Abstract
Targeting DNA repair pathways is a powerful strategy to treat cancers. To gauge efficacy in vivo, typical response markers include late stage effects such as tumor shrinkage, progression free survival, or invasive repeat biopsies. These approaches are often difficult to answer critical questions such as how a given drug affects single cell populations as a function of dose and time, distance from microvessels or how drug concentration (pharmacokinetics) correlates with DNA damage (pharmacodynamics). Here, we established a single-cell in vivo pharmacodynamic imaging read-out based on a truncated 53BP1 double-strand break reporter to determine whether or not poly(ADP-ribose) polymerase (PARP) inhibitor treatment leads to accumulation of DNA damage. Using this reporter, we show that not all PARP inhibitor treated tumors incur an increase in DNA damage. The method provides a framework for single cell analysis of cancer therapeutics in vivo.
Poly(ADP-ribose) polymerase 1 (PARP1) is a nuclear enzyme that utilizes nicotinamide-adenine-dinucleotide (NAD+) to catalyze the addition of poly(ADP-ribose) (PAR) moieties onto target proteins1,2. These bulky, negatively charged PAR polymers affect numerous cellular processes, most notably by changing the localization and activity of signaling proteins3,4,5. PARP1 promotes DNA repair by undergoing a conformational change in response to single-strand and double-strand DNA breaks6,7,8. PARylation by PARP1 then leads to the recruitment of early DNA damage response factors, such as XRCC1, among many others9,10,11,12,13,14,15. Small molecule PARP inhibitors were originally developed for combined use with DNA damaging agents upon discovery that PARP1 is activated following either ionizing radiation or treatment with DNA-methylating agents4,8,16. In addition, the synthetic lethal relationship between PARP1 and BRCA has been exploited in several clinical trials4.
Most research on the mechanism of action and failure of PARP inhibitors has been limited to cell culture or traditional pharmacodynamic (PD) assays. These PD assays include measurements on tissue samples and traditional pharmacodynamic measurements of tumor volume following repeated inhibitor treatment in mice bearing BRCA wild-type or BRCA mutant tumors17,18,19. While these approaches are important towards developing more efficient PARP inhibitors, they often do not allow one to study effects at the single cell level in vivo. For example, there are a number of questions related to the mechanism of in vivo action (PD) such as: is the mechanism of action the same for every cell within the tumor or is there heterogeneity? Are there differences in response mechanisms within tumor classes (i.e. different models of ovarian cancer)? Do host cells impact efficacy? Which combination therapies work synergistically (mechanistically within cells and between tumor and host cells)?
A critical aspect in understanding drug action in vivo are robust response markers which yield sufficiently high signal-to-noise to be imaged in real time and which can be imaged in semiautomated fashion. Here we created a double-strand break (DSB) DNA damage reporter based on truncated 53BP120 and using this approach, we measured the single-cell pharmacodynamics of olaparib in different xenografts of human tumor-derived ovarian, breast, and Ewing’s sarcoma cancers21 (Fig. 1a). Our results surprisingly show that i) DNA damage can be measured in vivo, ii) that, in the case of olaparib, in vitro measurements do not predict in vivo effects and iii) that there is no clear relationship between BRCA1 status and sensitivity to the PARP inhibitor olaparib, at least in the studied xenografts of ovarian, breast and Ewing’s sarcoma tumors. The studies described here provide a framework for testing the single-cell pharmacodynamics of other DNA damaging agents.
Figure 1. Single-cell pharmacodynamic imaging analysis of olaparib efficacy.
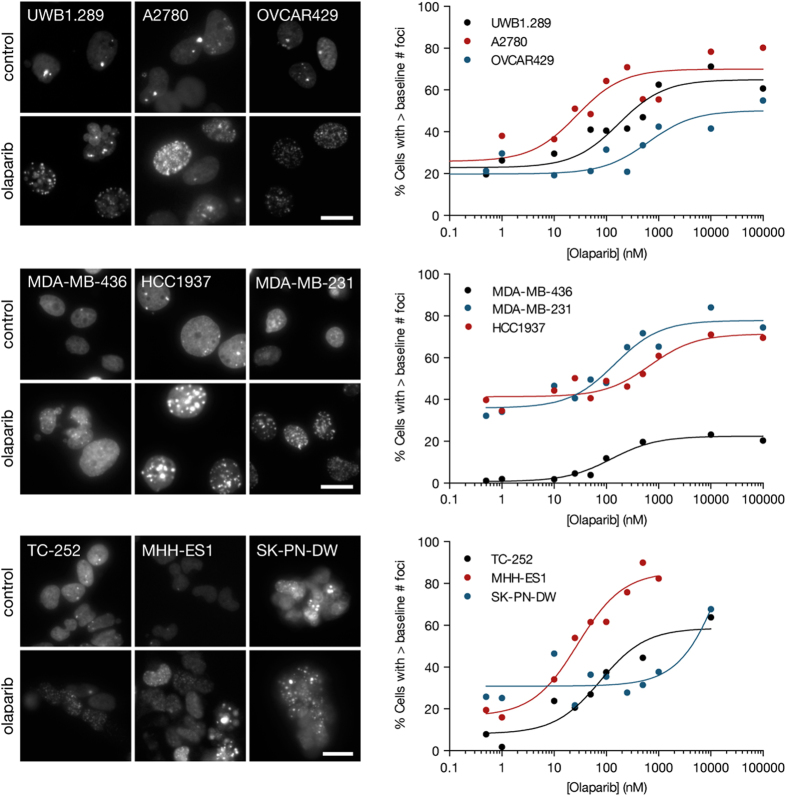
(a) Image stacks from 53BP1trunc-Apple/H2B-GFP in vivo imaging experiments were analyzed using MATLAB to determine the number of foci on a per nucleus basis. Individual non-overlapping stacks were selected from a region of tumor imaging for analysis (a, left). Nuclei of interest were identified manually (a, center, red squares) and then the number of foci per nucleus were counted (a, right). The resulting number of foci per nucleus were plotted on a single-cell basis to determine if olaparib treatment caused an increase in the number of 53BP1 foci. Scale bar = 20 μM. (b) Synthetic lethality in the context of PARPi and BRCA mutation. Treatment with PARP inhibitors leads to a block in the base excision repair pathway (BER-), which results in accumulation of single-strand DNA breaks. During cell division, these breaks accumulate as double-strand DNA breaks. One of the proteins that accumulates at sites of double-strand breaks is 53BP1 (red foci). In BRCA wild-type cells (homologous recombination positive, HR+), the double-strand breaks are repaired and the cell remains viable. However, in BRCA mutant cells (BRCA-), homologous recombination is blocked (HR-), which leads to sustained 53BP1 accumulation, unrepaired DNA damage, and eventual cell death. (c) Schematic of the 53BP1trunc-Apple (amino acids 1220–1711)20 reporter for in vivo single-cell pharmacodynamics. (d) Example of PARPi (olaparib) induced increase in 53BP1 foci in an ovarian cancer cell line (OVCAR429) after 24 hrs of 100 μM olaparib. Scale bar = 20 μM.
Results
Development of a single-cell pharmacodynamic DNA damage reporter
To develop an in vivo single-cell pharmacodynamic assay to measure DSBs following olaparib treatment, we chose 53BP1 as a DSB reporter, which has previously been used to measure DNA damage in live cells20,22,23,24,25,26(Fig. 1b). Specifically, we fused a truncated portion of 53BP1 (amino acids 1220-1711) to Apple fluorescent protein (53BP1trunc-Apple)20 (Fig. 1c). Apple fluorescent protein was used for in vivo imaging due to its increased brightness over mCherry, which is critical for successful imaging in live tissue. The truncated version of 53BP1 retains its ability to bind to sites of DSBs, but lacks the known functional domains of 53BP120 (Fig. 1d). Moreover, we show that 53BP1trunc-Apple localizes to sites of double-strand breaks with an antibody targeting the canonical marker of double-strand breaks, γH2A.X (Supplementary Fig. S1).
Relationship between BRCA1 status and PARP expression
We chose a panel of breast and ovarian cancer cell lines with either BRCA1 wild-type or mutant status, as well as several Ewing’s sarcoma cell lines that are BRCA1 wild-type, but had been shown to be sensitive to PARP inhibitors21 (Supplementary Table S1). Among these cell lines, we were first interested in whether or not BRCA1 status correlated with PARP1 expression and if PARP1 expression could therefore predict olaparib sensitivity. Western blot analysis revealed no obvious correlation between BRCA1 status and PARP expression (Supplementary Fig. S2). Some BRCA mutant cell lines had very low PARP1 expression (HCC1937), while others had high PARP1 expression (MDA-MB-436). Similarly, the wild-type cell lines ranged from low expressing (OVCAR429) to high expressing (A2780).
BRCA1 mutant cell lines are generally more sensitive to olaparib in vitro
We next compared cell viability following olaparib treatment to confirm the correlation between BRCA1 status and PARP inhibitor sensitivity. Cell lines were plated at low density and were treated with increasing concentrations of olaparib the following day. After six days treatment, viability was measured using a cell viability assay. As expected, the BRCA1 mutant cell lines generally exhibited increased sensitivity to olaparib compared to the wild-type cell lines (Table 1 and Supplementary Table S1, EC50 column; Supplementary Fig. S3), with the exception of the HCC1937 BRCA1−/− cell line, which has a relatively high EC50 of 3.7 μM. The Ewing’s sarcoma cell lines exhibited sensitivity comparable to that previously reported (MHH-ES1 is more sensitive than the SK-PN-DW cell line)21.
Table 1. Relationship between BRCA status and PARP inhibitor efficacy in breast and Ewing’s sarcoma cell lines.
| PD50 (μM) in vitro | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Model | BRCA1 | BRCA2 | PARP | EC50 (μM) in vitro | 24 hr | 48 hr | 6 day | Max fold change in vivo |
| Breast | MDA-MB-436 | −/− | WT | Hi | 0.032 | 0.15 | 0.19 | 0.05 | 3.9 |
| HCC1937 | −/− | WT | Low | 3.7 | 0.6 | 0.7 | 0.09 | 1.7 | |
| Ewing’s | MHH-ES1 | WT | WT | Hi | 0.16 | 0.02 | ND | ND | 2.5 |
Wild-type or BRCA mutant status is shown for each cell line (compiled from the COSMIC database and literature). Relative PARP expression is qualitatively compared from the Western blot in Supplementary Fig. S2. The EC50 value from viability assays in cell culture is also shown. The PD50 value (half-maximal pharmacodynamic response) was determined from imaging and quantifying the increase in foci formation following olaparib treatment and fitting the resulting curve to a dose response using GraphPad (Prism) (ND = not determined). The in vivo maximum fold change was determined by comparing the number of foci from pre-treatment and after daily IP injections of olaparib.
53BP1 foci generally increase more in BRCA1 mutant and Ewing’s sarcoma cell lines
To determine if DSB accumulation occurs in cell culture following olaparib treatment, all the cell lines listed in Table 1 and Supplementary Table S1 were transduced with lentivirus containing the 53BP1trunc-Apple pharmacodynamic reporter. Olaparib induction of DSBs was first measured in culture by single-cell imaging. Each cell line was plated and the next day, increasing concentrations of olaparib were added. Cells were then imaged at 24 hrs (or serially in selected experiments) following olaparib treatment (Fig. 2). For each cell line, the percent increase in 53BP1trunc-Apple foci was then determined as a function of time and dose. Drug responses were compared by determining the half-maximal pharmacodynamic response (PD50; percent increase in foci/nucleus) as a function of olaparib dose. Interestingly, the PD50 values - a measure of DSB effect - do not always correlate with the EC50 values, a measure of cell kill (Table 1, Supplementary Table S1, and Fig. 3A). For example, the A2780 ovarian cancer cell line had a high EC50 value, but a very low PD50 value. This is likely the case because the DSBs that appear after 24 hrs of olaparib treatment are later repaired rather than the cells undergoing cell death (Supplementary Table S1).
Figure 2. Single-cell pharmacodynamic imaging in cell culture.
The indicated cell lines from ovarian, breast, or Ewing’s sarcoma were treated with increasing concentrations of olaparib for 24 hrs, at which point the cells were imaged for 53BP1trunc-Apple foci (left). The percentage of cells with greater than the baseline (DMSO control) number of foci was plotted versus olaparib concentration and fit to a sigmoidal dose response curve using Prism (GraphPad) to obtain PD50 values (Table 1 and Supplementary Table S1). Data are representative of at least two independent experiments. Scale bar = 20 μM.
Figure 3. Correlation between pharmacodynamic parameters.
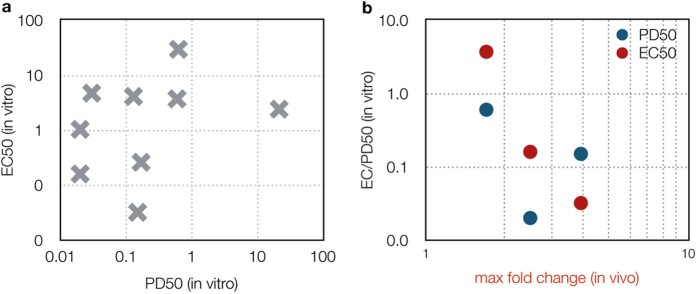
(a) The half-maximal pharmacodynamic response (PD50) from in vitro experiments was plotted versus the EC50 value from viability assays for the cell lines listed in Supplementary Table S1. (b) The maximum fold change in foci formation in vivo (x-axis) is plotted versus the half-maximal pharmacodynamic response (PD50, blue) or versus the EC50 (red) from viability assays.
In contrast to what the synthetic lethal model would predict for PARP inhibitors, the HCC1937 BRCA1−/− cell line had a high EC50 and PD50 value. We thus asked if this BRCA1 mutant cell line is either insensitive to olaparib or whether it simply required longer drug incubation to observe an increase in DSBs? To address this question, 53BP1trunc-Apple foci formation was imaged and analyzed again at 48 hrs and 6 days after olaparib treatment (this was done for both the HCC1937 and MDA-MB-436 cell lines). Not surprisingly, the PD50 values remain the same after 48 hrs of olaparib treatment (Table 1). However, after 6 days olaparib treatment, the PD50 values decreased 3-fold for the MDA-MB-436 cells and 7-fold for the HCC1937 cells (Table 1). The increase in DSBs (and resulting decrease in PD50) after 6 days of olaparib treatment is consistent with a need for the cells to go through several rounds of cell division to accumulate sufficient numbers of DSBs, particularly at lower concentrations of olaparib. However, for the BRCA1−/− HCC1937 cell line, the increase in DSBs at 6 days (PD50) still did not translate to a lower EC50 value. It is possible that in this cell line, the additional time required to accumulate DSBs translates into additional time required to observe a decrease in cell viability. Another possibility is that DNA repair pathways in the HCC1937 cell line still function at levels sufficient to repair the damaged DNA. In general, the data in Table 1 and Supplementary Table S1 show that the BRCA mutant cell lines and the expected Ewing’s sarcoma cell lines were sensitive to olaparib.
Development of an in vivo model system to image 53BP1 DNA damage
Ultimately our interest was in measuring DSB accumulation in vivo. As an initial validation of the 53BP1trunc-Apple reporter in vivo, we analyzed the number of foci per nucleus in HT1080 xenografts before and after vehicle or cisplatin (a robust DNA damaging agent) treatment. Supplementary Figure S4 shows that cisplatin treatment resulted in a statistically significant increase in the mean number of foci per nucleus, indicating the 53BP1trunc-Apple reporter functions as expected in vivo.
To examine the effects of olaparib in vivo we established xenograft tumors of cells stably expressing the 53BP1trunc-Apple reporter and imaged these tumors at high spatial resolution to resolve the nuclear phenotype of DSB. We focused on three representative models in which serial measurements were carried out at different doses (50 or 100 mg/kg olaparib): MDA-MB-436 (breast cancer), HCC1937 (breast cancer), and MHH-ES1 (Ewing’s sarcoma). Tumors were imaged daily for the first week and then every few days thereafter until imaging was no longer feasible. A MATLAB script was utilized for semi-automated analysis of the single cell data (Fig. 1A). Nuclei regions of interest were identified manually and then the number of foci per nucleus was counted for 200–600 single cells from each tumor for each day of imaging (Fig. 4).
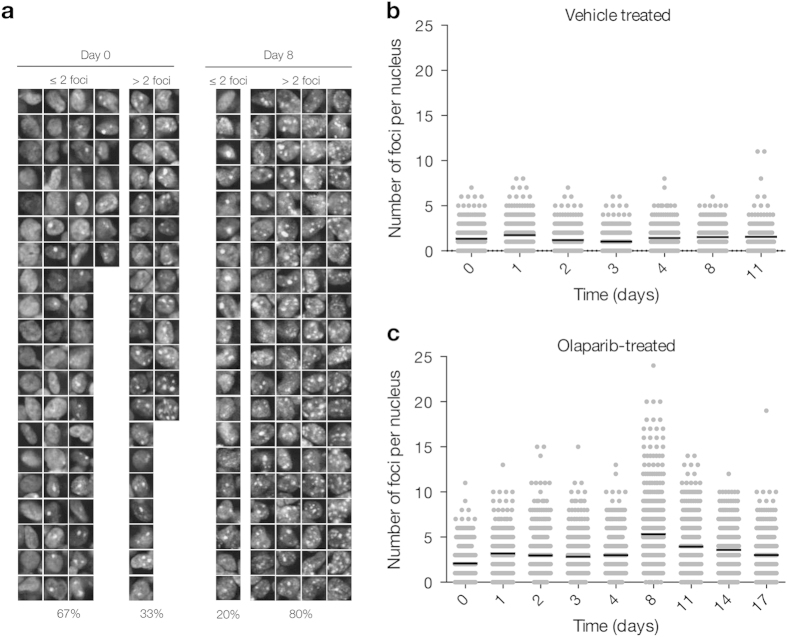
Figure 4. In vivo single-cell pharmacodynamic analysis of 53BP1 double-strand break formation following olaparib treatment in Ewing’s sarcoma tumors.
(a) Representative images of single cells from an MHH-ES1 tumor prior to treatment and after eight days of 100 mg/kg Olaparib. Images are separated into ≤2 foci per nucleus (2 is the average number of foci per nucleus pre-treatment) and >2 foci per nucleus. Percentage values at the bottom represent the percentage of the total number of nuclei that fall into either of these groups. (b and c) Single cell data for a nu/nu mouse with a Ewing’s sarcoma tumor xenograft (MHH-ES1 cells) that was treated for the duration of imaging with vehicle alone (b, 10% DMAC/Solutol in PBS) or with 100 mg/kg Olaparib (c). Note for b and c, each point represents the number of foci in a single nucleus, with 200–600 single cells analyzed for each day of imaging. The black line represents the mean number of foci per nucleus for each day of imaging.
Ewing’s sarcoma MHH-ES1 tumors exhibit more DNA damage following olaparib treatment
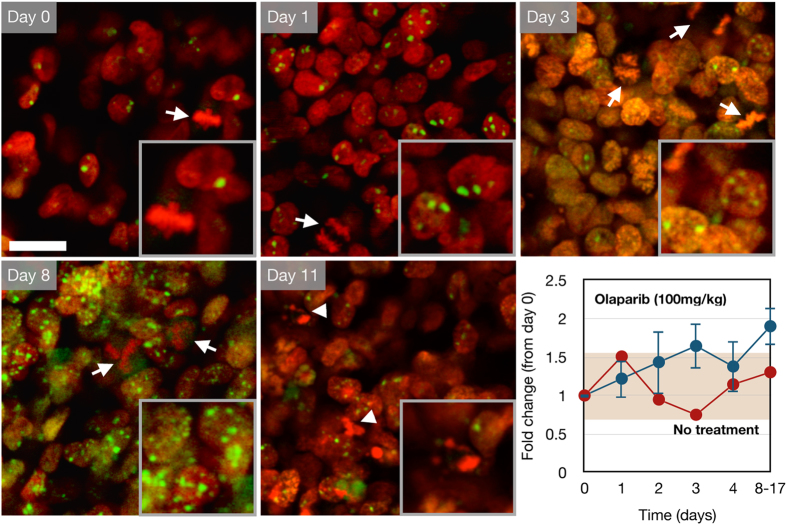
Figure 4 summarizes a set of experiments in the MHH-ES1 model. Examples of the regions of interest for the cells that were imaged each day is shown. Analysis of the fold change in number of foci per nucleus from pre-treatment (day 0) showed a gradual increase with a maximal fold change by day 8 of 100 mg/kg olaparib treatment (Fig. 4 and 5 and Table 1). Importantly, a tumor treated with vehicle (DMSO alone in DMAC/Solutol and PBS) did not show an increase in the average number of foci over time (Fig. 4B and Fig. 5). Interestingly, 50 mg/kg olaparib did not show any effects, but the higher dose of 100 mg/kg did (Fig. 6A and B). These data suggest that there is a dose dependence in the appearance of a pharmacodynamic DSB response. The MHH-ES1 cells in this particular tumor were also transfected with an H2B-GFP reporter that allows visualization of dividing cells, as well as cells that have recently undergone apoptosis27. Both of these events are crucial to the efficacy of olaparib treatment. Cell division indicates the ability of cells to accumulate double-strand DNA damage from the olaparib-induced single-strand DNA damage. This phenomenon was observed in the MHH-ES1 cell line following olaparib treatment (Fig. 5). Using the H2B-GFP reporter, we also observed the ultimate apoptotic effects of olaparib (Fig. 5).
Figure 5. Serial in vivo imaging of a nu/nu mouse with a MHH-ES1 Ewing’s sarcoma tumor expressing the 53BP1trunc-Apple (green) and H2B-GFP (red) reporter.
The mouse was dosed daily with 100 mg/kg olaparib by IP injection (blue curve, n = 3) or vehicle (10% DMAC/Solutol in PBS, n = 1). Days 8–17 indicate pooled data from mice imaged on different days after olaparib treatment. Dividing cells (arrow) and apoptotic cells (arrowhead) were observed using the H2B-GFP (red) reporter. Error bars represent the standard error of the mean. Scale bar = 20 μm (inset magnified 1.7-fold).
Figure 6. Effect of olaparib concentration on MHH-ES1 Ewing’s sarcoma tumors expressing the 53BP1trunc-Apple reporter in nu/nu mice.
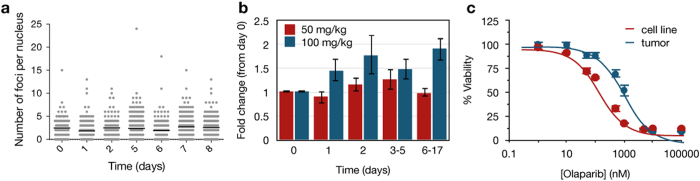
(a) Representative single-cell data showing the number of foci on a per nucleus basis for a mouse treated daily with 50 mg/kg olaparib (IP). Each point indicates the number of foci in a single nucleus, with black lines on each day representing the mean number of foci. (b) Comparison of the fold change in the number of foci for the 50 mg/kg (red, n = 2) and 100 mg/kg (blue, n = 3, same data as in Fig. 5 for comparison) olaparib doses (relative to day 0 prior to olaparib treatment). Error bars represent the standard error of the mean. (c) Comparison of cell viability following treatment with increasing concentrations of olaparib for 6 days in MHH-ES1 cells grown in culture alone (red) or grown in vivo and then dissociated and re-grown in cell culture at the endpoint of olaparib treatment/imaging (blue). Data were fit to a sigmoidal dose-response curve using Prism (GraphPad), with error bars representing the standard error of the mean.
At the end point of imaging, MHH-ES1 tumors treated with 100 mg/kg olaparib were isolated and the cells were dissociated and re-grown in vitro. Using these cell lines established from tumors treated with olaparib, we repeated the viability assays described earlier. Compared to the cell lines before growth in vivo, the tumor cell lines displayed a 6-fold increase in EC50 (Fig. 6C). These data suggest that growth and exposure to olaparib in vivo fundamentally alters the tumor cells, making them less sensitive to the inhibitor.
MDA-MB-436 tumors show increased 53BP1 foci after olaparib treatment
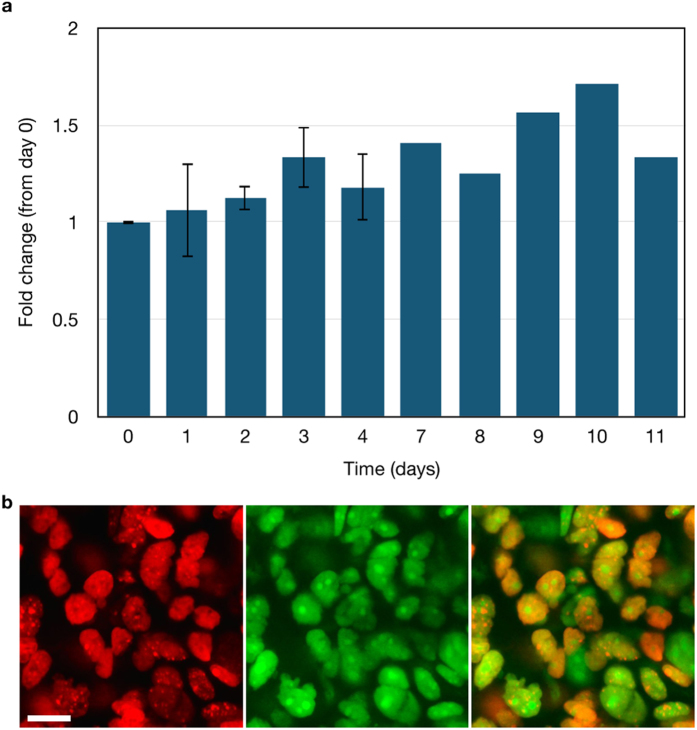
Similar analysis as above for Ewing’s sarcoma was also done for MDA-MB-436 BRCA1−/− tumors expressing the imaging reporter (Fig. 7). An increase in the number of foci per nucleus was observed by 21 days following daily olaparib treatment (Fig. 7 and Table 1). Both doses (50 and 100 mg/kg) of olaparib ultimately led to ~4-fold increase in the number of foci per nucleus. Olaparib-treated tumors were also irradiated with 10 gray at the experimental end point to demonstrate that the 53BP1trunc-Apple reporter was indeed functional, which was confirmed (Fig. 7A, IR).
Figure 7. In vivo quantification of the number of foci per nucleus in MDA-MB-436 cells expressing the 53BP1trunc-Apple reporter in a nu/nu mouse.
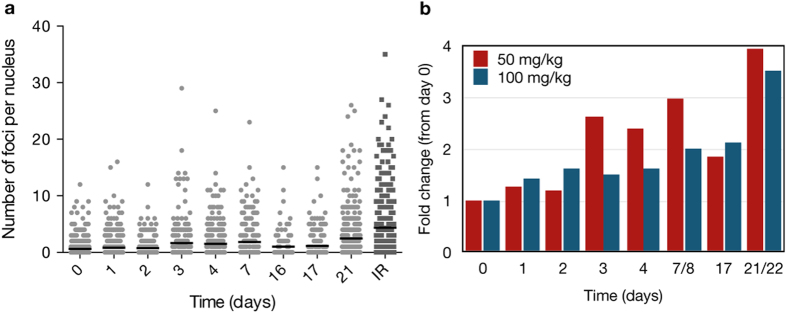
(a) Representative single cell data from a mouse treated with daily IP injections of 50 mg/kg olaparib. Each point represents an individual nucleus in vivo, with the y-value equal to the number of foci in that nucleus. 200–600 cells were analyzed each day following olaparib treatment and the black bar represents the average number of foci on each day. (b) Comparison of the fold change in the average number of foci per nucleus for daily IP injection of 50 mg/kg (n = 1) versus 100 mg/kg olaparib (n = 1).
HCC1937 BRCA mutant tumors do not undergo increased 53BP1 foci in vivo after olaparib treatment
Finally, two different HCC1937 BRCA1−/− tumors were studied, each dosed daily with 100 mg/kg olaparib (Fig. 8A). While the maximal fold change was slightly higher in cell culture for this line (~2.5-fold at 6 days olaparib treatment, data not shown), the single-cell tumor imaging showed essentially no fold change from pre-treatment over the 11 day treatment period. This lack of change is also consistent with the viability data for this cell line, which showed relatively poor sensitivity to olaparib (Table 1). The lack of a pharmacodynamic response from the HCC1937 cell line in vivo is not due to poor pharmacokinetics of olaparib on the tumor. Using a previously established olaparib-BODIPY FL companion imaging drug28 and an HCC1937 53BP1trunc-Apple tumor, we observed uptake of the drug in all the tumor cell nuclei (Fig. 8B). This suggests that not all BRCA1 mutant tumors undergo a DNA damage response following olaparib treatment. However, it is possible that the BRCA1 mutant tumor would shrink and respond to olaparib, perhaps through a different and yet unrealized mechanism.
Figure 8. In vivo quantification of the fold change in foci formation in HCC1937 53BP1trunc-Apple cells grown in nude mice.
(a) The fold change in the number of foci was determined relative to day 0 of treatment with 100 mg/kg olaparib. Error bars represent the standard error of the mean for n = 2 mice. (b) Olaparib-Bodipy FL nuclear uptake in a nu/nu mouse with an HCC1937 tumor. The 53BP1trunc-Apple reporter (red nuclei, left) was imaged 2 hrs after IV administration of olaparib-Bodipy FL (green, center). Images from the 53BP1trunc-Apple (red) channel and the olaparib-Bodipy FL (green) channel were overlaid (merge, right) to show that olaparib-Bodipy FL accumulates in all HCC1937 tumor cell nuclei. Scale bar = 20 μm.
Discussion
The synthetic lethal relationship between PARP1 and BRCA has been the firm basis of designing recent clinical trials4. These trials showed promising results early on in BRCA deficient breast and ovarian cancer with the inhibitor olaparib29. Based on these early advancements, the target patient population was expanded to include those that displayed BRCAness, where BRCA1 and BRCA2 are intact, but other HR defects exist. Iniparib showed early promise in a phase 2 trial in triple-negative breast cancer (TNBC) patients that had a BRCAness signature30. However, in a phase 3 trial, iniparib failed in the same patient population and at the same time olaparib showed no overall benefit in TNBC patients or overall survival in ovarian cancer patients31,32,33. While PARP inhibitor trials were temporarily halted, more recent studies show that iniparib was not a true PARP inhibitor34,35. Moreover, olaparib was reevaluated based on BRCA status, with results showing overall survival in patients with defective BRCA protein. Despite the sound scientific rationale36,37,38, extensive in vitro assay and preclinical studies17,18,19,39,40,41, the clinical results were mixed29,30,31,32,33,42,43,44. Furthermore, previous studies using fluorescent olaparib had shown that the drug clearly distributes throughout tumors and has a unique nuclear phenotype of accumulation in vivo28. Finally, mathematical modeling had shown that the human doses would clearly result in efficacy in cancer cells28. So, how can these discrepant results be explained?
In order to gain insight into the pharmacologic actions of PARP inhibitors, we have created tool sets to measure pharmacokinetics28 and now also pharmacodynamics at the single cell level in vivo and in real-time. The latter was accomplished by creating a sufficiently bright fluorescent 53BP1 reporter for confocal/multiphoton in vivo imaging. This reporter shows a remarkable phenotype (Fig. 1D) and allows one to derive quantitative measures of drug efficacy (EC50: concentration at which half of the cells die, PD50: concentration at which half of the cells show DNA damage in vitro, in vivo: maximum fold increase in DNA damage relative to pre-treatment). Our data in cell culture shows that the 53BP1trunc-Apple reporter generally agrees with the viability data for each cell line, with the major exception being the HCC1937 cell line. This line is BRCA1 mutant and has a low PD50 value after 6 days of olaparib treatment, indicating a strong increase in DSBs, but a high EC50 value over the same time frame, which indicates the cells do not die readily in response to olaparib. Perhaps this is owing to the relatively low expression of PARP1 in these cells compared to the other more responsive cell lines.
In vivo we show with three different tumor models, two of which are BRCA1 mutant, that there is a difference in the number of DSBs observed depending on the daily dose of olaparib. For one of the cell lines (MHH-ES1), a lower dose of olaparib (50 mg/kg) did not increase the number of DSBs. However, with a dose of 100 mg/kg a significant number of DSBs were observed. Surprisingly, the MDA-MB-436 cell line showed the greatest increase in the number of 53BP1trunc-Apple foci, even at the 50 mg/kg olaparib dose. This is in contrast to the cell culture data, which suggested that while the MDA-MB-436 cell line displayed high sensitivity to olaparib in a viability assay, there was only a moderate change in 53BP1trunc-Apple foci. Similarly, the HCC1937 BRCA1 mutant cell line showed little change in the number of foci in vivo, even at the higher 100 mg/kg daily dose of olaparib.
With certain caveats, the experiments of this study suggest two main conclusions: i) often there is no clear relationship between BRCA1 status and sensitivity to olaparib and ii) in vitro cell culture data do not predict in vivo efficacy for the PARP inhibitor studied here. While the MDA-MB-436 cell line is sensitive in vivo, the HCC1937 cell line is not, despite both lines being BRCA1−/−. This suggests that if HCC1937 tumors still decrease in size, it is likely independent of the synthetic lethal concept. PARP1 has a plethora of other functions in the cell ranging from promotion of transcription to chromatin modification to changes in signaling pathways. Modification of any of these pathways/proteins through PARP1 inhibition could in theory affect tumor growth. Recent studies demonstrate that olaparib induces a G2 phase arrest-like state, resulting in upregulation of p53 and p2145,46,47. Furthermore, these studies suggest that olaparib is most effective in cell lines with mutated p53. Here, the cell lines studied in vivo all have mutant p53 status48 (and canSAR database). Interestingly, the HCC1937 cell line has strong basal expression of p21, while the MDA-MB-436 and MHH-ES1 cell lines do not (Supplementary Fig. S5). Thus, it is possible that the HCC1937 cell line arrests during the cell cycle and does not proliferate at the same rate as the MDA-MB-436 and MHH-ES1 cell lines in vivo. Replication is essential to PARP inhibitor function in the context of synthetic lethality (Fig. 1B), suggesting that cell lines that may proliferate more slowly (such as HCC1937) may be less sensitive to olaparib. We also show that the Ewing’s sarcoma tumor is sensitive to olaparib in vivo, consistent with previous cell culture data21. These data also highlight the importance of in vivo measurements of single-cell pharmacodynamics. The cell culture data would predict that all the cell lines we tested in vivo would show an increase in DSBs following olaparib treatment, which is clearly not the case. We believe that the methodology we have established here to measure DSBs will be applicable not only to PARP inhibitors, but to other agents that also induce DSBs. Similar types of reporters could also be established for other cell processes so that one can gain a more complete view of drug action in vivo.
Materials and Methods
Cell lines and growth conditions
The UWB1.289, MDA-MB-436, MDA-MB-231, and SK-PN-DW cell lines were purchased from the American Type Culture Collection (ATCC). A2780 and OVCAR429 cell lines were kind gifts from Dr. Michael Birrer (Massachusetts General Hospital, Boston, MA). The HCC1937 cell line was a kind gift from Prof. Timothy Mitchison (Department of Systems Biology, Harvard Medical School, Boston, MA). The TC-252 cell line was a kind gift from Dr. Francis Hornicek (Massachusetts General Hospital, Boston, MA). MHH-ES1 cells were purchased from CLS Cell Lines Service (order # 300136).
MDA-MB-436, MDA-MB-231, A2780, OVCAR429, MHH-ES1, and HCC1937 cells were maintained in RPMI supplemented with 10% fetal bovine serum, 100 IU penicillin, 100 μg/ml streptomycin, and L-glutamine. UWB1.289 cells were maintained in 50% RPMI, 50% MEGM (consisting of MEBM basal medium and SingleQuot aliquots, excluding gentamycin-amphotericin B; Lonza CC-3150), 3% fetal bovine serum, 100 IU penicillin, 100 μg/ml streptomycin, and L-glutamine. SK-PN-DW, TC-252, and HT1080 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 IU penicillin, 100 μg/ml streptomycin, and L-glutamine. Cells expressing 53BP1trunc-Apple were maintained in 3 μg/ml puromycin and cells co-expressing H2B-GFP were also maintained in 200 μg/ml G418.
Constructs
Truncated 53BP1 (amino acids 1220–171120) was PCR amplified from 53BP1-YFP (kind gift from Prof. Galit Lahav, Harvard Medical School, Boston, MA). The PCR product was cloned into the pLVX lentiviral vector (632562, Clontech) containing a C-terminal Apple fluorescent protein (27698, Addgene), using In-Fusion cloning (639645, Clontech) and the BsrGI and XbaI restriction enzyme sites on the pLVX vector. The 53BP1trunc-Apple insert was sequenced in its entirety.
The H2B-GFP reporter was created by replacing Apple fluorescent protein in pTag-H2B-Apple49 (FP176, Evrogen) with pAcGFP1 (632492, Clontech). pTag-H2B-Apple was digested using the AgeI and NotI restriction enzymes, while pAcGFP1-HyG-C1 was digested using the AgeI and BspOMI restriction enzymes. pAcGFP1 was then ligated into the pTag-H2B vector and the resulting insert was sequenced in its entirety.
Transfection and lentiviral infection
Lentiviral particles of the 53BP1trunc-Apple reporter were produced in Lenti-X 293T cells (632180, Clontech) following the manufacturers’ instructions. Briefly, 2 × 106 Lenti-X 293T cells were plated in a 10 cm dish. The following day, cells were transfected with the pLVX 53BP1trunc-Apple reporter using Xfect (631317, Clontech). After 6 hrs, the media was changed to remove transfection complexes. 48 hrs after transfection the media was collected and filtered through 0.45 μm cellulose acetate. Viral particles were then concentrated using Lenti-X concentrator (631231, Clontech). Cells were plated at 50,000 cells per 12-well dish and the following day were transduced with 53BP1trunc-Apple lentiviral particles in media containing 4–10 μg/ml polybrene (depending on the cell line). Media was exchanged after 24 hrs and cells were split into media containing 3 μg/ml puromycin for selection after 48 hrs. Reporter expression was confirmed by fluorescence microscopy and cells with uneven expression were sorted by flow cytometry.
For co-expression of H2B-GFP, cells were plated 24 hrs before transfection and the following day, the pTag H2B-GFP plasmid was transfected into cells using Lipofectamine 2000 (11668-027, Life Technologies). Transfected cells were split into media containing 500–1000 μg/ml G418. Cells positive for both 53BP1trunc-Apple and H2B-GFP were obtained by FACS or by isolating individual colonies after selection.
Western blot
Cells were grown to confluence, washed twice with ice-cold PBS and then lysed in radioimmunoprecipitation buffer (RIPA, 9806S, Cell Signaling Technology) containing HALT protease inhibitor cocktail (87786, Pierce). Lysates were transferred to microfuge tubes, passed through a 23 g syringe, and incubated five minutes on ice. Lysates were sonicated for 40 s and then centrifuged at 14,000 × g for 15 min at 4 °C to remove cellular debris. Total protein was measured using the BCA assay (23227, Pierce) and equal protein was loaded on a 4–12% NuPAGE Bis-Tris gel (NP0322BOX, Life Technologies). Protein was then transferred to nitrocellulose and stained for total protein (for normalization) using the reversible protein stain kit for nitrocellulose membranes (24580, Pierce). Following staining and documentation, the stain was removed and the blot was blocked in SuperBlock T20 (TBS) (37536, Pierce), followed by brief washing in TBS containing 0.1% Tween-20 (TBST). Blots were incubated overnight at 4 °C in PARP1 (9532, Cell Signaling Technology), p53 (2527, Cell Signaling Technology), or p21 (2947, Cell Signaling Technology) primary antibody diluted 1:1000 in 10% SuperBlock/TBST. Blots were washed three times, 5 min each, followed by a one hour incubation in HRP-conjugated secondary antibody. Following secondary incubation, cells were again washed three times, 5 min each in TBST followed by detection using SuperSignal West Pico chemiluminescent substrate (34077, Pierce). PARP1 expression was quantified by densitometry using ImageJ (NIH) and normalized using the total protein stain and densitometry of ~80 kDa band in each lane. PARP1 expression was quantified from two independent experiments, with error calculated as standard error of the mean.
PrestoBlue viability assay
Cells were plated at 1000 to 3000 cells per well (depending on the cell line) 24 hrs prior to addition of olaparib (S1060, Selleckchem). Increasing concentrations of olaparib (0 to 100 μM) were added to each well for six day incubation. Cell viability was measured on the sixth day using the PrestoBlue cell viability reagent (A13261, Life Technologies) and a Tecan Safire2 plate reader in fluorescence mode (Männedorf, Switzerland). Data were plotted as percent viability versus olaparib concentration and the resulting curve was fit to a sigmoidal dose response equation using GraphPad (Prism). Error was calculated as standard error of the mean between triplicate measurements. Data are representative of at least two independent experiments.
Olaparib 53BP1trunc-Apple live cell imaging
Cells were plated at 2000 to 10,000 cells per well (depending on the number of days of olaparib treatment) in 96-well plates for microscopy (89626, iBidi). After 24 hrs, increasing concentrations of olaparib (0–100 μM) were added to the cells. Imaging was done on a DeltaVision (Applied Precision/GE Healthcare) microscope equipped with a 40X objective and a humidified environmental chamber 24 hrs after addition of olaparib (as well as 48 hrs and 6 days for select cell lines).
For colocalization with γH2A.X, cells were fixed with 4% paraformaldehyde after treatment for 24 hrs with 100 μM olaparib or 1 μM etoposide or 1 hr with 10 μM cisplatin (followed by 24 hr recovery). Cells were permeabilized using 100% MeOH, washed with PBST (PBS plus 0.1% Tween-20), blocked with odyssey blocking buffer (Li-COR), and incubated overnight at 4 °C in primary γH2A.X (phospho S139, abcam, ab26350) antibody diluted 1:100 in odyssey blocking buffer. Cells were washed with PBST and incubated 1 hr at room temperature in AlexaFluor 647 goat anti-mouse secondary antibody (1:100, Life Technologies). Cells were then imaged using a DeltaVision microscope equipped with a 40X objective.
In vivo single-cell pharmacodynamic imaging
All animal experiments were carried out in accordance with guidelines from the Institutional Subcommittee on Research Animal Care. Female nude mice (8–10 weeks old, Cox7, Massachusetts General Hospital) were implanted in the dorsal skinfold/subcutaneously with 2–4 × 106 cells that were suspended 1:1 in Hank’s balanced salt solution (HBSS) and Matrigel (356237, BD Biosciences). Tumors were allowed to grow (2 weeks to 2 months depending on the cell type) until they became vascularized and reached 1–2 mm in size. At this point, mice were surgically implanted with a dorsal skin fold window chamber. For the HT1080 experiments, window chambers and cells were implanted at the same time (without matrigel). Prior to olaparib treatment, mice were anesthetized with 2% isoflurane in 2 L/minute oxygen on a heated microscope stage. Day 0 images were collected from various regions of the tumor on an FluoView FV1000 confocal microscope (Olympus) equipped with a 20X water immersion objective. Mice were then treated daily by IP injection with 50 mg/kg olaparib (80 mg/ml stock in DMSO diluted in PBS with 10% DMAC/Solutol) or on a cycle of 5 days dosing, 2 days rest with 100 mg/kg olaparib. Initially, image stacks were collected daily and then every few days during olaparib treatment. For the HT1080 experiments, Cisplatin was injected as a single IV dose of 9 mg cisplatin per kilogram (in 10% solutol in saline)
Olaparib-Bodipy FL imaging in a nude mouse with a HCC9137 53BP1trunc-Apple tumor was done as previously described28. Briefly, 75nmol (7.5 μl of 10 mM stock in DMSO) olaparib-Bodipy FL was diluted in 30 μl of a 1:1 dimethylacetamide (DMAC): solutol solution. PBS (112.5 μl) was then slowly added with sonication to obtain a final injection volume of 150 μl. Olaparib-Bodipy FL was injected via a tail vein catheter.
Live cell and in vivo pharmacodynamic reporter analysis
Live cell imaging data and maximum intensity projections of select stacks from in vivo imaging were analyzed in 2D. Briefly, an in house produced MATLAB code was constructed to assist with foci counting. In this program, single cells were manually selected. Base level noise for the selected area was automatically filtered via Otsu’s method for thresholding. Foci were then automatically detected via incorporation of an automated algorithm for detecting and filtering local intensity clusters50, freely available from the Danuser group51. Results were verified by eye in each case. The average baseline number of foci prior to olaparib treatment was determined and the fold increase in foci formation (relative to day 0) was calculated for each concentration of olaparib (in vitro) or dose of olaparib (in vivo). The MATLAB code used to aid in image analysis is available upon request.
Author Contributions
K.Y. and R.W. wrote the main manuscript text and prepared Figures 1–8 and Table 1. K.Y. and R.K. collected the data. K.Y., M.L. and R.W. analyzed and interpreted the data. R.G. and M.L. provided an automated analysis method. All authors reviewed the manuscript.
Additional Information
How to cite this article: Yang, K. S. et al. Single cell resolution in vivo imaging of DNA damage following PARP inhibition. Sci. Rep. 5, 10129; doi: 10.1038/srep10129 (2015).
Supplementary Material
Acknowledgments
We would like to thank Alex Zaltsman for assistance in DeltaVision imaging and Matt Sebas for assistance in mouse surgical procedures. This study was supported by the following grants from the NIH: P50 CA086355 and R01CA164448. K.Y. was supported by T32 CA 79443.
References
- Kameshita I., Matsuda Z., Taniguchi T. & Shizuta Y. Poly (ADP-Ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. J Biol Chem 259, 4770–4776 (1984). [PubMed] [Google Scholar]
- Jagtap P. & Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov 4, 421–440 (2005). [DOI] [PubMed] [Google Scholar]
- D’Amours D., Desnoyers S., D’Silva I. & Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342, 249–268 (1999). [PMC free article] [PubMed] [Google Scholar]
- Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H. & Poirier G. G. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10, 293–301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B. A. & Kraus W. L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13, 411–424 (2012). [DOI] [PubMed] [Google Scholar]
- Juarez-Salinas H., Sims J. L. & Jacobson M. K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature 282, 740–741 (1979). [DOI] [PubMed] [Google Scholar]
- Benjamin R. C. & Gill D. M. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem 255, 10493–10501 (1980). [PubMed] [Google Scholar]
- Durkacz B. W., Omidiji O., Gray D. A. & Shall S. (ADP-ribose)n participates in DNA excision repair. Nature 283, 593–596 (1980). [DOI] [PubMed] [Google Scholar]
- Gagne J. P. et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36, 6959–6976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16, 923–929 (2009). [DOI] [PubMed] [Google Scholar]
- Kraus W. L. New functions for an ancient domain. Nat Struct Mol Biol 16, 904–907 (2009). [DOI] [PubMed] [Google Scholar]
- Ahel D. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A. J. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA 106, 13770–13774 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M. et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol 18, 3563–3571 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy S. F., Masutani M., Suzuki H. & Caldecott K. W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 31, 5526–5533 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori T., Yamanaka H. & Carson D. A. Poly(ADP-ribose) polymerase inhibits DNA synthesis initiation in the absence of NAD. Biochem Biophys Res Commun 163, 1113–1118 (1989). [DOI] [PubMed] [Google Scholar]
- Rottenberg S. et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA 105, 17079–17084 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann U. et al. Tumor growth inhibition by olaparib in BRCA2 germline-mutated patient-derived ovarian cancer tissue xenografts. Clin Cancer Res 17, 783–791 (2011). [DOI] [PubMed] [Google Scholar]
- Drew Y. et al. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J Natl Cancer Inst 103, 334–346 (2011). [DOI] [PubMed] [Google Scholar]
- Dimitrova N., Chen Y. C., Spector D. L. & de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S. & Boulton S. J. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15, 7–18 (2014). [DOI] [PubMed] [Google Scholar]
- Pryde F. et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci 118, 2043–2055 (2005). [DOI] [PubMed] [Google Scholar]
- Lukas C. et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13, 243–253 (2011). [DOI] [PubMed] [Google Scholar]
- Zgheib O., Pataky K., Brugger J. & Halazonetis T. D. An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol Cell Biol 29, 1050–1058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam K., Kafri R., Loewer A. & Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell 47, 320–329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J. D. et al. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res 71, 4608–4616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber G. M. et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat Commun 4, 1504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy J. et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med 364, 205–214 (2011). [DOI] [PubMed] [Google Scholar]
- Gelmon K. A. et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol 12, 852–861 (2011). [DOI] [PubMed] [Google Scholar]
- Kaye S. B. et al. Phase II, open-label, randomized, multicenter study comparing the efficacy and safety of olaparib, a poly (ADP-ribose) polymerase inhibitor, and pegylated liposomal doxorubicin in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer. J Clin Oncol 30, 372–379 (2012). [DOI] [PubMed] [Google Scholar]
- Ledermann J. et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 366, 1382–1392 (2012). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. Iniparib nonselectively modifies cysteine-containing proteins in tumor cells and is not a bona fide PARP inhibitor. Clin Cancer Res 18, 510–523 (2012). [DOI] [PubMed] [Google Scholar]
- Patel A. G., De Lorenzo S. B., Flatten K. S., Poirier G. G. & Kaufmann S. H. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res 18, 1655–1662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata C. M. & O’Shaughnessy J. Poly (ADP-ribose) polymerase as a novel therapeutic target in cancer. Clin Cancer Res 16, 4517–4526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 5, 387–393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangerich A. & Burkle A. How to kill tumor cells with inhibitors of poly(ADP-ribosyl)ation. Int J Cancer 128, 251–265 (2011). [DOI] [PubMed] [Google Scholar]
- Bryant H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- Menear K. A. et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phth alazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem 51, 6581–6591 (2008). [DOI] [PubMed] [Google Scholar]
- Evers B. et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin Cancer Res 14, 3916–3925 (2008). [DOI] [PubMed] [Google Scholar]
- Garber K. PARP inhibitors bounce back. Nat Rev Drug Discov 12, 725–727 (2013). [DOI] [PubMed] [Google Scholar]
- Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol 29, 373–374 (2011). [DOI] [PubMed] [Google Scholar]
- Tulin A. Re-evaluating PARP1 inhibitor in cancer. Nat Biotechnol 29, 1078–1079 (2011). [DOI] [PubMed] [Google Scholar]
- Jelinic P. & Levine D. A. New insights into PARP inhibitors’ effect on cell cycle and homology-directed DNA damage repair. Mol Cancer Ther 13, 1645–1654 (2014). [DOI] [PubMed] [Google Scholar]
- Nguyen D. et al. Poly(ADP-ribose) polymerase inhibition enhances p53-dependent and -independent DNA damage responses induced by DNA damaging agent. Cell Cycle 10, 4074–4082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota E. et al. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle 13, 2129–2137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M., Toillon R. A. & Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer 13, 293–325 (2006). [DOI] [PubMed] [Google Scholar]
- Reiner T. et al. Imaging therapeutic PARP inhibition in vivo through bioorthogonally developed companion imaging agents. Neoplasia 14, 169–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo-Marin J.-C., Extraction of spots in biological images using multiscale products. Pattern Recognition 35, 1989–1996 (2002). [Google Scholar]
- Starck J.-L., Murtagh F. & Bijaoui A. In Image processing and data analysis: the multiscale approach 1st edn Cambridge University Press1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.