Abstract
Drug discovery and development is a high-risk enterprise that requires significant investments in capital, time and scientific expertise. The studies of xenobiotic metabolism remain as one of the main topics in the research and development of drugs, cosmetics and nutritional supplements.
Antihypertensive drugs are used for the treatment of high blood pressure, which is one the most frequent symptoms of the patients that undergo cardiovascular diseases such as myocardial infraction and strokes. In current cardiovascular disease pharmacology, four drug clusters - Angiotensin Converting Enzyme Inhibitors, Beta-Blockers, Calcium Channel Blockers and Diuretics - cover the major therapeutic characteristics of the most antihypertensive drugs. The pharmacokinetic and specifically the metabolic profile of the antihypertensive agents are intensively studied because of the broad inter-individual variability on plasma concentrations and the diversity on the efficacy response especially due to the P450 dependent metabolic status they present.
Several computational methods have been developed with the aim to: (i) model and better understand the human drug metabolism; and (ii) enhance the experimental investigation of the metabolism of small xenobiotic molecules. The main predictive tools these methods employ are rule-based approaches, quantitative structure metabolism/activity relationships and docking approaches.
This review paper provides detailed metabolic profiles of the major clusters of antihypertensive agents, including their metabolites and their metabolizing enzymes, and it also provides specific information concerning the computational approaches that have been used to predict the metabolic profile of several antihypertensive drugs.
Keywords: Antihypertensive drugs, metabolism, computational approaches.
INTRODUCTION
Antihypertensive drugs are used for the treatment of high blood pressure, and more specifically, the primary and secondary hypertension. The complications of high blood pressure might trigger and/or cause several complex systematic cardiovascular disorders or syndromes such as stroke, unstable angina and myocardial infarction.
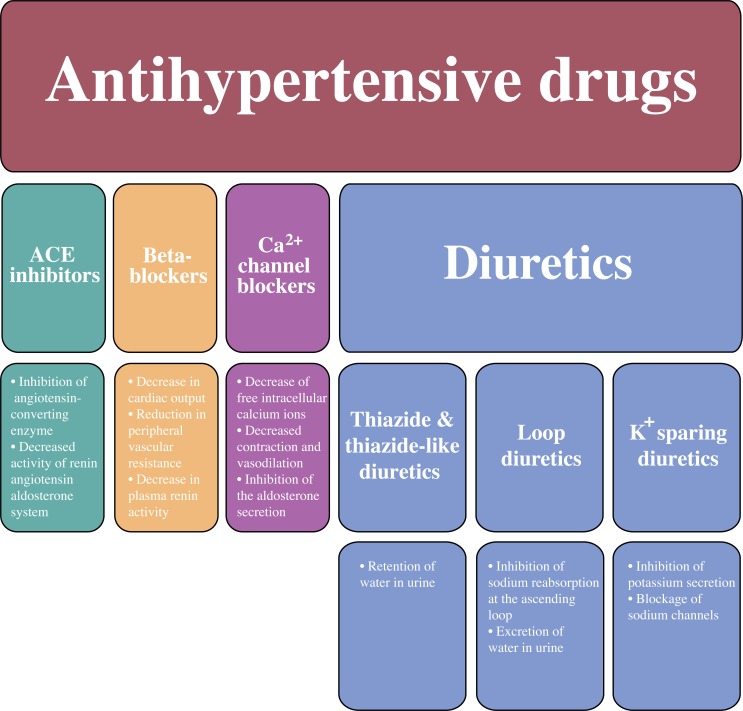
The most representative antihypertensive drugs are classified as: the Angiotensin Converting Enzyme inhibitors (ACE inhibitors), the beta-adrenergic blockers (β-blockers), the calcium (Ca+2) channel blockers and the diuretics. Additionally, the group of diuretics can be divided in four subgroups: potassium (K+) sparing diuretics, the loop diuretics, the thiazide and the thiazide-like diuretics (Fig. 1).
Fig. (1).
The classification of antihypertensive drugs and their mechanisms of actions.
It has been comprehensively described that extensive variability of anti-hypertensive drugs plasma concentrations in patients, due to the P450-mediated drugs metabolism, has a great impact and influence on the clinical outcome as well as on the drugs' response [1]. Among the antihypertensive drugs, the ACE inhibitor captopril, the beta blockers alprenolol, carvedilol, metoprolol and propranolol, the calcium channel blockers diltiazem, felodipine, nimodipine nifedipine and verapamil and the potassium sparing diuretic spironolactone are known either for their response variability due to the participation of P450-dependent metabolism or they are known for low and variable oral bioavailability and their first pass drug interactions. Usually, Single Nucleotide Polymorphisms (SNPs) are responsible for the drug response variability and they are able to modify the enzymatic activity of the P450 isoforms [2]. For this reason there is a great interest in the scientific community for further investigation and improved knowledge of antihypertensive drugs metabolism and all their potential biotransformations either by experimental or in silico studies.
Predictive tools for the drugs metabolic status are useful since insufficient knowledge about drug metabolism is one of the most common causes for failure and incompletion during clinical trials. The metabolizing activity of the most known phase I enzymes [3] such as cytochrome P450 [4], monoamine oxidases [5], alcohol dehydrogenases [6], hydrolases [7] and more specifically carbohydrolases [8, 9] and phase II enzymes such as UDP-glucuronosyl transferases [10], sulfotransferases [11], methyl-transferases [12] and glutathione S-transferases [13] has been investigated in great detail by computational techniques and can be successfully predicted by several approaches.
In the history of drug development, several different computational approaches have been developed for the prediction of human drug metabolism. The majority of these approaches employ databases, rule-based approaches, quantitative structure metabolism relationships (QSMR’s), quantitative structure activity relationships (QSAR’s), pharmacophore, statistical QSAR, electronic or homology models and crystal structures with docking approaches, and combinatorial methodologies where data and/or rules are used to predict all the possibilities of a molecule metabolism. For the improvement of the prediction of drug metabolism combined methods have been also developed and applied respectively.
The interaction between a substrate and P450 in terms of inhibition and induction of P450 enzymes has been also extensively studied. Pharmacophore models and three-dimensional quantitative structure-activity relationships (QSARs) have been used either alone or in combination with protein homology models to provide metabolic information for cytochrome P450 [14, 15]. Unlike the quantitative structure-activity relationships (QSARs), quantitative structure – property relationships methods have been developed in the drug discovery process [16]. These methods are used to generate by computational screening the ADME (Administration, Distribution, Metabolism, Excretion) profile components. Singh et al., have also developed a semi-quantitative model for evaluating the relative susceptibilities of different sites on drug molecules to metabolism by P450 [17].
Computational approaches have been employed for the scoring of compounds libraries in terms of their Km values to numerous P450 enzymes [18]. MetaDrug is a combinatorial method for predicting human drug metabolism. It has been used to predict some of the major metabolic pathways and to identify the role of P450 enzymes [19, 20]. Another computational method, called MetaSite, has been designed to identify the Site of Metabolism (SOM) for any human P450 acting on new substrates [21]. A statistical approach has also been applied to the prediction of aromatic hydroxylation sites for diverse sets of substrates [22]. Moreover, Jones et al., in 2002 suggested a combination of experimental data and semi-empirical molecular orbital calculations to predict activation energies for aromatic and aliphatic hydroxylation [23]. The development of model combined with analytical tools such as Mass Spectrometry (MS) or Liquid Chromatography LC/MS has facilitated the detection and the structural elucidation of chemical intermediates in drug metabolism [24, 25].
The META system is a commonly used drug metabolism prediction system. It is an expert system capable of predicting the potential for enzymatic attack sites and the nature of the chemical compounds formed by such metabolic transformations [26-28]. The model of PPS (Pathway Prediction System) also uses metabolic rules describing the transformation of chemical functional groups [29, 30]. On the other hand CATABOL is an approach that predicts chemical biodegradability in a quantitative manner [31]. It functions together with a probabilistic model that calculates probabilities of the individual transformation and matching substructure engine [31]. Furthermore, DEREK, StAR and METEOR prescribe rules to describe the relationship between chemical structure and either toxicity (DEREK and StAR) or metabolic fate (METEOR) [32].
In 2004, Hatzimanikatis et al. developed an alternative methodology called BNICE (Biochemical Network Integrated Computational Explorer) capable of generating every possible biochemical reaction based on enzyme reaction rules and starting or target compounds [33]. Recently, BNICE framework has been extended for the prediction of biodegradation pathways of xenobiotics, and has also been applied on xenobiotics compounds such as chlorinated aliphates, poly chlorinates biphenyls and polycyclic aromatic hydrocarbons [34]. BNICE was not only able to reproduce the proposed biodegradation routes but also to indicate new pathways consisting novel compounds and reactions.
ANGIOTENSIN-CONVERTING ENZYME (ACE) INHIBITORS
The angiotensin-converting enzyme (ACE) inhibitors are widely used in the management of essential hypertension, among the other cardiovascular diseases such as stable chronic heart failure, myocardial infarction and diabetic nephropathy. They include three categories: (a) captopril, (b) the prodrugs such as ramipril and enalapril and (c) lisinopril, which is water-soluble and the only ACE inhibitor that it is not undergoing metabolism. Their mechanism of action involves the inhibition of angiotensin-converting enzyme, the decreased activity of renin-angiotensin aldosterone system that permits and assists in sodium excretion and the increased formation of bradykinin and vasolidatory prostaglandins.
All of these antihypertensive agents are characterized as having carboxyl functional groups requiring hepatic activation to form pharmacologically active metabolites [35]. The majority of ACE inhibitors are prodrugs converted by hepatic esterolysis to a major active diacid metabolite. Lisinopril is an exception because it does not follow a prodrug biodegradation [36]. Table 1 presents the ACE inhibitors, their metabolites, and the known enzymes causing their metabolic biodegradation.
Table 1.
Detailed metabolic profile of ACE inhibitors.
| ACE inhibitors name | Metabolites | Enzymes | References |
|---|---|---|---|
| Benazepril | Benazeprilat | Carboxylesterases | [37] |
| Benazeprilat glucuronic acid | UDP-Glucuronosyltransferase | [37] | |
| Captopril | Cysteine-captopril disulfide | [38] | |
| Capropril dimer disulfide | [38] | ||
| Cilazapril | Cilazaprilat | Carboxylesterases | [39] |
| Enalapril | Enalaprilat | Carboxylesterases | [35, 40, 41] |
| Fosinopril | Fosinoprilat | Carboxylesterases | [42] |
| p-Hydroxy fosinopril metabolite | [42] | ||
| Glucuronides of fosinopril | UDP-Glucuronosyltransferase | [42] | |
| Lisinopril | Not metabolized | [36] | |
| Moexipril | Moexiprilat | Carboxylesterases | [43] |
| Perindopril | Perindoprilat | Carboxylesterases | [44] |
| Perindopril glucuronides | UDP-Glucuronosyltransferase | [44] | |
| Perindoprilat glucuronides | UDP-Glucuronosyltransferase | [44] | |
| Quinapril | Quinaprilat | Carboxylesterases | [45] |
| Ramipril | Ramiprilat | Carboxylesterases | [46] |
| Diketopiperazine ester | [46] | ||
| Diketopiperazine acid | [46] | ||
| Glucuronides of ramipril | UDP-Glucuronosyltransferase | [46] | |
| Glucuronides of ramiprilat | UDP-Glucuronosyltransferase | [46] | |
| Rescinnamine | No information available | ||
| Spirapril | Spiraprilat | Carboxylesterases | [35] |
| Trandolapril | Trandolaprilat | Carboxylesterases | [47] |
| Diketopiperazine ester | [47] | ||
| Diketopiperazine acid | [47] | ||
| Trandolapril glucuronides | UDP-Glucuronosyltransferase | [47] | |
| Trandolaprilat glucuronides | UDP-Glucuronosyltransferase | [47] |
More specifically, the prodrug of benazepril follows a hepatic carboxylation producing benazeprilat, which is the major active metabolite. Apart from the active metabolite only the glucuronides of benazepril and benazeprilat are found in the urine or in the feces [37].
Although the majority of ACE inhibitors belong to prodrugs and undergo hepatic carboxylation for releasing their active metabolites, the dominant urine metabolites of captopril are cysteine captopril and disulfide dimer captopril [38].
Cilazapril also belongs to the ACE inhibitors and it is rapidly converted to its active metabolite cilazaprilat, which is excreted unchanged through the kidneys [39].
Enalapril is extensively converted through hydrolysis to its active metabolite enalaprilat [35, 40]. Enalapril is also a prodrug, which is absorbed by the gastrointestinal tract and is extensively metabolized by hepatic esterases. In the elimination phase the enalaprilat is the only detectable metabolite [41].
Fosinopril, another ester prodrug, is metabolized in the liver by carboxylesterases to fosinoprilat. In addition to fosinoprilat, p-hydroxy fosinoprilat and the conjugated glucuronide fosinoprilat are the second generation detectable metabolites [42].
Lisinopril is the only ACE inhibitor that is not metabolized [36]. No information about the metabolism of rescinnamine, which belongs to the same drug group as lisinopril, is available.
Moexipril, an ACE inhibitor used for treatment of arterial hypertension, is also a prodrug, yielding moexiprilat, its active metabolite, by hydrolysis of an ethyl ester group [43].
Perindopril is hydrolysed in vivo to the active diadic metabolites perindoprilat and minor metabolic pathways lead to glucuronide-conjugates [44].
The prodrug of quinapril undergoes hepatic hydrolysis into its major active diacid metabolite quinaprilat [45].
Ramipramil is another ACE inhibitor that acts as a prodrug. Its main metabolite, the diacid metabolite, is a potent angiotensin converting enzyme inhibitor. The minor metabolites ramipril glucuronide, diacid, diacid-glucuronide, diketopiperazine and diketopiperazine acid are associated with the metabolism of ramipramil, and they are detectable in the urine and serum specimens [46].
Spirapril, is also a prodrug that has to be hydrolyzed by hepatic carboxylesterases to achieve its ACE inhibitory activity [35].
Similar to ramipril, trandolapril is metabolized to its main diacid metabolite. Trandolapril glucuronide, diacid, diacid-glucuronide, diketopiperazine and diketopiperazine acid are minor metabolites detectable in the urine and serum specimens from the metabolism of trandolapril [47].
BETA-BLOCKERS
The β-adrenergic blocking agents cause initially decrease in cardiac output, followed by reduction in peripheral vascular resistance and plasma renin activity decrease.
The majority of the beta-blockers are metabolized by P450 reactivity deriving dealkylated and hydroxylated metabolites and they appear to have similar metabolic characteristics. The elimination of the most of them occurs via hepatic metabolism and/or renal excretion of the unchanged drug [48]. Phase II glucuronidation reactions take place also in the most beta-blockers metabolic pathways. Atenolol and nadolol are the only beta-blockers that appear to be excreted in the unchanged form by the kidneys, while CYP1A2 and CYP2D6 seem to affect the propranolol biotransformation [49-68] (Table 2).
Table 2.
Detailed metabolic profile of beta-blockers.
| Beta-blocker name | Metabolites | Enzymes | References |
|---|---|---|---|
| Acebutolol | Diacetolol | CYP2D6 | [49] |
| Alprenolol | 4-Hydroxy-alprenolol | CYP2D6 | [50, 68] |
| Atenolol | Not metabolized | [51] | |
| Betaxolol | α-Hydroxy betaxolol | CYP1A2, CYP2D6 | [52, 53] |
| Bisoprolol | Demethyl carboxylic acid | CYP3A4, CYP2D6 | [54] |
| Deethyl carboxylic acid | [54] | ||
| Deethyl derivative | [54] | ||
| Carvedilol | 8-Hydroxy carvedilol | CYP1A2, CYP3A4, CYP1A1 | [55] |
| 4'-Hydroxy carvedilol | CYP2D6, CYP2E1, CYP2C9 | [55] | |
| 5'-Hydroxy carvedilol | CYP2D6, CYP2E1, CYP2C9 | [55] | |
| O-Desmethyl carvedilol | CYP2C9, CYP2D6, CYP1A2, CYP2E1 | [55] | |
| 1-Hydroxy carvedilol | Prostaglandin G/H synthase 1 | [55] | |
| 4'-Hydroxyphenyl carvedilol | [55] | ||
| Esmolol | Methanol | Esterases | [56] |
| Labetalol | 7 Glucuronides metabolites | UDP-Glucuronosyltransferase | [57, 58] |
| 3-Amino-1-phenyl butane | GI mucosa | [57, 58] | |
| 3-Amino-1-(4-hydroxyphenyl) butane | GI mucosa | [57, 58] | |
| Benzyl acetone | GI mucosa | [57, 58] | |
| Metoprolol | α-Hydroxy metoprolol | CYP2D6 | [59] |
| Nadolol | Not metabolized | [60] | |
| Nebivolol | Hydroxy alicyclic metabolites | CYP2D6 | [61] |
| Hydroxy aromatic metabolites | CYP2D6 | [61] | |
| 1st generation glucuronides | UDP-Glucuronosyltransferase | [61] | |
| N-dealkylation | CYP2D6 | [61] | |
| 2nd generation glucuronides (after the hydroxylation) | UDP-Glucuronosyltransferase | [61] | |
| Oxprenolol | Glucuronides of unchanged oxprenolol | UDP-Glucuronosyltransferase | [62, 63] |
| Hydroxylation to the aromatic ring of unchanged oxprenolol | CYP2D6 | [62, 63] | |
| Glucuronides of hydroxylated oxprenolol | UDP-Glucuronosyltransferase | [62, 63] | |
| N-dealkylation, producing the hydroxy carboxylic acid | CYP2D6 | [62, 63] | |
| Carbinol | CYP2D6 | [62, 63] | |
| Monoallyl ether of catechol | CYP2D6 | [62, 63] | |
| Penbutolol | Glucuronide metabolite | UDP-Glucuronosyltransferase | [64, 66] |
| 4-Hydroxy metabolite | [64, 66] | ||
| Pindolol | Hydroxylated metabolites | [67] | |
| Glucuronides of hydroxylated metabolites | UDP-Glucuronosyltransferase | [67] | |
| Sulfate metabolites | Sulfotransferases | [67] | |
| Propanolol | N-Desisopropyl propranolol | CYP1A2 | [68] |
| 4'-Hydroxy propanolol | CYP2D6 | [68] |
Acetubolol undergoes significant first pass metabolism. Its major metabolite, diacetolol, is formed during its first pass metabolism by CYP2D6 and contributes to beta blocking activity [49].
Both alprenolol and propanol, and their hydroxyl glucuronides, undergo ring hydrolated metabolites by P450 (CYP2D6 and CYP3A5/7) [50]. Propanolol is extensively metabolized with most metabolites appearing in the urine. It is metabolized through three primary routes: aromatic hydroxylation, N-dealkylation (CYP1A2), and direct glucuronidation [68].
The metabolism of atenolol is not extensive and the parent drug appears to be the only major radiolabelled component in blood [51]. Following similar metabolic profile, nadolol is a poorly lipid-soluble drug and is excreted unchanged in the urine [60].
According to betaxolol pharmacokinetics, approximately 15% of the administered dose is excreted as unchanged drug, the remainder being metabolites produced by the reactivity of CYP1A2 and CYP2D6 (α-hydroxy betaxolol and its acid) whose contribution to the clinical effect is negligible [52, 53].
Bisoprol is subject only to moderate hepatic metabolism. In the metabolism of bisoprolol only oxidative pathways have been detected, with no subsequent conjugation. It is metabolized primarily by CYP3A4 to inactive metabolites and it is also metabolized by CYP2D6, which does not seem to be clinically significant [54].
The product metabolites of carvedilol are excreted in the urine and only 0.3% is excreted as unchanged drug [55]. Carvedilol is metabolized by oxidation and glucuronidation in the aromatic ring. P450 is responsible for the carvedilol hydroxylations as well as for the demethylated metabolites to the aromatic ring [55].
Esmolol is rapidly metabolized by esterases mainly in red blood cells to a free acid metabolite and methanol [56].
Labetalol is identified in humans only by conjugated metabolites of the drug. Other detectable metabolites include the 3-amino-1-phenyl butane derivative as well as the D-hydroxy derivative of 3-amino-1-phenyl butane [57]. A family of catechol-like metabolites is formed by P450-dependent hydroxylation at the C3 position of the benzamide ring. These metabolites also undergo ring cyclization forming two novel indolic metabolites [58].
Approximately 70% to 80% of an oral metoprolol dose is metabolized by CYP2D6 in the liver. The α-hydroxylation reaction, which makes the formation of α-hydroxymetoprolol from metoprolol in the liver was mediated almost exclusively by CYP2D6 [59].
Nebivolol human metabolism is complex and is subject to debridoquine type genetic polymorphism. N-dealkylation mainly in combination with hydroxylation, acyclic monoxidation and aromatic hydroxylation mediated by hepatic CYP2D6, followed by glucuronidation. The glucuronidation of the parent drug is also one of the major metabolic pathways [61].
Oxprenolol is extensively metabolized with direct O-glucuronidation being the major metabolic pathway. Glucuronide conjugates of unchanged and hydroxylated oxprenolol have been experimentally identified [62]. Moreover, the hydroxyl carboxylic acid and the monoallyl ether of catechol have been detected as oxprenolol metabolites from reactions mediated by hepatic CYP2D6. [63].
Penbutolol is extensively metabolized, and less than 4% of administered dose is excreted unchanged in urine [64]. The most important known metabolites of penbutolol are penbutolol glucuronides, 4-hydroxy penbutolol and 4-hydroxy penbutolol glucuronide [65]. The 4-hydroxy penbutolol has been described as a semi-active metabolite [66].
The liver extensively metabolizes pindolol, forming several groups of metabolites: first-generation hydroxylated metabolites (Phase I), second-generation sulphate metabolites, and conjugations with glucuronic acid with the parent compounds or with its hydroxylated derivatives (Phase II). 35 to 40% of the administered dose is excreted unchanged in the urine [67].
CALCIUM CHANNEL BLOCKERS
There are two types of calcium channel blockers: (i) the dihydropyridine group calcium channel blockers, such as nifedipine, amlodipine and felodipine and (ii) the non-dihydropyridine group calcium channel blockers such as phenylalkylamine verapamil and benzothiazepine diltiazem. These drugs are able to decrease the concentration of free intracellular calcium ions leading to decreased contraction and vasodilation and they inhibit the aldosterone secretion. They also present diuretic activity through an increase in renal blood flow and glomerular filtration rate.
Most calcium channel blockers have low and variable oral bioavailability because of extensive first pass metabolism. Just as the other groups of antihypertensive drugs, calcium channel blockers have similar ways of biodegradation. Most of them act as substrates of CYP3A4 causing aromatic hydroxylation and N-dealkyation (Table 3). Moreover, those belonging to dihydropyridine group undergo dehydrogenation of the same group [69-79].
Table 3.
Detailed metabolic profile of calcium channel blockers.
| Calcium channel blocker name | Metabolites | Enzymes | References |
|---|---|---|---|
| Amlodipine | Pyridine derivative | CYP3A4 | [69] |
| De-amino amlodipine | CYP3A4 | [69] | |
| Ester hydrolysis of the S-methoxycarbonyl group | Carboxylesterases | [69] | |
| Amlodipine glucuronides | UDP-Glucuronosyltransferase | [69] | |
| Diltiazem | N-desmethyl diltiazem | CYP3A4 | [70] |
| O-desacetyl-N-desmethyl diltiazem | CYP3A4 | [70] | |
| Felodipine | Dehydro felodipine | CYP3A4 | [71] |
| Isradipine | Pyridine of isradipine | CYP3A4 | [72] |
| Carboxylic acid of isradipine | CYP3A4 | [72] | |
| Carboxylic acid of isradipine pyridine | CYP3A4 | [72] | |
| Nifedipine | Pyridine of nifedipine | CYP3A4 | [79] |
| Carboxylic acid of nifedipine | CYP3A4 | [79] | |
| Carboxylic acid of nifedine pyridine | CYP3A4 | [79] | |
| Nimodipine | Dehydro nimopidine | CYP3A4 | [74, 98] |
| Carboxylic acid of nimopidine | CYP3A4 | [74, 98] | |
| Cardoxylic acid of dehydronimopidine | CYP3A4 | [74, 98] | |
| Nisoldipine | Hydroxylation of the isobutyl moiety | CYP3A4 | [75] |
| Dehydrogenation of the 1, 4-dihydropyridine system | CYP3A4 | [75] | |
| Oxidative ester cleavage | CYP3A4 | [75] | |
| Hydroxylation of one of the methyl groups in 2-position and subsequent oxidation to the carboxylic acid | CYP3A4 | [75] | |
| Hydroxylation of one of the methyl groups in 6-position and subsequent oxidation to the carboxylic acid | CYP3A4 | [75] | |
| Oxidation of one of the methyl groups of the isobutyl moiety to the carboxyl group reduction of the aromatic nitro group | CYP3A4 | [75] | |
| Glucuronidation | UDP-Glucuronosyltransferase | [75] | |
| Nitrendipine | Dehydro nitredipine | CYP3A4 | [76] |
| Carboxylic acid of nitrendipine | CYP3A4 | [76] | |
| Cardoxylic acid of dehydronitrendipine | CYP3A4 | [76] | |
| Verapamil | O-Desmethyl verapamil | [77, 78] | |
| O-Desmethyl verapamil (D-702) | CYP2C8, CYP2C18, CYP2C9 | [77, 78] | |
| O-Desmethyl verapamil (D-703) | CYP2C8, CYP2C9 | [77, 78] | |
| Norverapamil | CYP2C8, CYP3A4, CYP3A5, CYP1A2 | [77, 78] | |
| D-617 | CYP2C8 | [77, 78] |
Amlodipine is extensively metabolized in the liver via cytochrome P4503A4 isozyme. Firstly, it undergoes oxidation to the pyridine derivative, and then oxidative deamination or ester hydrolysis [69].
CYP3A4 metabolizes also diltiazem via deacetylation, N-demethylation, O-demethylation, N-oxidation and oxidative deamination. However, diltiazem acts simultaneously as substrate and inhibitor of CYP3A4 [70].
Following oral administration, felodipine is almost completely absorbed and undergoes extensive first pass metabolism. Different metabolites have been detected in plasma from felodipine metabolism including pyridine analogue, carboxylic monoacids, ester lactones, hydroxyl acid forms and a lactonic compound after its P450-dependent metabolism [71].
Isradipine is completely metabolized prior to excretion, and no unchanged drug can be detected in the urine. The major metabolites of isradipine are its pyridine, which is the product of the ring oxidation of the dihydropyridine moiety, and the carboxylic acids followed the ester cleavage [72].
Nifedipine is eliminated from the human body by oxidative hepatic metabolism catalyzed mostly by CYP3A4 to two major corresponding carboxylic acids and the pyridine of the parent compound [79].
Nimodipine follows hepatic metabolism mediated by CYP3A4. Pharmacokinetic studies on both healthy volunteers and patients reveal large variability in drug disposition that may be implicated in clinical effects diversification [73]. It is well known that nimodipine undergoes oxidative ester cleavage, oxidative demethylation, subsequent oxidation resulting in the generation of primary alcohols and a carboxylic acid, methyl hydroxylations and isopropyl hydroxylation. Reduction of the aromatic nitro group as well as in phase II reactions by glucuronidation have also been described [74].
Nisoldipine is an extensively metabolized calcium channel blocker whereby the unchanged drug cannot be detected either in the urine or in the bile. As in the case of nimodipine, it also follows hydroxylation of isobutyl moiety, dehydrogenation of the dihydropyridine group, and hydroxylation of the methyl group, subsequent aminogroup and finally conjugation with glucuronate regarding the phase II reactions [75]. Cytochrome P450 enzymes are believed to play a major role in the metabolism of nisodilpine and especially the CYP3A4 isoenzyme.
Nitrendipine is another dihydropyridine calcium channel blocker that follows a pattern of metabolism similar to nimodipine and nisodilpine. They all follow the oxidation of the dihydropyridine structure, hydroxylation to the methyl group and derivation of the corresponding carboxylic acids by the cleavage of the ester bonds [76].
Verapamil is extensively metabolized primarily by cytochrome P450 enzymes with less than 5% of orally administered dose being excreted unchanged [77]. Verapamil metabolism follows O-demethylation, N-demethylation or N-dealkylation and the derivation of norverapamil [78].
DIURETICS
In general, all diuretics result initially in the reduction of plasma volume and cardiac output. In long-term usage, they decrease the total peripheral vascular resistance. More particularly, thiazide related diuretics lead to retention of water in urine. Loop diuretics result into the excretion of water in the urine by the inhibition of the sodium reabsorption at the ascending loop in the nephron. On the other hand, potassium-sparing diuretics do not promote the secretion of potassium into the urine and they block the sodium channels.
K+ Sparing Diuretics
P450 complex of enzymes also participate in the potassium sparing diuretics metabolism causing mostly hydroxylations. Amiloride is not metabolized by the liver but it is excreted unchanged by the kidneys [80]. On the other hand, triamterene is extensively metabolized to a major hydroxytriamterene sulfate metabolite and spironolactone is converted to several metabolites (Table 4).
Table 4.
Detailed metabolic profile of K+ sparing diuretics.
| K+ sparing diuretic name | Metabolites | Enzymes | References |
|---|---|---|---|
| Amiloride | Not metabolized | [80] | |
| Eplerenone | 21-Hydroxy eplererone | CYP3A4 | [81] |
| 6b-Hydroxy elpererone | CYP3A4 | [81] | |
| 6b, 21-Dihydroxy elpererone | CYP3A4 | [81] | |
| 3a, 6b-Dihydroxy elpererone | CYP3A4 | [81] | |
| 6b, 15a-Dihydroxy elpererone | CYP3A4 | [81] | |
| 3a, 6b, 21-Trihydroxy elpererone | CYP3A4 | [81] | |
| 2a, 3b, 6b-Trihydroxy elpererone | CYP3A4 | [81] | |
| Spironolactone | Spironolactone-thiole | [82] | |
| Spironolactone-dethioacetate | [82] | ||
| Canrenone | [82] | ||
| Thio-methyl-spironolactone | [82] | ||
| Hydroxyl-thiomethyl-spironolactone | [82] | ||
| Triamterene | 4-Hydroxy triamterene | CYP1A2 | [83] |
| 4-Hydroxy-sulfate-triamterene | Sulfotransferases | [83] |
Eplerenone is cleared predominantly by CYP3A4 metabolism. The major metabolic products excreted in the urine and the feces include all the hydroxylation and ketoreduction derivatives; however, no active metabolites have been identified in human plasma [81].
It is known that spironolactone is extensively metabolized and that its metabolites mediate its therapeutic activity. There are two metabolic pathways for spironolactone degradation. In the first one the drug undertakes dethioacetylation and in the second one the compound keeps its sulfur moiety [82].
The diuretic triamterene is an extensively metabolized potassium sparing diuretic. P450 enzymes predominantly catalyze its hydroxylation, and sulfotransferases catalyze Phase II conjugation reactions [83].
Loop Diuretics
Loop diuretics undergo hepatic metabolism in several different ways. Butamenide and torasemide are metabolized by cytochrome P450, whereas furosemide is only glucuronidated (Table 5). Only 20% of furosemide is metabolized in the liver. In contrast, almost 75% of torasemide is metabolized [84].
Table 5.
Detailed metabolic profile of loop diuretics.
| Loop diuretic name | Metabolites | Enzymes | References |
|---|---|---|---|
| Bumetanide | Debutylbumetanide | [85] | |
| Alpha-hydroxy bumetanide | [85] | ||
| Bumetanide carboxylic acid | [85] | ||
| Bumetanide glucuronides | UDP-Glucuronosyltransferase | [85] | |
| Ethacrynic acid | Glutathione conjugation | Gloutathione-S-transferases | [86] |
| Mercapturic Acid | [86] | ||
| Furosemide | 4-Chloro-5-sulfamoyl anthranilic acid | [87, 88] | |
| Torasemide | Hydroxylation of the methyl group of the benzyl ring | CYP2C8, CYP2C9 | [89] |
| M1 carboxylic acid | CYP2C8, CYP2C9 | [89] | |
| 4-Hydroxy torasemide | CYP2C8, CYP2C9 | [89] |
Pharmacokinetic studies of bumetanide indicate that more than 2/3 of urinary metabolites are unchanged bumetanide. The identified metabolites indicate that metabolism occurs on the butyl side chain, with the primary alcohol being the major metabolite. Conjugates of these metabolites were also found in the urine [85].
Pharmacokinetic studies have shown that approximately 60% to 70% of ethacrynic acid is excreted to the bile as an unchanged compound, while the remainder is biotransformation products. The glutathione conjugate as well as the mercapturate derivative are the two major final metabolites of ethacrynic acid [86].
On average, 70% of the oral furosemide dose is absorbed and only about 10% of the drug is eliminated from the body during hemodialysis. The liver appears to play a minor role in the metabolism of furosemide and an indirect evidence suggests that the kidneys could metabolize this compound [87]. The only known metabolite is the 4-chloro-5-sulfamoylantranilic acid, which has also a potential diuretic activity [88].
Torasemide is rapidly absorbed following oral administration and metabolized by hepatic cytochrome P450 enzymes. In the case of intravenous administration 25% of a dose is traced in the urine as unchanged drug. This diuretic undergoes different hydroxylations, oxidations and reductions mediated by CYP2C8 and CYP2C9 in order to produce its semi active metabolites [89].
Thiazide and Thiazide-Like Diuretics
Despite the differences in effectiveness between the thiazide diuretics, they have several common pharmacokinetic properties. Most of these compounds appear to be well absorbed from oral dosage and the kidneys or the feces actively secrete them as unchanged substances. The degree of metabolism of the thiazide diuretics is not variable. In the case of chlorothiazide, hydrochloro thiazide and hydroflumethiazide it is essentially zero and almost zero for the rest of them [90].
The major portion of chlorthalidone is excreted unchanged by the kidneys [90]. In a similar pattern, metalazone is excreted unchanged in urine via glomerular filtration and active tubular secretion, and the possibility of undergoing enterohepatic recycling exists [91].
Although most of thiazide-like diuretics are excreted unchanged, indapamide is extensively hepatic metabolized. The metabolic studies of indapamide indicate the significance of the cytochrome P450 enzymes. More particularly, CYP3A4 is responsible for the dehydrogenation of indapamide, its hydroxylation and carboxylation (Table 6). Glucuronides of the hydroxylated metabolites have also been identified [92].
Table 6.
Detailed metabolic profile of thiazide diuretics.
| Thiazide diuretic name | Metabolites | Enzymes | References |
|---|---|---|---|
| Chlorthalidone | Excreted unchanged in the urine | [90] | |
| Indapamide | Hydroxyl-indapamide | CYP3A4 | [92] |
| Dehydro-indapamide | CYP3A4 | [92] | |
| Hydroxyl-dehydro-indapamide | CYP3A4 | [92] | |
| Indapamide-epoxide | Microsomal epoxide hydrolase | [92] | |
| Dihydroxy-indapamide | CYP3A4 | [92] | |
| Indapamide-glutathione conjugation | Glutathione (non enzymatic) | [92] | |
| Metolazone | Excreted unchanged in the urine | [91] |
COMPUTATIONAL APPROACHES FOR THE PREDICTION OF THE METABOLISM OF ANTIHYPERTENSIVE DRUGS
METEOR is a computational system for the prediction of drug metabolism that contains a knowledge base of reactions and rules relating structure and biotransformation [32]. In 2005, Testa et al. provided an evaluation of this system. The ACE inhibitor omaprilat was one of the drugs used in this evaluation [93]. According to the results, two of omaprilat’s reactions, amide hydrolysis and S-methylation, were in full qualitative agreement with the in vivo experimental results in humans. The system predicted five more oxidative reactions, which were false positives. METEOR was also able to produce conjugations such as S-glucuronidation and taurine conjugation that appear to be realistic predictions. Among the predictions for the metabolism of omaprilat there was only one false negative prediction, due to a gap in the knowledge base of the program.
In the case of the beta-blocker betaxolol, a combined protein and pharmacophore model for cytochrome P450 2D6 (CYP2D6) has been developed with a second pharmacophore in order to explain the CYP2D6 catalyzed metabolic reactions [94]. This computational approach indicated that the most favorable metabolic pathways would result in the hydroxylation at the aliphatic carbon atoms. The experimentally observed metabolism in humans involved the formation of acid metabolite and α-hydroxy betaxolol [53]. This computational approach was able to confirm only the formation of α-hydroxy betaxolol.
CYP2C9 is a predominant member of the 2C family with a major contribution to human drug metabolism. More particularly, a lot of anti-inflammatory compounds, antithrombotic agents and antihypertensive drugs belong to CYP2C9 substrates. CYP2C9 is also one of the “typical” enzymes screened during the pre-clinical and clinical investigations of drugs hepatic metabolism. Apart from experimental investigation of the xenobiotics biotransformation by P450 enzyme, computational methods are required for the prediction and the further investigation of its enzymatic activity. These methods employ docking techniques, pharmacophore modeling QSAR or QSMR approaches, molecular dynamics (MD) and quantum mechanical (QM) and molecular mechanical (MM) studies.
In one of the studies of the metabolism of a novel cardio active agent 3, 4 methylenedioxybenzoyl-2-phenyl hydrazine, known to improve the intracellular Ca+2 regulation, docking studies were used to predict the sites of metabolism (SOM) within the active site of CYP2C9, molecular dynamics (MD) simulations of docked complexes were performed to validate their stability and QM calculations were done to study the energy profiles for the oxidations [95]. This study demonstrated the ability of used computational approaches to provide reliable qualitative predictions of the novel cardio active agent metabolites produced by the action of the P450 enzyme CYP2C9.
QM/MM calculations were used to model the phase I oxidation reactions of CYP2C9 by Lonsdale and coworkers [96]. Although this study does not include the metabolism of antihypertensive drugs, many antihypertensive drugs belong to CYP2C9 substrates. In this study, the authors provide mechanistic features of metabolizing reactions occurred by the catalytic activity of CYP2C9. In addition, they show that the active site is important for rationalizing the experimentally observed selectivity that is a likely significant feature for making predictions of novel pharmaceutics compounds metabolism.
Liu et al., based on a 2D substructure structure-based site of metabolism (SOM) prediction method called SMARTCyp [97] were able to predict the SOM of major drug metabolism enzymes belonging to the superfamily of P450 such as CYP1A2, CYP2C9 and CYP2C19 [98]. SMARTCyp is an alternative method that is able to estimate the reactivity of molecular fragments by high level quantum mechanical calculations on representative molecules and assign reactivity to different sites of a substrate by matching structural patterns [97]. In this study, the authors have also used some of the antihypertensive drugs catalyzed by CYP1A2 such as carvedilol, by CYP2C9, torasemide and carvedilol, and by CYP2C19 such as the beta-blocker of propranolol.
The CYP2C9 is not the only “prevalent” catalyzing enzyme of antihypertensive drugs. Several antihypertensive drugs are substrates of CYP2D6, which catalyzes some of the beta-blockers such as propranolol. Pharmacophore studies have been also developed to include the hydroxylation O- and N- dealkylation caused by CYP2D6 [94, 99-102].
The group of phase I enzymatic reactions does not include only the P450 reactivity but it can be expanded to other catalytic activities. Carboxylesterases is a major and important group of enzymes that catalyzes the metabolism of drugs and prodrugs including a great range of antihypertensive drugs such as ACE inhibitors. Most ACE inhibitors are prodrugs, depending on the action of carboxylesterases to express their pharmaceutical activity. In silico studies have been performed to predict the metabolic activity of carboxylesterase-1 and carboxylesterase-2 [8, 9]. These studies involve docking approaches as well as molecular dynamics simulations of known substrates including antihypertensive drugs to predict the catalytic activity of these two carboxylesterases.
Another study developed a structure-based hepatic metabolic clearance prediction model based on molecular descriptors [103]. In this study, the researchers used a set of 50 drugs to cover a range of molecular properties. This set also included a significant number of antihypertensive drugs such as beta-blockers (carvedilol, metoprolol, pindolol, propranolol), calcium channel blockers (diltiazem, nifedipine, verapamil) and loop diuretics (furosemide). The objectives of this study were the development of quantitative relationships between the molecular description and observed human hepatic clearance values, the estimation of predictability of this in silico model by comparing it with the in vitro hepatocytes model and the identification of important molecular descriptors influencing the hepatic metabolic clearance. In the prediction of hepatic metabolic clearance for the antihypertensive drugs the obtained in silico values were in good agreement with those values obtained from human hepatocytes incubations. More specifically, only the “fold error” corresponding value of propranolol appeared to be unexpected with respect to the observed hepatic metabolic clearance.
Biopharmaceutics Drug Disposition Classification System (BDDCS) is an alternative study that develops computational classification models and predicts BDDCS classes for molecular properties. This study also elaborates antihypertensive drugs such as the calcium channel blocker amlodipine [104]. The Biopharmaceutics Classification System (BCS) has had a significant impact on the drug regulatory process and practice, and requires knowledge of solubility and permeability data [105, 106]. According to BCS, as defined by FDA, the drugs can be classified into four different classes according to their permeability and solubility. The BDDCS takes into account the BCS classification system as well as several different drug parameters such as elimination routes, pharmacodynamics properties, post absorption effects and intravenous dosing, drug-drug interactions, and it develops a four class classification system divided into extensive metabolic profile and poor metabolic profile [104, 107]. Among the drugs used for this study, several antihypertensive drugs have been classified into four categories as extensive metabolizing or poor metabolizing drugs with high and low solubility respectively as they have been also described in the literature. For instance, the antihypertensive drugs which are known not to undergo metabolism (amiloride, atenolol, hydrochlorotiazide, nadolol, chlorthalidone, chlorothiazide) have been classified among the two classes described as the poor metabolizing drugs. Following the same pattern, the antihypertensive drugs used in the study which are known to undergo extensive metabolism (diltiazem, enalapril, nifedipine, verapamil, propranolol, carvedilol, spironolactone) have been classified by the system as extensive metabolizing drugs, belonging to class 1 and 2 according to their solubility.
Recently, the computational framework of BNICE has been used in a yet unpublished study for the characterization of known drugs metabolism. The conceptual framework introduced in BNICE for the systematic formulation of generalized enzyme reaction rules is based on the E.C. (Enzyme Commission) classification system [108]. These rules are built through manual curation and analysis of all the known biochemical reactions and each rule is associated with a 3rd level EC class. The application of the formulated generalized reaction rules allows the users to produce all possible metabolites. The algorithm applies iteratively the generalized reaction rules to a set of starting compounds and a set of enzyme cofactors. Each starting compound is evaluated to determine if it contains the functional group that will be transformed by the reaction rules, generating all possible products. The rules are then applied to the products from the previous generation, and this process is repeated until no new compounds are created or the maximum specified number of iterations has been reached. In the next step, the BNICE algorithm continues with the pathway search, which tries to find out all the possible enzymatic routes from a starting compound to a target compound through the system. Among the used databases was also the DrugBank database, which combines drug chemical, pharmacological and pharmaceutical data with drug target information [109-111].
In this study 222 drugs, found in the pathway browser of the database, have been used. The pathway browser consists of drugs that are very well described according to their biochemical mechanism. 75 of these drugs belong to antihypertensive agents. For those that undergo metabolism, BNICE was able to reproduce all the drug metabolites reported in the literature indicating that the method is able to identify the known biodegradation routes, and it is also able to indicate new metabolic routes by producing new enzymatic reactions. The method also provides also the third level E.C. number of the enzymes producing metabolites that have never been confirmed by pharmacokinetics studies. Table 7 shows the E.C. numbers suggested by the BNICE methodology that are able to reproduce the metabolites found in experimental studies.
Table 7.
Detailed description of antihypertensive drugs metabolism according to the 3rd level E.C. classification of BNICE frame-work generalized reaction rules. The + indicates sequential reactions given by generalized reaction rules and the indicates alternative generalized reaction rules given the same metabolite.
| Category | Drug name | Metabolites | Enzymes | 3rd level E.C. number of BNICE generalized reaction rules |
|---|---|---|---|---|
| ACE Inhibitors | Benazepril | Benazeprilat | Carboxylesterases | 3.1.1.- |
| Benazeprilat glucuronic acid | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Captopril | Cysteine-captopril disulfide | 1.8.1.-|1.11.1.- | ||
| Capropril dimer disulfide | 1.8.1.-|1.11.1.- | |||
| Cilazapril | Cilazaprilat | Carboxylesterases | 3.1.1.- | |
| Enalapril | Enalaprilat | Carboxylesterases | 3.1.1.- | |
| Fosinopril | Fosinoprilat | Carboxylesterases | 3.1.1.- | |
| p-Hydroxy fosinopril metabolite | 1.14.13.- | |||
| Glucuronides of fosinopril | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Lisinopril | Not metabolized | |||
| Moexipril | Moexiprilat | Carboxylesterases | 3.1.1.- | |
| Perindopril | Perindoprilat | Carboxylesterases | 3.1.1.- | |
| Perindopril glucuronides | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Perindoprilat glucuronides | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Quinapril | Quinaprilat | Carboxylesterases | 3.1.1.- | |
| Ramipril | Ramiprilat | Carboxylesterases | 3.1.1.- | |
| Diketopiperazine ester | 3.5.1.- | |||
| Diketopiperazine acid | 3.5.1.- + 3.1.1.- | |||
| Glucuronides of ramipril | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Glucuronides of ramiprilat | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Rescinnamine | No information available | |||
| Spirapril | Spiraprilat | Carboxylesterases | 3.1.1.- | |
| Trandolapril | Trandolaprilat | Carboxylesterases | 3.1.1.- | |
| Diketopiperazine ester | 3.5.1.- | |||
| Diketopiperazine acid | 3.5.1.- + 3.1.1.- | |||
| Trandolapril glucuronides | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Trandolaprilat glucuronides | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Beta-blockers | Acebutolol | Diacetolol | CYP2D6 | 2.1.1.- + 2.1.1.- |
| Alprenolol | 4-Hydroxy-alprenolol | CYP2D6 | 1.14.13.- | |
| Atenolol | Not metabolized | |||
| Betaxolol | α-Hydroxy betaxolol | CYP1A2, CYP2D6 | 1.14.13.- | |
| Bisoprolol | Demethyl carboxylic acid | CYP3A4, CYP2D6 | 2.7.8.- +1.1.1.- +1.2.1.- | |
| Deethyl carboxylic acid | 2.7.8.- +1.1.1.- +1.2.1.- | |||
| Deethyl derivative | 2.7.8.- +1.1.1.- +1.2.1.- | |||
| Carvedilol | 8-Hydroxy carvedilol | CYP1A2, CYP3A4, CYP1A1 | 1.14.13.- | |
| 4'-Hydroxy carvedilol | CYP2D6, CYP2E1, CYP2C9 | 1.14.13.- | ||
| 5'-Hydroxy carvedilol | CYP2D6, CYP2E1, CYP2C9 | 1.14.13.- | ||
| O-Desmethyl carvedilol | CYP2C9, CYP2D6, CYP1A2, CYP2E1 | 1.14.13.- | ||
| 1-Hydroxy carvedilol | Prostaglandin G/H synthase 1 | 1.14.13.- | ||
| 4'-Hydroxyphenyl carvedilol | 1.14.13.- | |||
| Esmolol | Methanol | Esterases | 1.14.13.- | |
| Labetalol | 7 Glucuronides metabolites | UDP-Glucuronosyltransferase | 2.4.1B1 | |
| 3-Amino-1-phenyl butane | GI mucosa | 1.4.1.-|1.4.3.-|1.5.99.-|4.3.1.- | ||
| 3-Amino-1-(4-hydroxyphenyl) butane | GI mucosa | 1.4.1.-|1.4.3.-|1.5.99.-|4.3.1.- | ||
| Benzyl acetone | GI mucosa | 1.4.1.-|1.4.3.-|1.5.99.-|4.3.1.- | ||
| Metoprolol | α-Hydroxy metoprolol | CYP2D6 | 1.14.13.- | |
| Nadolol | Not metabolized | |||
| Nebivolol | Hydroxy alicyclic metabolites | CYP2D6 | 1.14.13.- | |
| Hydroxy aromatic metabolites | CYP2D6 | 1.14.13.- | ||
| 1st generation glucuronides | UDP-Glucuronosyltransferase | 2.4.1B1 | ||
| N-dealkylation | CYP2D6 | 1.4.1.-|1.4.3.-|1.5.99.-|4.3.1.- | ||
| 2nd generation glucuronides (after the hydroxylation) | UDP-Glucuronosyltransferase | 1.14.13.- +2.4.1.- | ||
| Oxprenolol | Glucuronides of unchanged oxprenolol | UDP-Glucuronosyltransferase | 2.4.1.- | |
| Hydroxylation to the aromatic ring of unchanged oxprenolol | CYP2D6 | 1.14.13.- | ||
| Glucuronides of hydroxylated oxprenolol | UDP-Glucuronosyltransferase | 1.14.13.- +2.4.1.- | ||
| N-dealkylation, producing the hydroxy carboxylic acid | CYP2D6 | 1.4.1.-|1.4.3.- + 1.1.1.- | ||
| Carbinol | CYP2D6 | 1.4.1.-|1.4.3.- | ||
| Monoallyl ether of catechol | CYP2D6 | 3.3.1.- | ||
| Penbutolol | Glucuronide metabolite | UDP-Glucuronosyltransferase | 2.4.1.- | |
| 4-Hydroxy metabolite | 1.14.13.- | |||
| Pindolol | Hydroxylated metabolites | 1.14.13.- | ||
| Glucuronides of hydroxylated metabolites | UDP-Glucuronosyltransferase | 1.14.13.- + 2.4.1.- | ||
| Sulfate metabolites | Sulfotransferases | 1.14.13.- | ||
| Propanolol | N-Desisopropyl propranolol | CYP1A2 | 1.4.1.-|1.4.3.- | |
| 4'-Hydroxy propanolol | CYP2D6 | 1.14.13.- | ||
| Calcium Channel Blockers | Amlodipine | Pyridine derivative | CYP3A4 | 5.3.3.-|1.3.1.-+1.5.3.-|1.5.1.- |
| De-amino amlodipine | CYP3A4 | 1.4.3.-|1.5.99.- | ||
| Ester hydrolysis of the S-methoxycarbonyl group | Carboxylesterases | 3.1.1.- | ||
| Amlodipine glucuronides | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Diltiazem | N-desmethyl diltiazem | CYP3A4 | 1.4.3.-|1.5.99.- | |
| O-desacetyl-N-desmethyl diltiazem | CYP3A4 | 1.4.3.-|1.5.99.-+1.14.13.-|3.1.1.- | ||
| Felodipine | Dehydro felodipine | CYP3A4 | 5.3.3.-|1.3.1.-+1.5.3.-|1.5.1.- +3.1.1.-+.1.14.13.- | |
| Isradipine | Pyridine of isradipine | CYP3A4 | 5.3.3.-. + 1.5.1.-|1.5.3.- | |
| Carboxylic acid of isradipine | CYP3A4 | 3.1.1.- | ||
| Carboxylic acid of isradipine pyridine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.-+3.1.1.- | ||
| Nifedipine | Pyridine of nifedipine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- | |
| Carboxylic acid of nifedipine | CYP3A4 | 3.1.1.- | ||
| Carboxylic acid of nifedine pyridine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.-+3.1.1.- | ||
| Nimodipine | Dehydro nimopidine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- | |
| Carboxylic acid of nimopidine | CYP3A4 | 3.1.1.- | ||
| Cardoxylic acid of dehydro nimopidine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.-+3.1.1.- | ||
| Nisoldipine | Hydroxylation of the isobutyl moiety | CYP3A4 | 1.14.13.- | |
| Dehydrogenation of the 1, 4-dihydropyridine system | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- | ||
| Oxidative ester cleavage | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.-+3.1.1.- | ||
| Hydroxylation of one of the methyl groups in 2-position and subsequent oxidation to the carboxylic acid | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- + 1.14.13.- | ||
| Hydroxylation of one of the methyl groups in 6-position and subsequent oxidation to the carboxylic acid | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- + 1.14.13.- | ||
| Oxidation of one of the methyl groups of the isobutyl moiety to the carboxyl group reduction of the aromatic nitro group | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- + 1.1.1.- | ||
| Glucuronidation | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Nitrendipine | Dehydro nitredipine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.- | |
| Carboxylic acid of nitrendipine | CYP3A4 | 3.1.1.- | ||
| Cardoxylic acid of dehydro nitrendipine | CYP3A4 | 5.3.3.- + 1.5.1.-|1.5.3.-+3.1.1.- | ||
| Verapamil | O-Desmethyl verapamil | 1.14.13.- | ||
| O-Desmethyl verapamil (D-702) | CYP2C8, CYP2C18, CYP2C9 | 1.14.13.- | ||
| O-Desmethyl verapamil (D-703) | CYP2C8, CYP2C9 | 1.14.13.- | ||
| Category | Drug name | Metabolites | Enzymes | 3rd level E.C. number of BNICE generalized reaction rules |
| Norverapamil | CYP2C8, CYP3A4, CYP3A5, CYP1A2 | 1.4.3.-|1.5.99.- | ||
| D-617 | CYP2C8 | 1.4.1.-|1.4.3.-|1.5.99.- | ||
| K+ sparing diuretics | Amiloride | Not metabolized | ||
| Eplerenone | 21-Hydroxy eplererone | CYP3A4 | 1.14.13.- | |
| 6b-Hydroxy elpererone | CYP3A4 | 1.14.13.- | ||
| 6b, 21-Dihydroxy elpererone | CYP3A4 | 1.14.13.-+1.14.13.- | ||
| 3a, 6b-Dihydroxy elpererone | CYP3A4 | 1.1.1.- + 1.14.13.- | ||
| 6b, 15a-Dihydroxy elpererone | CYP3A4 | 1.14.13.-+1.14.13.- | ||
| 3a, 6b, 21-Trihydroxy elpererone | CYP3A4 | 1.1.1.- + 1.14.13.- +1.14.13.- | ||
| 2a, 3b, 6b-Trihydroxy elpererone | CYP3A4 | 1.1.1.- + 1.14.13.- +1.14.13.- | ||
| Spironolactone | Spironolactone-thiole | 1.2.1.-|2.3.1.-|3.1.2.- | ||
| Spironolactone-dethioacetate | 1.2.1.-|2.3.1.-|3.1.2.-+3.3.1.-+4.2.1.-+1.3.1.- | |||
| Canrenone | 1.2.1.-|2.3.1.-|3.1.2.-+3.3.1.- +4.2.1.- | |||
| Thio-methyl-spironolactone | 1.2.1.-|2.3.1.-|3.1.2.- + 2.1.1.- | |||
| Hydroxyl-thiomethyl-spironolactone | 1.2.1.-|2.3.1.-|3.1.2.-+2.1.1.-+1.14.13A1 | |||
| Triamterene | 4-Hydroxy triamterene | CYP1A2 | 1.14.13.- | |
| 4-Hydroxy-sulfate-triamterene | Sulfotransferases | 1.14.13.- + 2.8.2.- | ||
| Loop Diuretics | Bumetanide | Debutylbumetanide | 1.4.1.-|1.4.3.-|4.3.1.- | |
| Alpha-hydroxy bumetanide | 1.14.13.-|1.14.18.-|1.17.99.- | |||
| Bumetanide carboxylic acid | 1.13.11.- | |||
| Bumetanide glucuronides | UDP-Glucuronosyltransferase | 2.4.1.- | ||
| Ethacrynic acid | Glutathione conjugation | Glutathione-S-transferases | 2.5.1.- | |
| Mercapturic Acid | 2.5.1.-+3.5.1.- | |||
| Furosemide | 4-Chloro-5-sulfamoyl anthranilic acid | 2.4.1.- | ||
| Torasemide | Hydroxylation of the methyl group of the benzyl ring | CYP2C8, CYP2C9 | 1.14.13.-|1.17.99.- | |
| M1 carboxylic acid | CYP2C8, CYP2C9 | 1.13.11.- | ||
| 4-Hydroxy torasemide | CYP2C8, CYP2C9 | 1.14.13.- | ||
| Thiazide Diuretics | Bendroflumethiazide | No available information | ||
| Chlorothiazide | Chlorothiazide is not metabolized but is eliminated rapidly by the kidney. | |||
| Cyclothiazide | No available information | |||
| Hydrochlorothiazide | Hydrochlorothiazide is not metabolized. | |||
| Hydroflumethiazide | Essentially unchanged | |||
| Methyclothiazide | No information available | |||
| Polythiazide | No information available | |||
| Quinethazone | No information available | |||
| Trichlormethiazide | No information available | |||
| Thiazide-like Diuretics | Chlorthalidone | Excreted unchanged in the urine | ||
| Indapamide | Hydroxyl-indapamide | CYP3A4 | 1.14.13.- | |
| Dehydro-indapamide | CYP3A4 | 1.14.13.-|1.14.15.-|1.1.1.- | ||
| Hydroxyl-dehydro-Indapamide | CYP3A4 | 1.14.13.-|1.14.15.-|1.1.1.- + 1.14.13.- | ||
| Indapamide-epoxide | Microsomal epoxide hydrolase | 1.3.1.-|1.5.3.-|1.14.19.- | ||
| Dihydroxy-indapamide | CYP3A4 | 1.14.12.- | ||
| Indapamide-glutathione conjugation | Glutathione (non enzymatic) | 1.14.12.- + 2.5.1.- | ||
| Metolazone | Excreted unchanged in the urine |
CONCLUSION
Several experimental pharmacokinetics studies and studies of metabolism have shown that the antihypertensive drugs are metabolized by the human body through different pathways and processes. Although ACE inhibitors are mostly metabolized by the liver carboxylesterases in order to express their antihypertensive activity, the most of the beta-blockers, the calcium channel blockers and the diuretics follow metabolic pathways that undergo the P450 reactivity. Only thiazide diuretics and the majority of thiazide-like diuretics are used essentially unchanged and they are known not to undergo metabolism. As was expected due to the structural similarity, the antihypertensive drugs belonging to the same drug category and presenting structural similarity are metabolized in the same pattern.
In recent years, only a few computational methods have been used for the prediction of antihypertensive drugs metabolism and none of them has been developed exclusively for the prediction of the metabolism of the antihypertensive drugs. The above-described methods use mostly rule-based approaches, molecular dynamics, quantum mechanical and molecular mechanical studies, docking approaches and statistical approaches. Even though all of them present reliable results regarding the metabolism of antihypertensive drugs, we believe that there is still a need for further development of in silico methods and for the evolution of the existing methods that will allow the better mechanistic understanding of the drug metabolism that would result in better efficacy of treatments or more efficient patient stratification into antihypertensive treatment.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
The authors, Aikaterini Zisaki, Ljubisa Miskovic and Vassily Hatzimanikatis were supported by funding from SystemsX.ch, The Swiss Initiative in Systems Biology, and from the Ecole Polytechnique Fédérale de Lausanne (EPFL).
REFERENCES
- 1.Hocht C, Bertera FM, Mayer MA, Taira CA. Issues in drug metabolism of major antihypertensive drugs beta-blockers, calcium channel antagonists and angiotensin receptor blockers. Expert Opin Drug Metab Toxicol. 2010;6:199–211. doi: 10.1517/17425250903397381. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–21. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 3.Testa B, Kramer SD. The biochemistry of drug metabolism--an introduction Part 2. Redox reactions and their enzymes. Chem Biodivers. 2007;4:257–405. doi: 10.1002/cbdv.200790032. [DOI] [PubMed] [Google Scholar]
- 4.Stjernschantz E, Vermeulen NP, Oostenbrink C. Computational prediction of drug binding and rationalisation of selectivity towards cytochromes P450. Expert Opin Drug Metab Toxicol. 2008;4:513–27. doi: 10.1517/17425255.4.5.513. [DOI] [PubMed] [Google Scholar]
- 5.Edmondson DE, Mattevi A, Binda C, Li M, Hubalek F. Structure and mechanism of monoamine oxidase. Curr Med Chem. 2004;11:1983–93. doi: 10.2174/0929867043364784. [DOI] [PubMed] [Google Scholar]
- 6.Carballeira JD, Quezada MA, Alvarez E, Sinisterra JV. High throughput screening and QSAR-3D/CoMFA useful tools to design predictive models of substrate specificity for biocatalysts. Molecules. 2004;9:673–93. doi: 10.3390/90800673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Testa B, Krämer SD. v: Wiley-VCH Weinheim;; 2008. The biochemistry of drug metabolism : principles, redox reactions, hydrolyses. [Google Scholar]
- 8.Vistoli G, Pedretti A, Mazzolari A, Testa B. In silico prediction of human carboxylesterase-1 (hCES1):metabolism combining docking analyses and MD simulations. Bioorg Med Chem. 2010;18:320–9. doi: 10.1016/j.bmc.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 9.Vistoli G, Pedretti A, Mazzolari A, Testa B. Homology modeling and metabolism prediction of human carboxylesterase-2 using docking analyses by GriDock a parallelized tool based on AutoDock 40. J Comput Aided Mol Des. 2010;24:771–87. doi: 10.1007/s10822-010-9373-1. [DOI] [PubMed] [Google Scholar]
- 10.Smith PA, Sorich MJ, Low LS, McKinnon RA, Miners JO. Towards integrated ADME prediction past, present and future directions for modelling metabolism by UDP-glucuronosyltransferases. J Mol Graph Model. 2004;22:507–17. doi: 10.1016/j.jmgm.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Hirose S, Shimizu K, Kanai S, Kuroda Y, Noguchi T. POODLE-L a two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics. 2007;23:2046–53. doi: 10.1093/bioinformatics/btm302. [DOI] [PubMed] [Google Scholar]
- 12.Sipila J, Taskinen J. CoMFA modeling of human catechol O-methyltransferase enzyme kinetics. J Chem Inf Comput Sci. 2004;44:97–104. doi: 10.1021/ci034189k. [DOI] [PubMed] [Google Scholar]
- 13.Soffers AE, Ploemen JH, Moonen MJ, et al. Regioselectivity and quantitative structure-activity relationships for the conjugation of a series of fluoronitrobenzenes by purified glutathione S-transferase enzymes from rat and man. Chem Res Toxicol. 1996;9:638–46. doi: 10.1021/tx9501804. [DOI] [PubMed] [Google Scholar]
- 14.de Groot MJ, Ekins S. Pharmacophore modeling of cytochromes P450. Adv Drug Deliv Rev. 2002;54:367–83. doi: 10.1016/s0169-409x(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 15.Ekins S, de Groot MJ, Jones JP. Pharmacophore and three-dimensional quantitative structure activity relationship methods for modeling cytochrome p450 active sites. Drug Metab Dispos. 2001;29:936–44. [PubMed] [Google Scholar]
- 16.Shen M, Xiao Y, Golbraikh A, Gombar VK, Tropsha A. Development and validation of k-nearest-neighbor QSPR models of metabolic stability of drug candidates. J Med Chem. 2003;46:3013–20. doi: 10.1021/jm020491t. [DOI] [PubMed] [Google Scholar]
- 17.Singh SB, Shen LQ, Walker MJ, Sheridan RP. A model for predicting likely sites of CYP3A4-mediated metabolism on drug-like molecules. J Med Chem. 2003;46:1330–6. doi: 10.1021/jm020400s. [DOI] [PubMed] [Google Scholar]
- 18.Balakin KV, Ekins S, Bugrim A, et al. Kohonen maps for prediction of binding to human cytochrome P450 3A4. Drug Metab Dispos. 2004;32:1183–9. doi: 10.1124/dmd.104.000356. [DOI] [PubMed] [Google Scholar]
- 19.Ekins S, Andreyev S, Ryabov A, et al. Computational prediction of human drug metabolism. Expert Opin Drug Metab Toxicol. 2005;1:303–24. doi: 10.1517/17425255.1.2.303. [DOI] [PubMed] [Google Scholar]
- 20.Ekins S, Kirillov E, Rakhmatulin EA, Nikolskaya T. A novel method for visualizing nuclear hormone receptor networks relevant to drug metabolism. Drug Metab Dispos. 2005;33:474–81. doi: 10.1124/dmd.104.002717. [DOI] [PubMed] [Google Scholar]
- 21.Cruciani G, Carosati E, De Boeck B, et al. MetaSite understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem. 2005;48:6970–9. doi: 10.1021/jm050529c. [DOI] [PubMed] [Google Scholar]
- 22.Borodina Y, Sadym A, Filimonov D, Blinova V, Dmitriev A, Poroikov V. Predicting biotransformation potential from molecular structure. J Chem Inf Comput Sci. 2003;43:1636–46. doi: 10.1021/ci034078l. [DOI] [PubMed] [Google Scholar]
- 23.Jones JP, Mysinger M, Korzekwa KR. Computational models for cytochrome P450: a predictive electronic model for aromatic oxidation and hydrogen atom abstraction. Drug Metab Dispos. 2002;30:7–12. doi: 10.1124/dmd.30.1.7. [DOI] [PubMed] [Google Scholar]
- 24.Anari MR, Baillie TA. Bridging cheminformatic metabolite prediction and tandem mass spectrometry. Drug Discov Today. 2005;10:711–7. doi: 10.1016/S1359-6446(05)03445-8. [DOI] [PubMed] [Google Scholar]
- 25.Anari MR, Sanchez RI, Bakhtiar R, Franklin RB, Baillie TA. Integration of knowledge-based metabolic predictions with liquid chromatography data-dependent tandem mass spectrometry for drug metabolism studies application to studies on the biotransformation of indinavir. Analytical Chem. 2004;76:823–32. doi: 10.1021/ac034980s. [DOI] [PubMed] [Google Scholar]
- 26.Klopman G, Dimayuga M, Talafous J. META 1. A program for the evaluation of metabolic transformation of chemicals. J Chem Inf Comput Sci. 1994;34:1320–5. doi: 10.1021/ci00022a014. [DOI] [PubMed] [Google Scholar]
- 27.Klopman G, Tu M, Talafous J. META 3. A genetic algorithm for metabolic transform priorities optimization. J Chem Inf Comput Sci. 1997;37:329–34. doi: 10.1021/ci9601123. [DOI] [PubMed] [Google Scholar]
- 28.Talafous J, Sayre LM, Mieyal JJ, Klopman G. META 2. A dictionary model of mammalian xenobiotic metabolism. J Chem Inf Comput Sci. 1994;34:1326–33. doi: 10.1021/ci00022a015. [DOI] [PubMed] [Google Scholar]
- 29.Ellis LB, Roe D, Wackett LP. The University of Minnesota Biocatalysis/Biodegradation Database the first decade. Nucleic Acids Res. 2006;34:D517–21. doi: 10.1093/nar/gkj076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou BK, Wackett LP, Ellis LB. Microbial pathway prediction a functional group approach. J Chem Inf Comput Sci. 2003;43:1051–7. doi: 10.1021/ci034018f. [DOI] [PubMed] [Google Scholar]
- 31.Jaworska J, Dimitrov S, Nikolova N, Mekenyan O. Probabilistic assessment of biodegradability based on metabolic pathways catabol system. SAR QSAR Environ Res. 2002;13:307–23. doi: 10.1080/10629360290002794. [DOI] [PubMed] [Google Scholar]
- 32.Greene N, Judson PN, Langowski JJ, Marchant CA. Knowledge-based expert systems for toxicity and metabolism prediction DEREK, StAR and METEOR. SAR QSAR Environ Res. 1999;10:299–314. doi: 10.1080/10629369908039182. [DOI] [PubMed] [Google Scholar]
- 33.Hatzimanikatis V, Li C, Ionita JA, Broadbelt LJ. Metabolic networks enzyme function and metabolite structure. Curr Opin Struct Biol. 2004;14:300–6. doi: 10.1016/j.sbi.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Finley SD, Broadbelt LJ, Hatzimanikatis V. Computational framework for predictive biodegradation. Biotechnol Bioeng. 2009;104:1086–97. doi: 10.1002/bit.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song JC, White CM. Clinical pharmacokinetics and selective pharmacodynamics of new angiotensin converting enzyme inhibitors an update. Clin Pharmacokinet. 2002;41:207–24. doi: 10.2165/00003088-200241030-00005. [DOI] [PubMed] [Google Scholar]
- 36.Kelly JG, O'Malley K. Clinical pharmacokinetics of the newer ACE inhibitors A review. Clin Pharmacokinet. 1990;19:177–96. doi: 10.2165/00003088-199019030-00003. [DOI] [PubMed] [Google Scholar]
- 37.Waldmeier F, Kaiser G, Ackermann R, et al. The disposition of [14C]-labelled benazepril HCl in normal adult volunteers after single and repeated oral dose. Xenobiotica. 1991;21:251–61. doi: 10.3109/00498259109039467. [DOI] [PubMed] [Google Scholar]
- 38.Creasey WA, Morrison RA, Singhvi SM, Willard DA. Pharmacokinetics of intravenous captopril in healthy men. Eur J Clin Pharmacol. 1988;35:367–70. doi: 10.1007/BF00561366. [DOI] [PubMed] [Google Scholar]
- 39.Begg EJ, Bailey RR, Lynn KL, Robson RA, Frank GJ, Olson SC. The pharmacokinetics of angiotensin converting enzyme inhibitors in patients with renal impairment. J Hypertens suppl. 1989;7:S29–32. [PubMed] [Google Scholar]
- 40.Marzo A, Dal Bo L, Mazzucchelli P, et al. Pharmacokinetic and pharmacodynamic comparative study of zofenopril and enalapril in healthy volunteers. Arzneimittelforschung. 2002;52:233–42. doi: 10.1055/s-0031-1299886. [DOI] [PubMed] [Google Scholar]
- 41.Kubo SH, Cody RJ. Clinical pharmacokinetics of the angiotensin converting enzyme inhibitors. A review. Clin Pharmacokinet. 1985;10:377–91. doi: 10.2165/00003088-198510050-00001. [DOI] [PubMed] [Google Scholar]
- 42.Singhvi SM, Duchin KL, Morrison RA, Willard DA, Everett DW, Frantz M. Disposition of fosinopril sodium in healthy subjects. Br J Clin Pharmacol. 1988;25:9–15. doi: 10.1111/j.1365-2125.1988.tb03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalasz H, Petroianu G, Tekes K, Klebovich I, Ludanyi K, Gulyas Z. Metabolism of moexipril to moexiprilat determination of In vitro metabolism using HPLC-ES-MS. Med Chem. 2007;3:101–6. doi: 10.2174/157340607779317490. [DOI] [PubMed] [Google Scholar]
- 44.Verpooten GA, Genissel PM, Thomas Jr, De Broe ME. Single dose pharmacokinetics of perindopril and its metabolites in hypertensive patients with various degrees of renal insufficiency. Br J Clin Pharmacol. 1991;32:187–92. doi: 10.1111/j.1365-2125.1991.tb03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cetnarowski-Cropp AB. Quinapril a new second-generation ACE inhibitor. DICP. 1991;25:499–504. doi: 10.1177/106002809102500510. [DOI] [PubMed] [Google Scholar]
- 46.Meyer BH, Muller FO, Badian M, et al. Pharmacokinetics of ramipril in the elderly. Am J Cardiol. 1987;59:33D–37D. doi: 10.1016/0002-9149(87)90050-6. [DOI] [PubMed] [Google Scholar]
- 47.Conen H, Brunner HR. Pharmacologic profile of trandolapril, a new angiotensin-converting enzyme inhibitor. Am Heart J. 1993;125:1525–31. doi: 10.1016/0002-8703(93)90450-n. [DOI] [PubMed] [Google Scholar]
- 48.Mehvar R, Brocks DR. Stereospecific pharmacokinetics and pharmacodynamics of beta-adrenergic blockers in humans. J Pharm Pharm Sci. 2001;4:185–200. [PubMed] [Google Scholar]
- 49.Sankey MG, Gulaid A, Kaye CM. Preliminary study of the disposition in man of acebutolol and its metabolite, diacetolol, using a new stereoselective hplc method. J Pharm Pharmacol. 1984;36:276–7. doi: 10.1111/j.2042-7158.1984.tb04370.x. [DOI] [PubMed] [Google Scholar]
- 50.Bai SA, Walle T. Isolation, purification, and structure identification of glucuronic acid conjugates of propranolol and alprenolol and their ring-hydroxylated metabolites. Drug Metab Dispos. 1984;12:749–54. [PubMed] [Google Scholar]
- 51.Reeves PR, McAinsh J, McIntosh DA, Winrow MJ. Metabolism of atenolol in man. Xenobiotica. 1978;8:313–20. doi: 10.3109/00498257809060956. [DOI] [PubMed] [Google Scholar]
- 52.Ludden TM, Boyle DA, Gieseker D , et al. Absolute bioavailability and dose proportionality of betaxolol in normal healthy subjects. J Pharm Sci. 1988;77:779–83. doi: 10.1002/jps.2600770913. [DOI] [PubMed] [Google Scholar]
- 53.Wong YW, Ludden TM. Determination of betaxolol and its metabolites in blood and urine by high-performance liquid chromatography with fluorimetric detection. J Chromatogr. 1990;534:161–72. doi: 10.1016/s0378-4347(00)82158-1. [DOI] [PubMed] [Google Scholar]
- 54.Leopold G. Balanced pharmacokinetics and metabolism of bisoprolol. J Cardiovasc Pharmacol. 1986;8(suppl 11):S16–20. doi: 10.1097/00005344-198511001-00003. [DOI] [PubMed] [Google Scholar]
- 55.Neugebauer G, Akpan W, von Mollendorff E, Neubert P, Reiff K. Pharmacokinetics and disposition of carvedilol in humans. J Cardiovasc Pharmacol. 1987;10(suppl 11):S85–8. [PubMed] [Google Scholar]
- 56.Wolman RL, Fiedler MA. Esmolol and beta-adrenergic blockade. AANA J. 1991;59:541–8. [PubMed] [Google Scholar]
- 57.Gal J, Zirrolli JA, Lichtenstein PS. Labetalol is metabolized oxidatively in humans. Res Commun Chem Pathol Pharmacol. 1988;62:3–17. [PubMed] [Google Scholar]
- 58.Alton KB, Chan TM, Pramanik BN. Urinary metabolites of (R), (R)-labetalol. Drug Metab Dispos. 1994;22:866–72. [PubMed] [Google Scholar]
- 59.Li Q, Wang R. Simultaneous analysis of tramadol, metoprolol and their metabolites in human plasma and urine by high performance liquid chromatography. Chin Med J (Engl) 2006;119:2013–7. [PubMed] [Google Scholar]
- 60.Riddell JG, Harron DW, Shanks RG. Clinical pharmacokinetics of beta-adrenoceptor antagonist, sAn update. Clin Pharmacokinet. 1987;12:305–20. doi: 10.2165/00003088-198712050-00001. [DOI] [PubMed] [Google Scholar]
- 61.Kamali F, Howes A, Thomas SH, Ford GA, Snoeck E. A pharmacokinetic and pharmacodynamic interaction study between nebivolol and the H2-receptor antagonists cimetidine and ranitidine. Br J Clin Pharmacol. 1997;43:201–4. doi: 10.1046/j.1365-2125.1997.54212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dieterle W, Faigle JW, Kung W, Theobald W. The disposition and metabolism of 14C-oxprenolol Cl in man. Xenobiotica. 1986;16:181–91. doi: 10.3109/00498258609043521. [DOI] [PubMed] [Google Scholar]
- 63.Riess W, Huerzeler H, Raschdorf F. The metabolites of oxprenolol (Trasicor) in man. Xenobiotica. 1974;4:365–73. doi: 10.3109/00498257409052112. [DOI] [PubMed] [Google Scholar]
- 64.Vallner JJ, Jun HW, Needham TE, et al. Plasma level studies of penbutolol after oral dose in man. J Clin Pharmacol. 1977;17:231–6. doi: 10.1177/009127007701700407. [DOI] [PubMed] [Google Scholar]
- 65.Spahn H, Kirch W, Hajdu P, Mutschler E, Ohnhaus EE. Penbutolol Pharmacokinetics the influence of concomitant administration of cimetidine. Eur J Clin Pharmacol. 1986;29:555–60. doi: 10.1007/BF00635892. [DOI] [PubMed] [Google Scholar]
- 66.Muller FO, Hundt HK, Bromley PA, Torres J, Vanderbeke O. Single and divided doses of penbutolol. Clin Pharmacol Ther. 1979;25:528–35. doi: 10.1002/cpt1979255part1528. [DOI] [PubMed] [Google Scholar]
- 67.Maurer G, Donatsch P, Galliker H, Kiechel Jr, Meier J. Pindolol disposition and metabolism in rhesus monkeys after chronic treatment. Xenobiotica. 1981;11:33–41. doi: 10.3109/00498258109045269. [DOI] [PubMed] [Google Scholar]
- 68.Paterson JW, Conolly ME, Dollery CT. The pharmacodynaics and metabolism of propanolol in Man. Pharmacologia Clinica. 1970;2:127–133. [Google Scholar]
- 69.Suchanova B, Kostiainen R, Ketola RA. Characterization of the In vitro metabolic profile of amlodipine in rat using liquid chromatography-mass spectrometry. Eur J Pharm Sci. 2008;33:91–9. doi: 10.1016/j.ejps.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 70.Yeung PK, Mosher SJ, Pollak PT. Pharmacokinetics and metabolism of diltiazem in rabbits after a single intravenous or single oral administration. Eur J Drug Metab Pharmacokinet. 1991;16:69–74. doi: 10.1007/BF03189877. [DOI] [PubMed] [Google Scholar]
- 71.Baarnhielm C, Backman A, Hoffmann KJ, Weidolf L. Biotransformation of felodipine in liver microsomes from rat, dog, and man. Drug Metab Dispos. 1986;14:613–8. [PubMed] [Google Scholar]
- 72.Jean C, Laplanche R. Assay of isradipine and of its major metabolites in biological fluids by capillary gas chromatography and chemical ionization mass spectrometry. J Chromatogr. 1988;428:61–9. doi: 10.1016/s0378-4347(00)83890-6. [DOI] [PubMed] [Google Scholar]
- 73.Liu XQ, Ren YL, Qian ZY, Wang GJ. Enzyme kinetics and inhibition of nimodipine metabolism in human liver microsomes. Acta Pharmacol Sin. 2000;21:690–4. [PubMed] [Google Scholar]
- 74.Scherling D, Buhner K, Krause HP, Karl W, Wunsche C. Biotransformation of nimodipine in rat, dog, and monkey. Arzneimittelforschung. 1991;41:392–8. [PubMed] [Google Scholar]
- 75.Scherling D, Karl W, Ahr G, Ahr HJ, Wehinger E. Pharmacokinetics of nisoldipine III Biotransformation of nisoldipine in rat, dog, monkey, and man. Arzneimittelforschung. 1988;38:1105–10. [PubMed] [Google Scholar]
- 76.Mast V, Fischer C, Mikus G, Eichelbaum M. Use of pseudoracemic nitrendipine to elucidate the metabolic steps responsible for stereoselective disposition of nitrendipine enantiomers. Br J Clin Pharmacol. 1992;33:51–9. doi: 10.1111/j.1365-2125.1992.tb04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pauli-Magnus C, von Richter O, Burk O, et al. Characterization of the major metabolites of verapamil as substrates and inhibitors of P-glycoprotein. J Pharmacol Exp Ther. 2000;293:376–82. [PubMed] [Google Scholar]
- 78.Borlak J, Walles M, Levsen K, Thum T. Verapamil metabolism in cultures of primary human coronary arterial endothelial cells. Drug Metab Dispos. 2003;31:888–91. doi: 10.1124/dmd.31.7.888. [DOI] [PubMed] [Google Scholar]
- 79.Patki KC, Von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes p450: role of cyp3a4 and cyp3a5. Drug Metab Dispos. 2003;31:938–44. doi: 10.1124/dmd.31.7.938. [DOI] [PubMed] [Google Scholar]
- 80.Yip MS, Coates PE, Thiessen JJ. High-performance liquid chromatographic analysis of amiloride in plasma and urine. J Chromatogr. 1984;307:343–50. doi: 10.1016/s0378-4347(00)84105-5. [DOI] [PubMed] [Google Scholar]
- 81.Cook CS, Berry LM, Bible RH, Hribar JD, Hajdu E, Liu NW. Pharmacokinetics and metabolism of [14C]eplerenone after oral administration to humans. Drug Metab Dispos. 2003;31:1448–55. doi: 10.1124/dmd.31.11.1448. [DOI] [PubMed] [Google Scholar]
- 82.Los LE, Pitzenberger SM, Ramjit HG, Coddington AB, Colby HD. Hepatic metabolism of spironolactone. Production of 3-hydroxy-thiomethyl metabolites. Drug Metab Dispos. 1994;22:903–8. [PubMed] [Google Scholar]
- 83.Fuhr U, Kober S, Zaigler M, Mutschler E, Spahn-Langguth H. Rate-limiting biotransformation of triamterene is mediated by CYP1A2. Int J Clin Pharmacol Ther. 2005;43:327–34. doi: 10.5414/cpp43327. [DOI] [PubMed] [Google Scholar]
- 84.Hagos Y, Bahn A, Vormfelde SV, Brockmoller J, Burckhardt G. Torasemide transport by organic anion transporters contributes to hyperuricemia. J Am Soc Nephrol. 2007;18:3101–9. doi: 10.1681/ASN.2007010106. [DOI] [PubMed] [Google Scholar]
- 85.Halladay SC, Carter DE, Sipes IG, Brodie BB, Bressler R. Evidence for the metabolism of bumetanide in man. Life Sci. 1975;17:1003–9. doi: 10.1016/0024-3205(75)90455-5. [DOI] [PubMed] [Google Scholar]
- 86.Klaassen CD, Fitzgerald TJ. Metabolism and biliary excretion of ethacrynic acid. J Pharmacol Exp Ther. 1974;191:548–56. [PubMed] [Google Scholar]
- 87.Pichette V, du Souich P. Role of the kidneys in the metabolism of furosemide its inhibition by probenecid. J Am Soc Nephrol. 1996;7:345–9. doi: 10.1681/ASN.V72345. [DOI] [PubMed] [Google Scholar]
- 88.Prandota J, Witkowska M. Pharmacokinetics and metabolism of furosemide in man. European Journal of Drug Metabolism and Pharmacokinetics. 1976;4:177–181. [Google Scholar]
- 89.Barroso MB, Alonso RM, Jimenez RM. Simultaneous determination of torasemide and its major metabolite M5 in human urine by high-performance liquid chromatography-electrochemical detection. J Chromatogr Sci. 2001;39:491–6. doi: 10.1093/chromsci/39.11.491. [DOI] [PubMed] [Google Scholar]
- 90.Welling PG. Pharmacokinetics of the thiazide diuretics. Biopharm Drug Dispos. 1986;7:501–35. doi: 10.1002/bdd.2510070602. [DOI] [PubMed] [Google Scholar]
- 91.Babu B MS, Meyyanathan SN, Suresh B. Pharmacokinetic Evaluation of Metolazone Tablets using Healthy Human Volunteers. journal of Bioequivalence and Bioavailability. 2010;2:15–17. [Google Scholar]
- 92.Sun H, Moore C, Dansette PM, Kumar S, Halpert Jr, Yost GS. Dehydrogenation of the indoline-containing drug 4-chloro-N-(2-methyl-1-indolinyl)-3-sulfamoylbenzamide (indapamide) by CYP3A4: correlation with in silico predictions. Drug Metab Dispos. 2009;37:672–84. doi: 10.1124/dmd.108.022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Testa B, Balmat AL, Long A, Judson P. Predicting drug metabolism--an evaluation of the expert system METEOR. Chem Biodivers. 2005;2:872–85. doi: 10.1002/cbdv.200590064. [DOI] [PubMed] [Google Scholar]
- 94.de Groot MJ, Ackland MJ, Horne VA, Alex AA, Jones BC. A novel approach to predicting P450 mediated drug metabolism. CYP2D6 catalyzed N-dealkylation reactions and qualitative metabolite predictions using a combined protein and pharmacophore model for CYP2D6. J Med Chem. 1999;42:4062–70. doi: 10.1021/jm991058v. [DOI] [PubMed] [Google Scholar]
- 95.Braga RC, Alves VM, Fraga CA, Barreiro EJ, de Oliveira V, Andrade CH. Combination of docking, molecular dynamics and quantum mechanical calculations for metabolism prediction of 3, 4-methylenedioxybenzoyl-2-thienylhydrazone. J Mol Model. 2012;18:2065–78. doi: 10.1007/s00894-011-1219-9. [DOI] [PubMed] [Google Scholar]
- 96.Lonsdale R, Houghton KT, Zurek J, et al. Quantum mechanics/molecular mechanics modeling of regioselectivity of drug metabolism in cytochrome P450 2C9. J Am Chem Soc. 2013;135:8001–15. doi: 10.1021/ja402016p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rydberg P, Gloriam DE, Olsen L. The SMARTCyp cytochrome P450 metabolism prediction server. Bioinformatics. 2010;26:2988–9. doi: 10.1093/bioinformatics/btq584. [DOI] [PubMed] [Google Scholar]
- 98.Liu R, Liu J, Tawa G, Wallqvist A. 2D SMARTCyp reactivity-based site of metabolism prediction for major drug-metabolizing cytochrome P450 enzymes. J Chem Inf Model. 2012;52:1698–712. doi: 10.1021/ci3001524. [DOI] [PubMed] [Google Scholar]
- 99.de Groot MJ, Bijloo GJ, Martens BJ, van Acker FA, Vermeulen NP. A refined substrate model for human cytochrome P450 2D6. Chem Res Toxicol. 1997;10:41–8. doi: 10.1021/tx960129f. [DOI] [PubMed] [Google Scholar]
- 100.de Groot MJ, Ackland MJ, Horne VA, Alex AA, Jones BC. Novel approach to predicting P450-mediated drug metabolism development of a combined protein and pharmacophore model for CYP2D6. J Med Chem. 1999;42:1515–24. doi: 10.1021/jm981118h. [DOI] [PubMed] [Google Scholar]
- 101.de Groot MJ, Bijloo GJ, Hansen KT, Vermeulen NP. Computer prediction and experimental validation of cytochrome P4502D6-dependent oxidation of GBR 12909. Drug Metab Dispos. 1995;23:667–9. [PubMed] [Google Scholar]
- 102.de Groot MJ, Bijloo GJ, van Acker FA, Fonseca Guerra C, Snijders JG, Vermeulen NP. Extension of a predictive substrate model for human cytochrome P4502D6. Xenobiotica. 1997;27:357–68. doi: 10.1080/004982597240514. [DOI] [PubMed] [Google Scholar]
- 103.Li H, Sun J, Sui X, et al. First-principle, structure-based prediction of hepatic metabolic clearance values in human. Eur J Med Chem. 2009;44:1600–6. doi: 10.1016/j.ejmech.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 104.Wu CY, Benet LZ. Predicting drug disposition via application of BCS transport/absorption/ elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res. 2005;22:11–23. doi: 10.1007/s11095-004-9004-4. [DOI] [PubMed] [Google Scholar]
- 105.Blume HH, Schug BS. The biopharmaceutics classification system (BCS): class III drugs - better candidates for BA/BE waiver? Eur J Pharm Sci. 1999;9:117–21. doi: 10.1016/s0928-0987(99)00076-7. [DOI] [PubMed] [Google Scholar]
- 106.Polli JE, Yu LX, Cook JA , et al. Summary workshop report biopharmaceutics classification system--implementation challenges and extension opportunities. J Pharm Sci. 2004;93:1375–81. doi: 10.1002/jps.20064. [DOI] [PubMed] [Google Scholar]
- 107.Takagi T, Ramachandran C, Bermejo M, Yamashita S, Yu LX, Amidon GL. A provisional biopharmaceutical classification of the top 200 oral drug products in the United States, Great Britain, Spain, and Japan. Mol Pharm. 2006;3:631–43. doi: 10.1021/mp0600182. [DOI] [PubMed] [Google Scholar]
- 108.McDonald AG, Tipton KF. Fifty-five years of enzyme classification advances and difficulties. FEBS J. 2014;281:583–92. doi: 10.1111/febs.12530. [DOI] [PubMed] [Google Scholar]
- 109.Knox C, Law V, Jewison T , et al. DrugBank 30: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39:D1035–41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wishart DS, Knox C, Guo AC, et al. DrugBank a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–6. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wishart DS, Knox C, Guo AC, et al. DrugBank a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]