | 3 (3.4) | 6 (6.7) | |
| Sex | ||||||||
| Male | 15 | 5 (33.3) | 3 (20.0) | 0 (0) | 1 (6.7) | 0 (0) | 4 (26.7) | |
| Female | 74 | 44 (59.5) | 4 (5.4) | 4 (5.4) | 4 (5.4) | 3 (4.1) | 2 (2.7) | |
| Age | ||||||||
| ≥65 | 56 | 32 (57.1) | 5 (8.9) | 2 (3.6) | 4 (12.5) | 3 (5.4) | 2 (3.6) | |
| <65 | 33 | 17 (51.5) | 2 (6.1) | 2 (6.1) | 1 (3.0) | 0 (0) | 4 (12.1) | |
| Staging | ||||||||
| I | 57 | 33 (57.9) | 6 (10.5) | 4 (7.0) | 4 (7.0) | 1 (1.8) | 2 (3.5) | |
| II | 9 | 4 (44.4) | 0 (0) | 0 (0) | 0 (0) | 1 (11.1) | 0 (0) | |
| III | 19 | 10 (52.6) | 1 (5.3) | 0 (0) | 0 (0) | 1 (5.3) | 4 (21.1) | |
| IV | 4 | 2 (50.0) | 0 (0) | 0 (0) | 1 (25.0) | 0 (0) | 0 (0) | |
1Adenocarcinoma, Adeno with bronchioloalveolar carcinoma, Bronchioloalveolar carcinoma and Adenosquamous carcinoma.
2Includes BRAF point mutations and SND1-BRAF transcript fusions.
3Includes ALK+, EZR-ROS1 and KIF5B-RET.
*No statistically significant difference in mutation distributions for the clinical variables shown.
Transcript fusion identification and validation
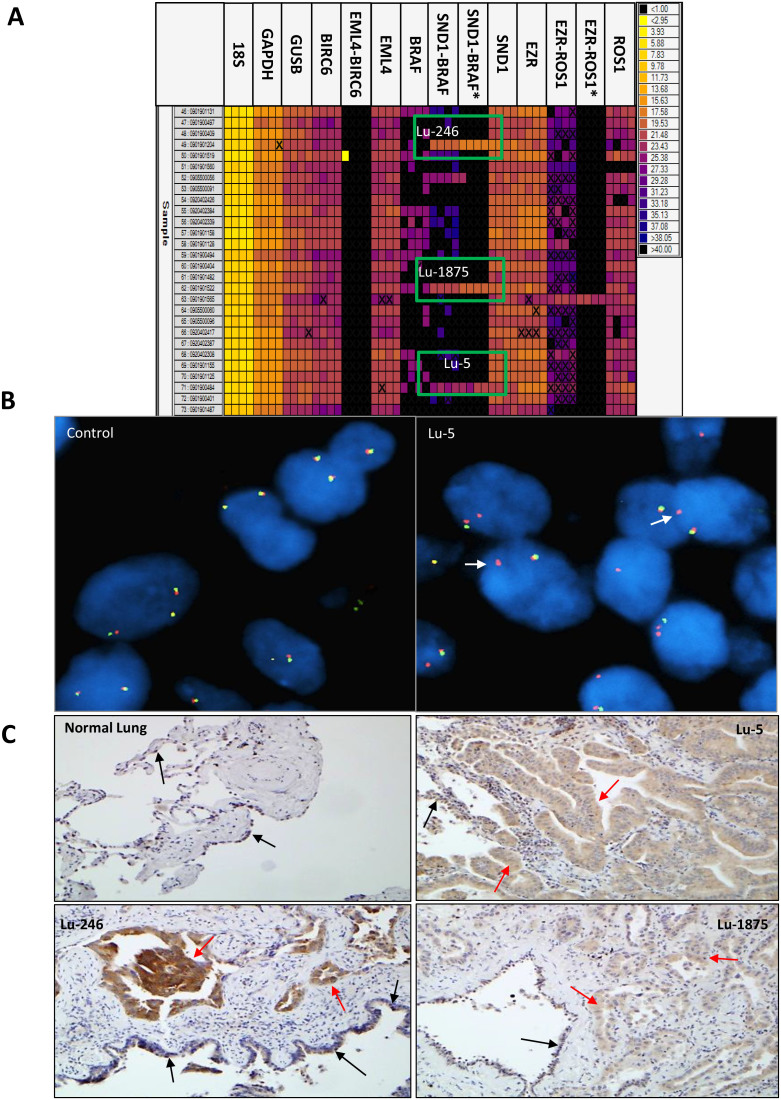
Among the 20 tumors analyzed by RNA-Seq, 74 unique fusion transcripts were called and all 20 tumors had at least one fusion (Supplementary Table S3). Of those, 2 of 13 tumors without an oncogene mutation by the Mass Array assay, samples Lu-1566 and Lu-1995, had transcript fusions involving EZR-ROS1 and KIF5B-RET, respectively (Table 2 and Supplementary Fig. S1). Among the 7 of 20 tumors having at least one oncogene mutation, a novel fusion transcript involving SND1-BRAF gene was identified in Lu-5 (Table 2 and Fig. 1). Quantitative PCR by the Fluidigm Dynamic Array confirmed the fusion in sample Lu-5 and also identified two additional tumors, Lu-246 and Lu-1875, carrying the same SND1-BRAF fusion (Fig. 2 and Table 3). FISH confirmed all three fusions identified by RNA-Seq involving ROS1, RET and BRAF (Supplementary Fig. S1 and Fig. 3B).
Table 2. Fusion transcripts identified by RNA-Seq that involved known or potential targetable genes.
| Sample | Fusion gene directional | Type | Fusion Strand | Encompassing Read Pairs | Total SplitReads | Exon Boundary Fusion | 5′ gene Information | 3′ Gene Information |
|---|---|---|---|---|---|---|---|---|
| Lu-5 | SND1->BRAF | intra-chr | + | 29 | 4 | YES | E9:chr7:SND1 | E2:chr7:BRAF |
| Lu-1566 | EZR->ROS1 | intra-chr | − | 108 | 153 | YES | E10:chr6:EZR | E34:chr6:ROS1 |
| Lu-1995 | KIF5B->RET | intra-chr | − | 144 | 82 | YES | E15:chr10:KIF5B | E12:chr10:RET |
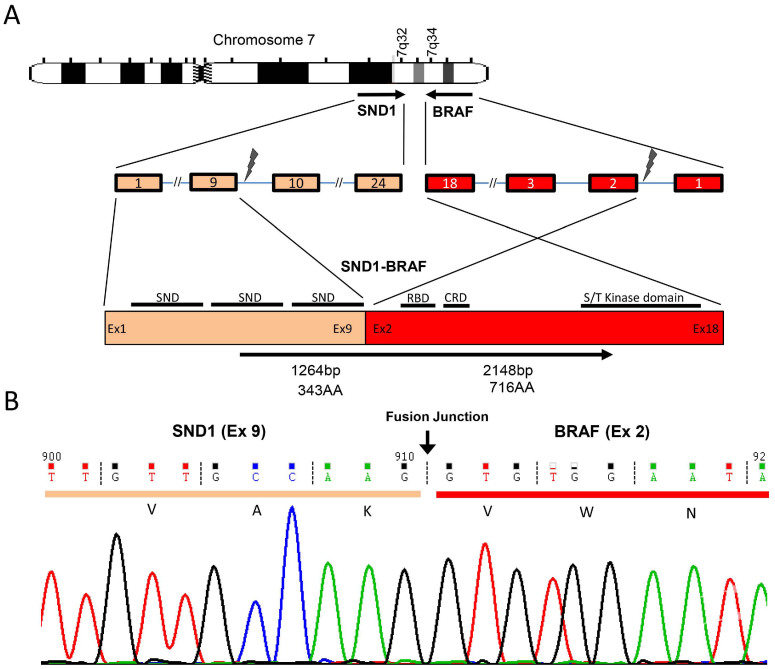
Figure 1. SND1-BRAF transcript fusion in a never smoker lung adenocarcinoma.
(A). Chromosomal rearrangement at 7q32 and 7q34 results in exons 1–9 of SND1 fused to the exon 2 to 3′ end of an inverted BRAF. SND, Staphylococcal nuclease domain; Ex, exon; RBD, Ras-binding domain; CRD, cysteine-rich domain; S/T, serine-threonine. (B). Sequencing traces of the RT-PCR product at fusion junction spanning exon 9 of SND1 (orange) and exon 2 of BRAF (red). The dashed lines show the the reading frame.
Figure 2. Validation and identification of additional SND1-BRAF fusions in lung adenocarcinomas.
(A). SND1-BRAF fusion genes in 3 of 89 samples by qRT-PCR using Fluidigm Dynamic Array. Samples are loaded on each row and the customized assays from ABI or IDT (*) are loaded in each column as quadruplets. The targets amplified are shown at the top and the positives are indicated by boxes. (B). FISH split-apart probe flanking BRAF in a control and Lu-5 samples. Arrows indicate the break-apart probe adjacent to the BRAF gene. Magnification ×1,000. (C). IHC using anti-BRAFwt antibody in normal lung and indicated tumor samples. Overexpression of BRAF in tumor cells (red arrows) was observed when comparing staining intensity with those in the adjacent bronchial epithelium cells having a basal expression level of the protein (black arrows). Magnification ×200.
Table 3. Lung adenocarcinomas having SND1-BRAF fusion genes.
| Sample ID | Sample Type | Methods of Detection | Supporting Reads# | Sanger Sequencing* | 5' Gene | Break Point | 3' Gene | Break Point | Other Mutations |
|---|---|---|---|---|---|---|---|---|---|
| Lu-5 | FF, RNA/DNA | RNA-Seq, qRT-PCR, WGSeq, FISH | 29 | + | Ex9, SND1 | Chr.7, 127347701 | Ex2, BRAF | Chr7. 140550012 | ERBB2, M774_A775insAYVM |
| Lu-1875 | FF, RNA | qRT-PCR, IHC | − | + | Ex9, SND1 | Chr.7, 127347701 | Ex2, BRAF | Chr7. 140550012 | EGFR, S768I, PIK3CA, E542K |
| Lu-246 | FF, RNA | qRT-PCR, IHC | − | + | Ex9, SND1 | Chr.7, 127347701 | Ex2, BRAF | Chr7. 140550012 | EGFR S768I, H773_V774insNPH |
| Y59 | FFPE, DNA/RNA | Gene Capture, qRT-PCR | 17 | + | Ex9, SND1 | Chr.7, 127347703 | Ex2, BRAF | Chr7. 140550015 | EML4-ALK |
| Y69 | FFPE, DNA/RNA | Gene Capture, qRT-PCR | 20 | + | Ex9, SND1 | Chr.7, 127347704 | Ex2, BRAF | Chr7. 140550014 | N.F. |
#NextGen sequencing reads that either span the fusion junction of the transcript in Lu-5 or mapped to the corresponding genomic regions in Y59 and Y69.
*Sanger sequencing was performed using cDNA. N.F., Not Found.
Figure 3. Functional Analyses of SND1-BRAF.
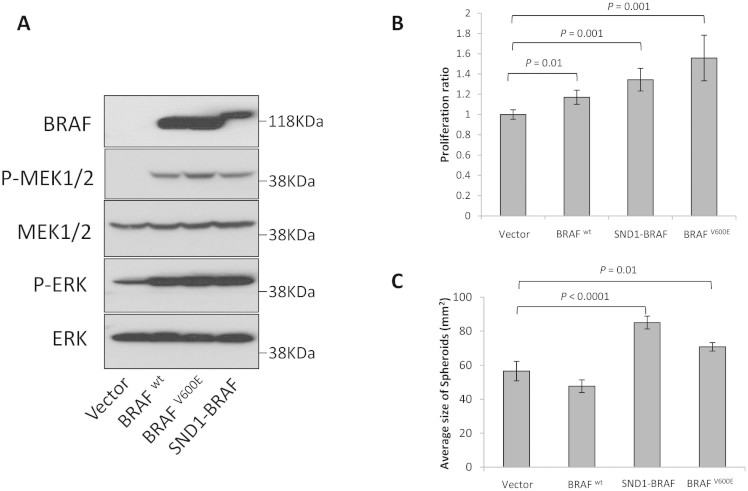
(A). Western blots of lysates from H1299 cells transfected with pCMV6-AC-GFP vector, pCMV6-AC-GFP-BRAFwt, pCMV6-AC-GFP-BRAFV600E, or pCMV6-AC-GFP-SND1-BRAF were probed with antibodies to C-terminus of BRAF, phospho-MEK1/2 (pMEK), total MEK (MEK), phospho-ERK1/2(pERK), or total ERK (ERK). (B). Proliferation assay of H1299 cells transfected as in A. Error bars represent the average ratio from triplicates. (C). Spheroid formation assay of H1299 cell transfected with indicated constructs. Error bars represent the standard error of the mean (SEM). Statistical significances are as indicated.
Genomic structure of the SND1-BRAF transcript fusion in lung cancer
Sequencing analysis of the SND1-BRAF fusion transcript in sample Lu-5 revealed an intra-chromosomal rearrangement between the exons 1–9 of SND1 and the exons 2-3′ end of the inverted BRAF on chromosome 7 (Fig. 1). The resulting fusion protein is predicted to include 1- 343AA of SND1 and 50 - 766AA of BRAF. Interestingly, all three tumors with SND1-BRAF fusion also carried at least one other oncogene mutation. Lu-5 had an additional M774_A775insAYVM mutation in the ERBB2 gene; Lu-246 had a mutation in the EGFR gene (S768I and H773_V774insNPH); while Lu-1875 had compound mutations in both EGFR (S768I) and PIK3CA (E542K) genes.
IHC in tumor samples with SND1-BRAF fusion protein expression
We stained whole tissue sections using an antibody to the wild type of the BRAF protein by immunohistochemistry. Because SND1 is fused to exon 2 of BRAF, we expected IHC to be positive using an antibody against the wild type of BRAF if the fusion transcript generated a protein product. All three tumors with the SND1-BRAF fusion showed an increased expression of BRAF in cancer cells compared to the adjacent non-involving bronchial epithelium present in the same section which had a basal level staining of the protein (Fig. 2C).
DNA rearrangements involving SND1-BRAF fusion
We assessed the genetic rearrangement that led to the SND1-BRAF fusion by whole genome sequencing using a shot-gun approach (Complete Genomics, Inc, Mountain View, CA) using sample Lu-5. At 40x read depth coverage, whole genome analysis revealed multiple genomic rearrangements, consistent with chromothripsis13, at the region surrounding SND1 and BRAF genes on chromosome 7 involving several different junction points of both genes and a third TBXAS1 gene located between the two fusion genes (Supplementary Fig. S2). Therefore, it seems that while many DNA level changes were observed by the whole genome shotgun sequencing of the same tumor, only one productive fusion transcript was formed between SND1-BRAF as a result of the genomic rearrangement in the region.
Using a whole gene capture strategy for formalin-fixed, paraffin-embedded (FFPE) DNA samples, we identified two additional never smoker lung cancer cases from the same cohort also carrying the SND1-BRAF fusion (Table 3, Y59 and Y69). We then confirmed the presence of the same fusion in these samples by direct sequencing of the cDNA product using RNA isolated from the matched FFPE tumor samples (Table 3 and Supplementary Fig. S3).
SND1-BRAF transcript increases MEK/ERK phosphorylation and oncogenic functions
In order to investigate the role of SND1-BRAF fusion protein, we examined the Raf signaling pathway by transiently expressing the fusion gene product and analyzed the phosphorylation status of the downstream proteins by western blot in the H1299 lung cancer cell line. Fig. 3A shows that the SND1-BRAF fusion protein significantly increased phosphorylation of its downstream effectors, MEK and ERK, compared to the vector control. In addition, the phosphorylation level of ERK in the SND1-BRAF fusion protein was increased similarly to BRAFWT and BRAFV600E, an activating mutation most commonly observed in melanomas (Fig. 3A). We also observed that the SND1-BRAF fusion protein increased cell proliferation and spheroid formation compared to the vector control or BRAFWT (Fig. 3B and 3C). The fusion protein demonstrated the highest spheroid formation activity in this assay, followed by BRAFV600E and BRAFWT.
Discussion
In this study, we used a customized Mass Array assay panel to determine the presence of oncogene mutations that are frequently observed and potentially targetable in lung adenocarcinoma from never smokers. We recently demonstrated the robustness of this customized Mass Array assay for detecting oncogene mutations in FFPE tumor samples12. In the 89 tumors examined in this study, 62 had a mutation in at least one of the tested genes. Furthermore, our study showed that mutations in the EGFR gene were the most frequent and occurred in 55% of all tested tumors (49 of 89 cases) from a population of European descent. Mutations in KRAS, BRAF, and ERBB2 were much less frequent, non-overlapping, and present in tumors without with EGFR mutations. These results are consistent with previous findings and further supporting the dependence on EGFR signaling in lung adenocarcinoma from never smokers8,14,15.
In addition to identifying two previously reported fusions involving EZR-ROS1 and KIF5B-RET, the unique finding of our study is the identification of a novel, in-frame fusion involving exons 1–9 of SND1 and exons 2-3′ end of BRAF gene. Through a combined analysis that began with RNA-Seq and followed by validation through RT-PCR and Sanger sequencing, we additionally utilized methods such as whole genome shot-gun sequencing, whole gene-targeted capture of the entire genomic region of both genes, as well as FISH and IHC (when possible) to confirm and to identify additional cases with this new fusion. In total, we observed the SND1-BRAF fusion in five lung adenocarcinomas from never smoker patients. Each fusion was validated by at least two independent methods using different sample preps (Summary in Table 3).
BRAF mutations such as BRAFV600E and its fusion transcripts lead to constitutive activation of its Ser/Thr kinase activity and down-stream activation of RAF/MEK/ERK pathway in lung, melanoma, thyroid, and colon cancers7,16,17,18,19. Recently, Hutchinson, et al.18 utilized a similarly integrative approach to identify new fusions involving BRAF in melanomas potentially sensitive to MEK inhibition by Trametinib using ectopically expressed BRAF fusion gene in the embryonic kidney HEK293 cells. In all reported BRAF fusions to date, the 3′ portion of the gene from exon 9 or later was involved16,17,18,19,20 and excluded the RAS binding domain (RBD) known to play a role in BRAF dimerization associated with the wild-type BRAF. In contrast, the SND1-BRAF fusion we identified in this study includes all but the first exon of BRAF. Therefore, our unique fusion is expected to retain all functional domains as the wild-type BRAF and whether it is targetable in cancer remains to be determined.
SND1 is a component of the RNA-induced silencing (RISC) complex and plays a role as a regulator for transcription of specific mRNAs through mediating RNA interference21. Overexpression of SND1 is associated with colon and prostate cancer and hepatocellular carcinoma progression22,23. Recently, Lee et al.20 reported observing a 979 AA SND1-BRAF fusion transcript between exons 1–16 of SND1 providing the promoter and exons 9–18 of BRAF in a gastric cancer cell line after treatment with MET inhibitor and provides the driver of the overexpressed BRAF in a fusion. They further demonstrated that the SND1-BRAF transcript increased phosphorylation of ERK that was susceptible to MEK inhibition20. Similarly, our functional analysis using the 1,059 AA fusion protein demonstrated constitutive BRAF activity and down-stream activation of MEK and ERK (Fig. 3A). The preservation of all but the first exon of BRAF in the SND1-BRAF fusion protein identified in this study suggests a possible biological activity similar to the wild-type BRAF. Thus, RAF and MEK inhibitors such as Vemurafenib and Trametinib needs to be evaluated for their effectiveness in tumors having the SND1-BRAF fusion7,18,19,20.
In summary, several major findings from our study are worth noting. 1) The frequency of a targetable oncogene mutation in the EGFR gene is similarly high in our cohort of NS patients of European descent compared to Asian populations1,2,3. 2) We identified a novel transcript fusion involving exons 1–9 of SND1 and exons 2 to 3′ end of the BRAF gene. 3) The fusions transcript was detected at ~3%, in five cases of the 153 (89 + 64) cases, a rate similar to that of fusions involving ALK, ROS1, and RET in lung adenocarcinoma. 4) Based on read evidence that span the fusion junction by RNA-Seq in Lu-5 (Table 2) and the relative peak heights by direct sequencing in Y59 and Y69 (Figure S3), the fusion transcript appeared to be present in a relatively small population of tumor cells compared to the > 60% tumor percentage in the primary tissues. 5) Four of the five tumors having the SND1-BRAF fusion also harbored mutations in another known driver genes suggesting a heterozygous nature of the fusion alleles in these samples (Table 3). 6) Whole genome sequencing in sample Lu-5 where sufficient DNA was available revealed complex genomic rearrangements at the region surrounding the fusion gene partners (Supplementary Fig. S2).
Finally, the relative fraction of the concurring mutations in genes known to be oncogenic within the same tumor and its clinical implications are worth further investigation. Biologically, the oncogene induced senesces is known to be associated with the active BRAF and may therefore permit only a small fraction of tumor cells to carry the fusion in most cases unless a secondary event occurs that enables the precursor tumor cells to progress through the RAS/RAF/ERK pathway via an overexpressed SND1-BRAF fusion. In this study, we identified a new SND1-BRAF fusion that appeared to be present in a subpopulation of tumor cells. Further studies will be needed to understand the biological and oncogenic implications of this novel fusion; how it contributes to tumor development and influence therapeutic outcome, particularly those based on targeted strategy.
Methods
Samples
Primary lung adenocarcinomas of never smokers (<100 cigarettes lifetime) of European descent were obtained from 89 patients who had surgery between January 1997 and September 2008. These patients were a subset of a cohort that had been previously described24 and chosen solely based on the availability of fresh frozen (FF) tissues having greater than 60% tumor cell purity (Table 1). Four cases were ALK positive based on immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH)24. Genomic DNA was extracted with the QIAamp DNA Mini Kit (Qiagen Inc, Valencia, USA) by the Biospecimen Accessioning and Processing (BAP) Core at Mayo Clinic. Total RNA was extracted using the miRNAeasy Kit. The study was approved by the Institutional Review Board of Mayo Clinic, and signed informed consent was obtained from all participants or from patients' representatives if direct consent could not be obtained. All experiments were performed in accordance with relevant guidelines and regulations, and the Institutional Review Board of Mayo Clinic had approved all study protocols.
Mutation identification by Mass Array assays
We first surveyed the performance of the commercially available OncoCartaTM Panel that interrogates 19 oncogenes and 238 mutations. We then selected for genes having a detectable mutation in at least one tested sample and used the COSMIC database and literature to include other known somatic oncogene mutation hotspots potentially targetable for therapy. As a result, our customized panel included 187 mutations of which 134 were in OncoCartaTM while 53 were new. The custom panel examines 10 oncogenes: EGFR, K-RAS, BRAF, ERBB2, MET, PIK3CA, AKT1 and AKT2, KIT, and JAK2. All primers for PCR amplification and for single base extension (SBE) were designed using the Sequenom MassARRAY Design software and synthesized using standard purification protocols (Integrated DNA Technology, Coralville, IA). Amplicons were consolidated when possible to reduce their overlap with each other after PCR amplification. Primer sequences are available upon request. Each single extension probe mixture was individually evaluated by MassArray and optimized based on the Primer Adjustment Report from the MassArray Typer Analyzer 4.0 software. All individual calls were made by the Typer Analyzer 4.0 software provided by Sequenom based on peak height at the expected molecular weight; they were then manually reviewed as previously described12. A mutation was called positive when the peak height was ≥5% of that observed for the wild type allele. MET oncogene mutations known to be germ line changes were excluded from further analysis. All 89 samples were screened independently by the custom panel and all mutations were validated by Sanger sequencing (Applied Biosystems).
Fusion transcript identification by RNA-Seq
We used RNA-Seq to analyze samples from 13 of 20 tumors without any mutation and 7 tumors with at least one mutation by the MassArray assay where sufficient total RNA were available. For each sample, 100 ng total RNA extracted from FF tumor samples were used to construct libraries using TruSeqTM v2 RNA sample preparation protocol (Illumina, San Diego). Sequencing was performed on a HiSeq 2000 instrument with a 50 cycles pair-end read. FASTQ formatted raw files for each sample were mapped and aligned in reference to hg19. Fusion transcripts were identified as previously described using SnowShoes-FTD version 2.025. A fusion was called when there were at least two reads that span the fusion junction and three reads that encompassing the two fusion transcripts26. Candidate fusion transcripts involving an oncogene were confirmed by RT-PCR and Direct sequencing.
Fusion identification by RT-PCR, direct sequencing, and Fluidigm Dynamic Array
For reverse transcription, 100 ng total RNA was used to generate cDNA using oligo(dT) primers and the Superscript III First-Strand Synthesis kit following the manufacturer's protocol (Life Technologies, Carlsbad, CA). PCR was performed using cDNA diluted by 1:10 with RNAase free water and FastStart Taq DNA polymerase (Roche, Basel, Switzerland). The products were separated on 2% agarose gels to detect potential fusion products, then gel-extracted and sequenced to determine the exact break point sequences for each fusion.
For Fluidigm Dynamic Array analysis, TaqMan probes for 5′ and 3′ partners of the identified fusion genes and controls (GAPDH, GUSB and 18S) were purchased from Life Technologies. The detection probes for identified fusion transcripts, such as SND1-BRAF, were custom designed to span fusion junctions and were obtained from both Life Technologies (Carlsbad, CA) and Integrated DNA Technologies (Coralville, IA). The sequences for all primers and probes are listed in Supplementary Table S4. Real-time PCR was performed in quadruplets using Fluidigm dynamic array as previously described27. The presence of a fusion by Fluidigm was considered when RT-PCR for the expected fusion transcript was uniformly positive by both assays (Life Technology and IDT).
Fluorescence in situ Hybridization (FISH)
Rearrangements of ROS1 (6q22) and RET (10q11) were independently detected using a laboratory developed dual-color break-apart probe (BAP) strategy probe set (FROS1 and FLRET, Mayo Medical Labs, Rochester, MN). 5′ and 3′ of probes were labeled with either green or red fluorescence, respectively. For detecting BRAF (7q34) rearrangement, bacterial artificial chromosome (BAC) clones flanking BRAF were obtained from the Children's Hospital Oakland Research Institute (Oakland, CA). DNA isolation, nick translation, and hybridization were performed as described previously28. The 5′ BRAF BACs were labeled with Green dUTP (Abbott Molecular, Des Plaines, IL) and included clones RP11-767F15, RP11-73H23, and RP11-715H9. The 3′ BAC RP11-577C22 was labeled with Orange dUTP (Abbott Molecular, Des Plaines, IL). Tumor samples were considered positive if ≥10% of 200 cells showed split signals.
Immunohistochemical staining for BRAF expression
Immunohistochemical (IHC) staining was performed using a Leica Bond III Stainer (Leica, Buffalo, IL) at the Mayo Pathology Research Core. First, tissue slides were dewaxed and retrieved for 20 minutes with Bond Dewax (Leica, Buffalo, IL) and Epitope Retrieval 2 (Leica, Buffalo, IL) reagents. Then, slides were incubated for 15 minutes with a 1:400 dilution of BRAF antibody (Clone EP152Y, recognizes 70-86AA of human BRAF, Abcam). For detection, a Polymer Refine Detection System (Leica, Buffalo, IL) was used that includes the hydrogen peroxidase block, secondary antibody polymer, DAB, and Hematoxylin. Once completed, slides were rinsed for 5 minutes in tap water, dehydrated in increasing concentrations of ethyl alcohol, and xylene-treated prior to permanent coverslipping in xylene-based media.
Whole genome shot-gun sequencing and whole gene capture sequencing
Genomic DNA from one of the tumors, Lu-5, and its uninvolved adjacent normal lung was subjected to whole genome sequencing using the shot-gun method (Complete Genomics, Inc., Mountain View, CA). Fifteen micrograms of DNA were provided and the genome was sequenced to a 40x coverage on average. The reads were mapped and reassembled by CGI tools version 1.11.0.18 to generate SNP, CNV, and structure variants relative to NCBI Build 37.1 genome29. SNPs were mapped to dbSNP release 132 and Cosmic v48 known SNPs. Concordance was greater than 98.5% in all calls when compared against a total of 2.5 × 106 SNPs. A custom designed whole gene capture kit (SureSelect, Agilent, Santa Clara, CA) that covered the entire genomic regions of both SND1 and BRAF was used to assess gene rearrangements in the targeted regions in 64 additional never smoker lung adenocarcinoma cases from the same cohort24. Up to 500 ng total DNA isolated from FFPE sections were used to prepare DNA libraries followed by targeted whole gene capture and sequencing using HighSeq 2000 with a 101 paired end reads protocol. The sequence reads were aligned to hg19 using Novoalign (version 2.08.01, http://www.novocraft.com/main/index.php), and realignment was subsequently carried out by GATK30. The structural variants of translocations were detected using CREST (v1.0) as described previously31.
Plasmid, cell culture, and transfection
pCMV6-AC-GFP plasmid containing cDNA of a wild type BRAF and SND1-BRAF fusion gene were synthesized and purchased from Origene (Rockville, MD, USA). The BRAFV600E construct was generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, CA, USA) and verified by Sanger sequencing. The wild type BRAF construct was used as a BRAFV600E mutagenesis template. The NCI-H1299 large cell lung cancer cell line with wild type of EGFR from a male Caucasian was purchased from ATCC (USA). H1299 cells were maintained in RPMI1640 (Life Technologies, USA) supplemented with 10% FBS (Life Technologies) and 1% antibiotics (Life Technologies). Transfection was performed by using Lipofectamine 2000 (Life Technologies) as instructed by the manufacturer.
Western blotting
Western blotting was performed as described32. Briefly, cells were lysed in 10 mM Tris (pH 7.4) containing 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 10 mM Na4P2O7, 10 H2O, 1 mM PMSF, and protease inhibitors (Santa Cruz). Lysates were quantified with BCA (Pierce). Ten micrograms of protein were separated with SDS-PAGE gel and transferred onto nitrocellulose membranes (Amersham,Hybond ECL). The membranes were blocked with 5% skim milk in TBS-Tween-20 and incubated with the diluted at 1:3000 anti-BRAF (#ab33899, Abcam), anti-MEK (#4694, Cell Signaling), anti-pMEK(#2338, Cell Signaling), anti-ERK (#4370, Cell Signaling), or anti-pERK(#4370, Cell Signaling) antibodies overnight at 4°C with gentle shaking. Protein bands were visualized with Pierce ECL substrates (Pierce).
Cell proliferation and spheroid formation assays
Transfected H1299 cells were trypsinized and seeded in a 96-well plate at 5,000 cells/well. After 3 days in culture, cell proliferation was assayed using CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega, USA) with measurement absorbance at 490 nm (Tecan, Switzerland). For spheroid formation assay33, transfected cells were prepared as single cells by trypsinization and cell number was adjusted at 2.5 × 104 cells/ml. Twenty microliters of single cell suspensions was dropped on the lid of a 100 mm culture dish (500 cells/drop), and the drops were placed sufficiently apart so as not to touch. The lid was then inverted onto the PBS-filled bottom dish and cultured for 5 days. The size of spheroids was measured under a microscope (Zeiss, Germany).
Additional Information
How to cite this article: Jang, J.S. et al. Common Oncogene Mutations and Novel SND1-BRAF Transcript Fusion in Lung Adenocarcinoma from Never Smokers. Sci. Rep. 5, 9755; doi: 10.1038/srep09755(2015).
Funding: Supported by funding from the American Cancer Society, the National Foundation for Cancer Research Hillsberg Lung Cancer Translational Research Grant, the Mayo Clinic Cancer Center,the Mayo Center for Individualized medicine, and start-up founds to Dr. Yanan Yang from the Mayo Clinic Foundation.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Dr. Serban San Marina for critical review and helpful revision to the manuscript; Drs. JinHui Zhang and Ji Wen for discussions regarding the genomic rearrangements surrounding the SND1 fusion genes; Dr. Wilma Lingle and Dr. Doo-Sup Choi for technical assistance, and Dr. Rodney Feddersen for the RNA-Seq library construction. We also thank Ms. Heather Flynn Gilmer and Darlene Knutson at the Mayo Cytogenetics Core for assistance and discussions with the FISH analysis.
Footnotes
: HL is an employee of Sequenom. All other authors have no competing interests exist.
Author Contributions J.S.J. performed the research, analyzed the data, and drafted the paper. A.L. helped develop the Mass Array panel. H.L. supported the MassArray primer design and assay optimization but had no role in study design or its conclusions. L.X.G., Y.Y., H.J. and H.H. contributed to the functional assays. Y.A., P.W.L. and Z.S. performed bioinformatics analysis. M.E.J., Y.S., A.O. and E.Y. performed and analyzed the FISH and IHC studies. A.B., E.E., M.C.A. and P.Y. provided all samples and clinical data used in the study. V.S. carried out the qPCR screening. J.L. helped validate all mutations by Sanger sequencing. B.E. and E.D.W. directed the NextGen Sequencing. K.K. supervised tissue sectioning and DNA extraction. D.W. and R.B.D. provided helpful discussions. J.J. designed and directed the research, reviewed the study results, and revised the manuscript.
References
- Wakelee H. A. et al. Lung cancer incidence in never smokers. J Clin Oncol 25, 472–478 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Schiller J. H. & Gazdar A. F. Lung cancer in never smokers--a different disease. Nat Rev Cancer 7, 778–790 (2007). [DOI] [PubMed] [Google Scholar]
- Subramanian J. & Govindan R. Lung cancer in never smokers: a review. J Clin Oncol 25, 561–570 (2007). [DOI] [PubMed] [Google Scholar]
- Paik P. K. et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 29, 2046–2051 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese A. M. et al. Lungs don't forget: Comparison of the KRAS and EGFR mutation profile and survival of collegiate smokers and never smokers with advanced lung cancers. J Thorac Oncol 8, 123–125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America 101, 13306–13311 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W. & Girard N. New driver mutations in non-small-cell lung cancer. The lancet oncology 12, 175–180 (2011). [DOI] [PubMed] [Google Scholar]
- Gazdar A. F., Shigematsu H., Herz J. & Minna J. D. Mutations and addiction to EGFR: the Achilles 'heal' of lung cancers? Trends Mol Med 10, 481–486 (2004). [DOI] [PubMed] [Google Scholar]
- Massarelli E. et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res 13, 2890–2896 (2007). [DOI] [PubMed] [Google Scholar]
- Pao W. et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2, e17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. S. et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer discovery 1, 44–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland J. M. et al. MET and EGFR mutations identified in ALK-rearranged pulmonary adenocarcinoma: molecular analysis of 25 ALK-positive cases. J Thorac Oncol 8, 574–581 (2013). [DOI] [PubMed] [Google Scholar]
- Forment J. V., Kaidi A. & Jackson S. P. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer 12, 663–670 (2012). [DOI] [PubMed] [Google Scholar]
- Ding L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A. et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol 23, 857–865 (2005). [DOI] [PubMed] [Google Scholar]
- Ciampi R. et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest 115, 94–101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cin H. et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta neuropathologica 121, 763–774 (2011). [DOI] [PubMed] [Google Scholar]
- Hutchinson K. E. et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 19, 6696–6702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy N. et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 16, 793–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N. V. et al. A novel SND1-BRAF fusion confers resistance to c-Met inhibitor PF-04217903 in GTL16 cells though MAPK activation. PloS one 7, e39653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Drapkin R., Yalamanchili R., Mosialos G. & Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol 15, 4735–4744 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruma H. et al. Staphylococcal nuclease domain-containing protein 1 as a potential tissue marker for prostate cancer. Am J Pathol 174, 2044–2050 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhekadur P. K. et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J Biol Chem 287, 13952–13958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi E. S. et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol 6, 459–465 (2011). [DOI] [PubMed] [Google Scholar]
- Asmann Y. W. et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res 39, e100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmann Y. W. et al. Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer research 72, 1921–1928 (2012). [DOI] [PubMed] [Google Scholar]
- Spurgeon S. L., Jones R. C. & Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PloS one 3, e1662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson-Johnson M. R. et al. Nodular fasciitis: a novel model of transient neoplasia induced by MYH9-USP6 gene fusion. Lab Invest 91, 1427–1433 (2011). [DOI] [PubMed] [Google Scholar]
- Drmanac R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010). [DOI] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods 8, 652–654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest 121, 1373–1385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland R. M. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240, 177–184 (1988). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data