Abstract
Staphylococcal superantigen-like (SSL) proteins, which are encoded by a cluster of eleven ssl genes, contribute to the Staphylococcus aureus virulence. Recently we reported ssl8 expression profiles in seven clinically important strains—MW2, USA300FPR3757, MSSA476, Newman, RN6390, Mu50, and N315—and showed the differential expression of ssl8 in Newman, RN6390, and USA300FPR3757 strains, despite harboring identical allelic forms of ssl8, suggesting the roles for different regulatory elements for this gene in different S. aureus strains. In this communication, using RN6390, a common laboratory S. aureus strain and its isogenic knockout mutant strains of agr, sae, sarA, sigB, rot, and the agr-/sigB− double mutant, we showed that SarA and Rot are inducer and repressor, respectively, for ssl8 expression in RN6390. This is in contrast to the Newman strain, where ssl8 is positively regulated by Sae but negatively by Agr, indicating the variable expression of ssl8 in clinical strains is more likely due to strain-specific regulatory elements.
Keywords: Staphylococcus aureus, ssl8, SarA, Rot, MRSA, Gene regulation
Pathogenesis of Staphylococcus aureus is complex and involves the coordinate expression of multiple toxin genes that are tightly controlled by global regulators at the transcription level.1–3 These toxins are synthesized preferentially during the transition between exponential and stationary growth phases.4 Among its large number of secreted toxins, is a group of staphylococcal superantigen-like (SSL) proteins encoded by 11 ssl (formerly known as staphylococcal exotoxin-like or set) genes.5,6 Genes for ssl are located on a highly variable region called RD13 that corresponds to the ssl containing genomic islands (vSaα, SaPIn2, or SaPIm2) in S. aureus strains.5,7 SSL proteins have limited overall sequence homology to the staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin 1 (TSST-1) and, thus, represent a novel family of exotoxin-like proteins.6
ssl genes are present in all S. aureus strains studied so far, suggesting a non-redundant role in bacterial survival.5,8,9 The overall order of ssl genes is conserved, and the allelic variants of the same gene in different strains share 85% to 100% sequence similarity,7 indicating the evidence of selective pressure during the infection. SSL proteins in the same strain have 36% to 67% sequence homology.6,7 Although the SSL proteins have sequence homology and similar predicted structures,5,6,8 each protein studied so far has a related but distinct function in host-pathogen interactions.6,9–12 SSL proteins have been shown to help S. aureus to escape from the protective adaptive immune response of the host and thus may contribute to bacterial pathogenicity.6,9–12 SSL5 interferes with the interaction between PSGL-1 (P-selectin glycoprotein ligand-1) and P-selectin and prevents neutrophil extravasations toward the site of infection.11 SSL7 inhibits IgA dependent cellular effector functions mediated by FcαRI, such as phagocytosis, degranulation, and respiratory burst.12 SSL7 and SSL9 interact with human antigen presenting cells (monocytes and dendritic cells) at independent active sites.10 SSL11 induces proinflammatory cytokines from peripheral blood mononuclear cells.13 SSL8 binds to tenascin C (TNC), a glycoprotein and inhibits TNC and fibronectin interaction and cell motility in keratinocytes.14 ssl8 gene seem to be more frequently present in S. aureus isolates causing atopic dermatitis.15
The success of S. aureus as a pathogen lies in its ability to exploit multiple virulence factors regulated by complex network of global gene regulators that includes but not limited to Agr(accessory gene regulator), SarA(staphylococcal accessory regulator), Sae (S. aureus exoprotein expression), SigB (sigma factor B) and Rot (repressor of toxins).2–4,9,16,17 Recently, we reported the differential expression of ssl8 in seven clinically relevant S. aureus isolates and showed that ssl8 expression in the Newman strain is regulated by Sae and Agr.16 In this brief communication, we show that the ssl8 expression in the common laboratory strain, RN6390 is regulated differently than the Newman strain suggesting ssl8 regulation is strain dependent.
Materials and Methods
Bacterial Strains, Media and Culture Conditions
The laboratory S. aureus strain RN6390 and its isogenic mutants used in this study are listed in table 1. For each experiment, S. aureus was grown either in tryptic soy broth (TSB) or on tryptic soy agar (TSA) plates (Beckton Dickinson) as needed. For gene expression studies, an overnight shaking culture, grown at 37°C in TSB was used to inoculate 50 ml of fresh TSB (dilution, 1:200). Bacterial growth was subsequently monitored by incubating the flasks at 37°C with constant shaking and measuring the turbidity of the culture every 30 min at OD600 using a spectrophotometer (Beckman Coulter, Inc., CA) until the cultures reached the stationary phase. There were no growth differences between RN6390 and mutant strains (data not shown).
Table 1.
S. aureus wild type and mutant strains used in this study.
Construction of an agr/sigB Knockout Mutant of RN6390
A agr/sigB mutant in RN6390 background was generated using a phage transduction procedure as described previously.9
RNA Extraction
For gene expression studies, both the RN6390 and the mutant strains were grown up to early-stationary phase (5.5 hours) and harvested immediately for RNA isolations. RNA was extracted using a Qiagen RNeasy Mini kit (Qiagen, Valencia, CA) by following the manufacturer’s recommendations with a few modifications. Briefly, a 1.5 mL of bacterial culture was dispensed into 3 mL of RNA Protect (Qiagen Inc, CA) in a 15-mL tube and mixed by vortexing for 5 seconds followed by 5 minutes incubation at room temperature. The cells were pelleted by centrifuging at 1252g for 5 minutes followed by a wash with DEPC treated water and re-suspended into 1.0 mL of TRIzol reagent. The suspension was then transferred to a 2-mL Lysing Matrix tube (MP Biomedicals, Irvine, CA) and homogenized by shaking three times in the mini bead beater for 20 seconds at a speed of 6 m/s. The cells were incubated on ice for 1 minute in between each run. To the homogenate, 200 μL of RNA-grade chloroform was added and vortexed for 15 seconds, followed by a centrifugation at high speed for 5 minutes and carefully aspirated the upper layer. To this supernatant, 500 μL of RNA-grade isopropyl alcohol was added and mixed by inverting the tube several times. This sample (~700 μL) was then transferred to an RNeasy column and centrifuged for 15 seconds at ≥8000g and followed the manufacturer’s guidelines for RNeasy cleanup protocol (Qiagen Inc, CA). Finally, RNA was eluted with 50 μL of DEPC-treated water. RNA concentration was determined by using Bio-Rad SmartSpec Plus Spectrophotometer (Analytical Instruments, LLC, MN). An aliquot of each RNA sample was electrophoresed on a 1.0% agarose gel to assess its integrity and quality.
Quantitative Real-Time PCR Analysis of ssl8 Expression
To quantify the ssl8 transcript levels in RN6390 and its mutant strains, the isolated RNA samples were subjected to DNase treatment with Turbo DNA-free kit (Ambion, Austin, TX) and confirmed to be DNA free by PCR before cDNA synthesis. The first strand cDNA was prepared with 2 μg of total RNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Roche, Foster City, CA) according to the manufacturer’s instructions.
From the above reaction mix, 2 μL (~100 to 200 ng) of cDNA template was further used for quantitative real-time PCR in the Light Cycler (Roche Diagnostics Corp, Indianapolis, IN). The reaction was carried out using TaqMan Universal PCR Master Mix (2X) (Applied Biosystems Inc, CA) and the TaqMan Gene Expression Assays [which included PCR forward and reverse primers (900 nM/μL) and 6-FAM dye-labeled MGB probe (250 nM/μL) specific for ssl8, RNAIII and gmk individually] (Applied Biosystems Inc, CA) by following the manufacturer’s instructions. The PCR primers and probes are listed in table 2.
Table 2.
Primers and probes used in this study.
| Gene | Primer | Primer sequence (5′ → 3′) | Probe sequence | Reference |
|---|---|---|---|---|
| ssl8 | Forward | CCAACAAAATCAAAAGTTCCA | FAM-CACATGGTTTAGATGT CTTTGCGGTACC-NFQ | This study |
| Reverse | CACCACTAACACTAAATATTCTTCCA | This study | ||
| Gmk | Forward | ACTAGGGATGCGTTTGAAGCTTTAA | FAM-AAAGATGACCAATTT ATAG AATATG-NFQ | This study |
| Reverse | ACCATAATAGTTGCCTACAT | This study | ||
| RNAIII | Forward | TCCATTTTACTAAGTCACCGATTGT | FAM-ATCTTGTGCCATTGAAATCACTCCTTCCTT-NFQ | 33 |
| Reverse | TGTGATGGAAAATAGTTGATGAGTTGT | 33 |
Relative quantification of ssl8, RNAIII (target genes), and gmk (guanylate kinase; endogenous control) was performed using 2−ΔΔCT calculation according to the manufacturer’s guidelines (Roche Diagnostics Corp., Indianapolis, IN). This method compensates for the factors such as variability in cDNA synthesis and template concentration, and calculates transcript ratios (ssl8/gmk and RNAIII/gmk) rather than absolute values. However, the reliability of 2−ΔΔCT calculation depends on the PCR efficiency. By validation experiment, we confirmed that the PCR efficiency for ssl8, RNAIII and gmk was approximately two, as expected. To quantify the ssl8 and RNAIII expressions in our experiments, we used gmk as a reference gene.13,19. All the results were obtained from two independent cultures.
Results
RN6390 ssl8 Regulation by SarA and Rot
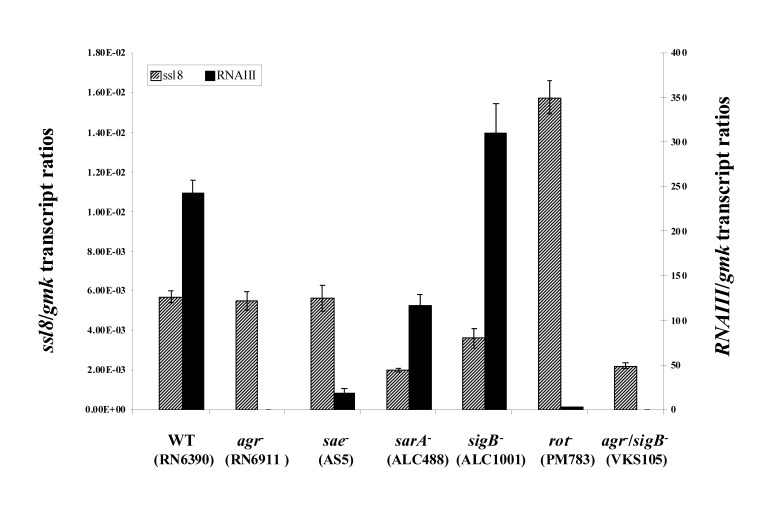
We have quantified the ssl8 and RNAIII transcript levels in RN6390 and its mutant strains: RN6911 (RN6390 agr::tetM), AS5 (RN6390 sae::Tn917), ALC488 (RN6390 ΔsarA::ermC), ALC1001 (RN6390 ΔsigB::ermC), PM783 (RN6390 Δrot::tetM) and VKS105 (Δagr::tetM/sigB::kanr double mutant of RN6390).
The ssl8/gmk transcript ratios in RN6390 (5.68 × 10−03) were comparable to the ratios in sae− strain, AS5 (5.64 × 10−03) and agr− strain, RN6911 (5.48 × 10−03). The ssl8/gmk transcript ratios in sarA− strain, ALC488 and sigB− strain, ALC1001 were 1.98 × 10−03 and 3.63 × 10−03, respectively. In case of rot− strain, PM783 the ratio was 1.57 × 10−02 and in the double mutant, agr−/sigB− strain VKS105, the ratio was 2.20 × 10−03 (figure 1).
Figure 1.
Transcript ratios of ssl8/gmk and RNAIII/gmk, quantified at early-stationary phase from RN6390 and its isogenic knockout mutants: RN6911 (RN6390 agr::tetM), AS5 (RN6390 sae::Tn917), ALC488 (RN6390 ΔsarA::ermC), ALC1001 (RN6390 ΔsigB::ermC), PM783 (RN6390 Δrot::tetM) and VKS105 (Δagr::tetM/sigB::kanr double mutant of RN6390). The striped and dark bars indicate the ssl8 (on primary axis) and RNAIII (on secondary axis) expression levels, respectively. Data represent the mean values of two independent measurements.
The sarA and rot null mutations down-regulated (~3 fold) and up-regulated (~3 fold) ssl8 expression, respectively, suggesting that the SarA is a positive regulator, whereas Rot is a negative regulator of ssl8 expression in RN6390 (figure 1). However, the sae and agr mutations did not change the levels of ssl8 transcripts, in contrast to sigB mutation, which caused ~1.5 fold decrease compared to the RN6390, suggesting SigB could be another positive regulator of ssl8. The double mutant, agr/sigB (VKS105) showed ~2.5 fold repression of ssl8 compared to the RN6390, suggesting agr and sigB act in conjunction to regulate the ssl8 expression.
Since Agr is one of the master regulators of exotoxins, we determined the transcript levels of RNAIII in these strains as well. The RNAIII/gmk transcript ratio in sigB− strain ALC1001 showed 1.2 fold increases compared to its wild type strain, RN6390. However, the RNAIII/gmk transcript ratios in sarA− (ALC488), sae− (AS5), and rot− (PM783) strains showed 2-, 13-, and 85-fold decreases, respectively, compared to the RN6390. As expected, the RNAIII levels were not detectable in agr− strains RN6911 (agr−) and VKS105 (agr−/sigB−). Since Agr does not seem to have a role in regulation of ssl8 in RN6390, the increase and decrease in RNAIII levels did not show any corresponding up or down regulation of ssl8 expression (figure 1).
Discussion
Staphylococcal exotoxins and SSL proteins significantly contribute to the S. aureus pathogenicity.1,6,9–11,19–21 Their production is controlled by a network of global regulators, which act both positively and negatively depending on the virulence genes and host strains.2–4,9,16,17 We showed here that sarA and rot mutations down-regulated and up-regulated ssl8 expression, respectively, in RN6390, suggesting these two were the positive and negative regulators of ssl8 in this strain (figure 1). SarA is a known positive regulator of a number of genes such as α- and δ-hemolysins, Agr response genes, agrA-D, fnbA and fnbB (fibronectin binding proteins), splA, splB, splD and splF (serine proteases), ssl4, and negative regulator of surface protein A (spa).22 Similarly, a sarA homolog, rot is a known master regulator of a large number of genes as well. Using Affymetrix GeneChip based approach, Said-Salim et al showed that rot up regulates the expression of more than 80 genes (eg, coagulase, clumping factor B) and down regulates ~60 genes (eg, β-hemolysin, serine proteases).9 In addition, we observed that sigB mutation also caused reduction in ssl8 transcript levels but not to the extent of sarA mutant, indicating that SigB is a relatively weak positive regulator of ssl8 in RN6390. In the agr−/sigB− double mutant, the expression of ssl8 was down regulated. Similar down regulation of exotoxin genes has been shown in sar−/agr− double mutants compared to their parental and single mutant strains.23 Our results indicated that Agr and SigB possibly act in concert to regulate ssl8 expression. It was interesting to note that ssl8 in the Newman strain was positively regulated by Sae and SigB and negatively by Agr, which is in contrast to RN6390.18 Indeed, others have shown that ssl7 and ssl11 are positively regulated by SaeRS system in Newman strain.24 All of the Sae regulated genes were also found to be influenced by SigB but not always in the same direction.25
RNAIII levels do not seem to have any effect on ssl8 expression in RN6390. However, it was interesting to note that RNAIII levels were highest in RN6390 compared to MW2, Newman, FPR3757, MSSA476, Mu50, and N315 due to deletion in rsbU gene of sigB operon,18 suggesting the sigB has negative effect on RNAIII expression as has also been shown by others. Lauderdale et al26 had reported that the agr RNAIII levels were elevated in the sigB mutants causing the anti-biofilm phenotypic effect. Unlike previously reported, we observed reduced RNAIII transcript levels in sae− strain of RN6390.27 Curiously, RNAIII levels were also reduced in rot− strain of RN6390 (figure 1). Indeed, RNAIII interacts with rot mRNA and is involved in inhibition of rot mRNA translation.28,29
In summary, it appears that the expression and regulation of ssl in S. aureus is complicated and is determined by the strain specificity and not necessarily the common genetic background which is probably due to unrelated regulatory pathways. We showed here that the regulatory pathways of ssl8 in RN6390 are different from our findings for the ssl8 in the Newman strain18 suggesting regulation of many individual virulence factors in S. aureus are strain-specific. This observation further highlights the complexity in regulation of virulence in S. aureus strains and poses challenges in considering all S. aureus as equal.
Acknowledgements
We thank Drs. Peter J. McNamara, Christiane Wolz and Ambrose Cheung for providing some of the strains essential to this work.
Footnotes
Grant Support: This study was funded, in part, by research grant RO1 AI061385 from the National Institutes of Allergy and Infectious Diseases to SKS and Marshfield Clinic Research Foundation.
References
- 1.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol 2004;40:1–9. [DOI] [PubMed] [Google Scholar]
- 2.Cheung AL, Zhang G. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front Biosci 2002;7:d1825–d1842. [DOI] [PubMed] [Google Scholar]
- 3.Giraudo AT, Cheung AL, Nagel R. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol 1997;168:53–58. [DOI] [PubMed] [Google Scholar]
- 4.Kornblum JB, Kreiswirth BN, Projan SJ, Ross H, Novick RP. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick RP, (ed.) Molecular biology of the staphylococci. New York, NY: VCH Publishers; 1990. 373 402. [Google Scholar]
- 5.Fitzgerald JR, Reid SD, Ruotsalainen E, Tripp TJ, Liu M, Cole R, Kuusela P, Schlievert PM, Jarvinen A, Musser JM. Genome diversification in Staphylococcus aureus: Molecular evolution of a highly variable chromosomal region encoding the Staphylococcal exotoxin-like family of proteins. Infect Immun 2003;71:2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams RJ, Ward JM, Henderson B, Poole S, O’Hara BP, Wilson M, Nair SP. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect Immun 2000;68:4407–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 2001;357:1225–1240. [DOI] [PubMed] [Google Scholar]
- 8.Arcus VL, Langley R, Proft T, Fraser JD, Baker EN. The Three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J Biol Chem 2002;277:32274–32281. [DOI] [PubMed] [Google Scholar]
- 9.Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 2003;185:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Shangiti AM, Nair SP, Chain BM. The interaction between staphylococcal superantigen-like proteins and human dendritic cells. Clin Exp Immunol 2005;140:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bestebroer J, Poppelier MJ, Ulfman LH, Lenting PJ, Denis CV, van Kessel KP, van Strijp JA, de Haas CJ. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood 2007;109:2936–2943. [DOI] [PubMed] [Google Scholar]
- 12.Langley R, Wines B, Willoughby N, Basu I, Proft T, Fraser JD. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J Immunol 2005;174:2926–2933. [DOI] [PubMed] [Google Scholar]
- 13.Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol 2001;183:7094–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh S, Yamaoka N, Kamoshida G, Takii T, Tsuji T, Hayashi H, Onozaki K. Staphylococcal superantigen-like protein 8 (SSL8) binds to tenascin C and inhibits tenascin C-fibronectin interaction and cell motility of keratinocytes. Biochem Biophys Res Commun. 2013;433:127–132. [DOI] [PubMed] [Google Scholar]
- 15.Rojo A, Aguinaga A, Monecke S, Yuste JR, Gastaminza G, España A. Staphylococcus aureus genomic pattern and atopic dermatitis: may factors other than superantigens be involved? Eur J Clin Microbiol Infect Dis. 2014;33:651–658. [DOI] [PubMed] [Google Scholar]
- 16.Bronner S, Monteil H, Prevost G. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 2004;28:183–200. [DOI] [PubMed] [Google Scholar]
- 17.McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol 2000;182:3197–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pantrangi M, Singh VK, Wolz C, Shukla SK. Staphylococcal superantigen-like genes, ssl5 and ssl8 are positively regulated by sae and negatively regulated by agr FEMS Microbiol Lett 2010;308:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nieto PA, Covarrubias PC, Jedlicki E, Holmes DS, Quatrini R. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol Biol 2009;10:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 2000;13:16–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowy FD. Staphylococcus aureus infections. N Engl J Med 1998;339:520–532. [DOI] [PubMed] [Google Scholar]
- 22.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 2001;183:7341–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booth MC, Cheung AL, Hatter KL, Jett BD, Callegan MC, Gilmore MS. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun 1997; 65:1550–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol 2006;188:7742–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goerke C, Fluckiger U, Steinhuber A, Bisanzio V, Ulrich M, Bischoff M, Patti JM, Wolz C. Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect Immun 2005;73:3415–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 2003;149:2709–2717. [DOI] [PubMed] [Google Scholar]
- 28.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–1048. [DOI] [PubMed] [Google Scholar]
- 30.Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol Microbiol 2001;40:1439–1447. [DOI] [PubMed] [Google Scholar]
- 31.Cheung AL, Eberhardt K, Heinrichs JH. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun 1997;65:2243–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung AL, Chien YT, Bayer AS. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect Immun 1999;67:1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loughman JA, Fritz SA, Storch GA, Hunstad DA. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J Infect Dis 2009; 199:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]