Abstract
The presence of polymorphisms in the CYP2D6 gene may modulate enzyme level and activity, thereby affecting individual responses to pharmacological treatment. Here, we compared the prevalence of the CYP2D6*10, *4, and 14* alleles in an Iranian population of different ethnicities with those of other populations. Allele and genotype frequency distributions of CYP2D6*10 variants and predicted phenotypes including extensive metabolizers, intermediate metabolizers, and poor metabolizers were analysed in blood samples of 300 unrelated healthy individuals in an Iranian population using polymerase chain reaction (PCR)-restriction fragment length polymorphism, PCR-single-strand conformation polymorphism, and direct genomic DNA sequencing. The CYP2D6*4 (G1846A) and *14 (G1758A) allelic frequencies were not detected in different ethnicities, demonstrating the absence of a significant contribution of these alleles in Iranian populations. However, the T/T, C/T, and C/C genotype frequencies of the CYP2D6*10 allele were significantly different (P<0.01) in all Iranian ethnic groups. Additionally, the frequency of the homozygous T/T variant of the CYP2D6*10 allele was significantly high in the Lure (P<0.017) and low in the Kurd (P<0.002) ethnicities. The frequency of the T/T variant of the CYP2D6*10 allele in central Iran was the highest (P<0.001), while the south of Iran had the lowest frequency (P<0.001). The frequency of the C/T variant of the CYP2D6*10 allele was significantly a bit high (P<0.001) in females compare to males, while the frequencies of the T/T variant in females is similar to males, which are 24.4% and 24.3%, respectively. In contrast to absence of the CYP2D6*4 (G1846A) and *14 (G1758A) alleles in Iranian populations of different ethnicities, the prediction of the CYP2D6*10 allele is required in drug research and routine treatment, where the information would be helpful for clinicians to optimize therapy or identify persons at risk of adverse drug reactions before clinical trials. Approximately 39.3% of subjects (24.3% homozygous T/T CYP2D6*10 as poor metabolizers and 15% heterozygous C/T CYP2D6*10 as intermediate metabolizers) had this allele; therefore, the harmful effects of drugs are relatively common among Iranians.
Keywords: pharmacogenetics, polymorphism, cytochrome P450 genes, CYP, Iranian population, antipsychotics, antidepressants
Introduction
Pharmacogenetics is the investigation of different responses of people, based on changes in their genetic structures, towards treatment and drugs. Liver and small intestine microsomal enzymes metabolize the majority of drugs, in which the activity of drugs are determined based on their interaction with enzymatic activity of the cytochrome P450 (CYP) system.1
More than 50 human CYP isozymes have been identified to date,2 and more than 20 are functionally encoded by polymorphic CYP2A6, CYP2C9, CYP2C19, and CYP2D6 genes.2,3 CYP2D6 is of great importance for the metabolism of clinically used drugs, metabolizing approximately 20%–25% of these drugs, including antipsychotics, antidepressants, antiarrhythmic and beta-adrenoceptor antagonists (beta blockers).4
The CYP2D6 is a highly polymorphic gene locus with more than 75 allelic variants, and subjects can be classified into poor metabolizers (PM), extensive metabolizers (EM), or ultra-rapid metabolizers of a given CYP2D6 substrate.5,6 The CYP2D6*9, CYP2D6*10, CYP2D6*17, and CYP2D6*41 are the most important intermediate metabolizer (IM) alleles,7 in which CYP2D6*10 (49.5%),8 CYP2D6*1 (40.1%–43.5%),9,10 CYP2D6*2 (11.9%–26.2%),11 and CYP2D6*4 (0.2%–2.8%)8,10 are the most common CYP2D6 alleles in Asians. Approximately 13%–50% of Asian populations may be homozygous for the CYP2D6*10 allele.12
A cytosine-to-thymine substitution (C>T) at nucleotide 100 in the CYP2D6*10 allele, resulting in a modification of proline-to-serine at codon 34, is common among Asian populations13 and is associated with reduced metabolism of various CYP2D6 substrates.12 There is a highly conserved proline-rich area among microsomal P450s. The proline at codon 34 probably acts as a center between the lipophilic membrane anchor and the active site (heme moiety) of the enzyme. The change of an amino acid in this region may well decrease the activity of the enzyme.14,15 The CYP2D6*4 (G1846A) allele results in a splicing defect with nonfunctioning enzyme activity due to replacement of guanine (G) by adenine (A) at position 1,846. This single base substitution in the intron 3/exon 4 boundary causes a shift of the consensus acceptor splice site, yielding a spliced messenger RNA with additional base, thereby resulting in a premature termination codon.16 Some studies indicated that the CYP2D6*14 allele may play an important role in contributing to the PM phenotype, in which the CYP2D6*14 allele frequency in Chinese and Japanese populations is approximately 2%.17,18 In this study, the prevalence of the CYP2D6*10, *4, and *14 alleles was investigated in Iranian populations of different ethnicities and in comparison with other countries.
Materials and methods
Sample collection and ethical statement
Three hundred blood samples were obtained from unrelated and healthy donors from Iranian populations of different ethnicities through the Special Medical Center, Tehran, Iran, including those from 118 Fars, 79 Turk, 42 Mazani (Mazandarani), 37 Lure, and 24 Kurd. Blood samples (2 mL) were taken and collected into tubes with ethylenediaminetetraacetic acid. All the individuals who participated in this study belonged to various ethnicities (Fars, Turk, Mazani, Lure, and Kurd) from different provinces of Iran and were referred for genetic counseling at the Special Medical Center of Tehran between 2006 and 2008. Written informed consent, including consent to participate in the study for genetic analysis and consent to publish, was obtained from the individuals, and the Medical Ethics Committee of the Special Medical Center specifically approved this study. The exclusion criterion for the control healthy group was any history of cancer, metabolic diseases, and nuclear and mitochondrial DNA-related diseases that may affect the DNA.
DNA extraction and primer sequences
Genomic DNA was extracted using a MBST kit (salting-out method) from 2 mL of whole blood. Primer sequences for the CYP2D6*10 alleles were ordered as described in a previous study.6 The oligonucleotide forward (F) and reverse (R) primers that were used to amplify the CYP2D6*10, *4, and *14 alleles are as follows: F: 5′–TCAACACAGCAGGTTCA–3′, R: 5′–CTGTGGTTTCACCCACC–3′ [6] F: 5′–TGCCGCCTTC GCCAACCACT–3′, R: 5′–TCGCCCTGCAGAGACTCCTC– 3′19 and F: 5′–GTGGATGGTGGGGCTAATGCCTT–3′, R: 5′–CAGAGACTCCTCGGTCTCTCGCT–3′,20 respectively.
The CYP2D6 metabolism phenotypes
The classification of CYP2D6 phenotypes is based on the assumption of dominance, in which the predicted phenotype is defined as EM, with one or two active alleles; IM, with two reduced activity alleles or one reduced activity and one inactive allele; and PM, with two inactive alleles.16,21
Restriction fragment length polymorphism
Genotyping of the CYP2D6*10 alleles was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The PCR amplification was carried out with 20–60 ng of genomic DNA, 0.3 U of Taq DNA polymerase, 20 mM of each primers, 10× PCR buffer, 2.5 mM magnesium chloride, and 10 mM deoxynucleotide. The reaction mixture was initially denatured at 95°C for 3 minutes, followed by 35 cycles of 95°C for 1 minute, 52.8°C for 1 minute, 72°C for 2 minutes, and a final extension at 72°C for 7 minutes. The 433 bp amplified fragment was analyzed on 1.5% agarose gel stained with ethidium bromide before restriction digestion. One unit of HphI restriction enzyme was added to each PCR product (5 μL) and incubated at 37°C for 16 hours. Digested products were analyzed on polyacrylamide gel. The PCR product with a length of 433 bp was subsequently digested with the restriction endonuclease HphI. Only the amplicon of the allele carrying the 100 C>T single-nucleotide polymorphism was cut by HphI between positions 91 and 92 of the CYP2D6 gene, which is between position 258 and 259 of the PCR product (Figure 1). As HphI also cuts at position 362 of the product, irrespective of the amplified allele, the digestion leads to a 362 bp fragment for the wild-type allele and two fragments of 262 bp and 100 bp for the allele carrying the 100 C>T substitution. Additionally, the BSTN1 and MSP1 restriction enzymes were separately added to PCR products in order to cut the amplicon of the CYP2D6*419 and *1420 alleles carrying the G1846A and G1758A single-nucleotide polymorphism, respectively.
Figure 1.
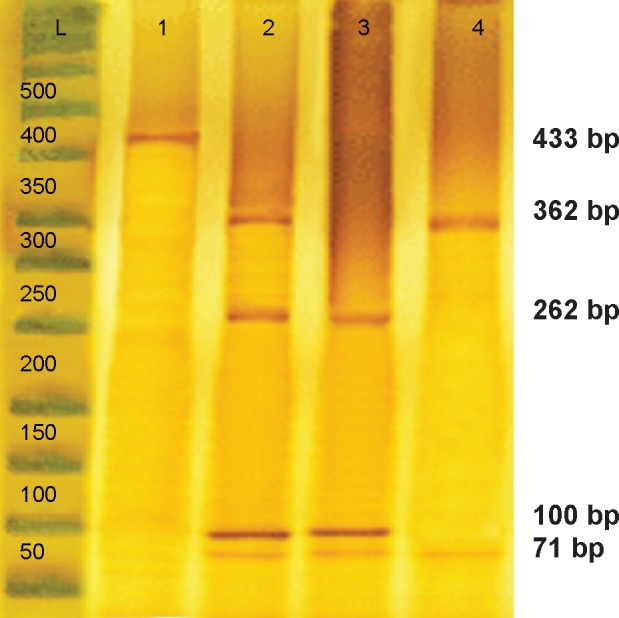
PCR-RFLP analysis of the CYP2D6*10 allele using the HphI enzyme.
Notes: Lane 1 indicates the PCR product; This enzyme generated four fragments of 362 bp, 262 bp, 100 bp, and 71 bp (lane 2) in the heterozygous C/T genotype; three fragments of 262 bp, 100 bp, and 71 bp (lane 3) in the homozygous T/T genotype; and two fragments of 362 bp and 71 bp (lane 4) in length in the wild-type C/C genotype.
Abbreviation: PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Single-strand conformation polymorphism
The double-stranded, PCR-amplified DNA fragment was denatured before loading on agarose gel. For this purpose, 10 μL of PCR mixture was mixed with 7 μL of freshly prepared denaturation buffer (990 μL of 100% formamide, 10 μL of 1M sodium hydroxide, and a few granules of bro-mophenol blue) in a 500 μL PCR eppendorf tube. Samples were placed at least 30 minutes in 45°C, and after 23 minutes, 12 μL of the single-strand conformation polymorphism (SSCP) color was added to the sample. After 30 minutes, the samples were analyzed on a 6% acrylamide gel for 16 hours at 110 V. To identify samples, the SSCP gel was stained, and the samples with the C/C, T/T, and C/T genotypes were observed as two, three, and four bands, respectively (Figure 2).
Figure 2.
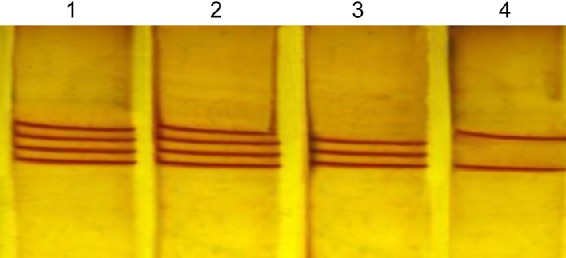
PCR-SSCP analysis of the CYP2D6*10.
Notes: The two, three, and four bands in each lane represent the homozygous wild-type (C/C), homozygous (T/T), and heterozygous (C/T) alleles of the CYP2D6*10, respectively; Lanes 1 and 2 represent the heterozygous C/T genotype (four fragments), lane 3 represents the homozygous T/T genotype (three fragments), while lane 4 represents the wild-type C/C genotype (two fragments).
Abbreviation: PCR-SSCP, polymerase chain reaction-single-strand conformation polymorphism.
Sequencing analysis
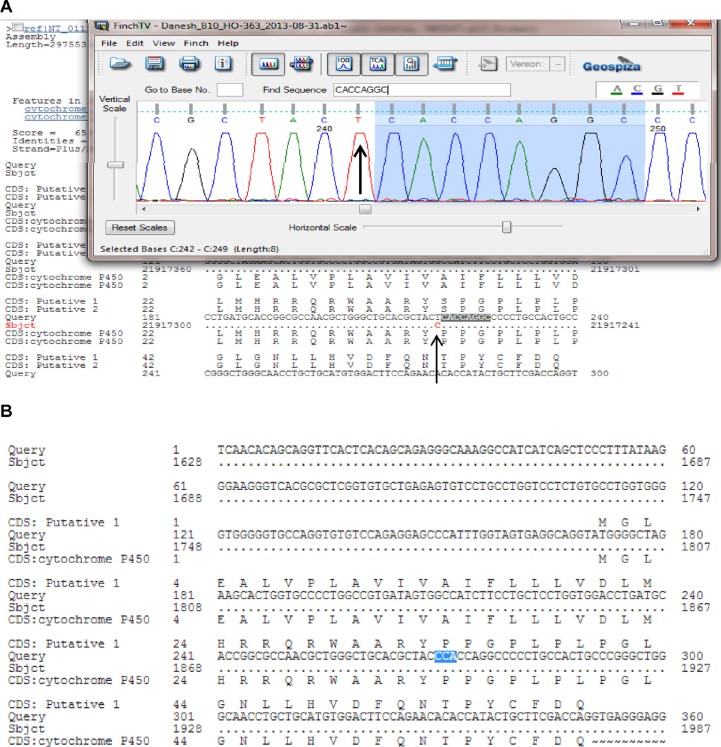
The PCR products were examined for specificity using 1.5% agarose gel electrophoresis. Double-stranded DNA automated sequencing was performed using an Applied Biosystems ABI 3100 sequencing machine (Thermo Fisher Scientific, Waltham, MA, USA). All fragments were sequenced in both the F and R directions for confirmation of any nucleotide variation. Sequence variants were analyzed using a FinchTV program (PerkinElmer Inc., Waltham, MA, USA) (Figure 3).
Figure 3.
PCR-sequencing of the CYP2D6*10 allele (C100T).
Notes: The chromatogram shows the mutant allele (A) compared to the wild-type allele as highlighted in blue color (B); The arrows mark the polymorphism.
Abbreviation: PCR, polymerase chain reaction.
Statistical analysis
The statistical analysis was conducted using SPSS 17.0 (for Windows) to perform the chi-square and 95% confidence interval (CI) tests to calculate the CYP2D6*10 frequencies in different ethnicities and different geographical regions of Iran. A P-value of <0.01 was considered statistically significant.
Results
Allele and genotype frequency distribution of the CYP2D6*10, *4 (G1846A), and *14 (G1758A) variants was analyzed in blood samples of 300 unrelated healthy individuals in an Iranian population of different ethnicities. The allele frequencies of the homozygous (T/T), heterozygous (C/T), and wild-type (C/C) variants of CYP2D6*10 were significantly different (χ2=90.02, P<0.01) in all Iranian ethnic groups. Genotype combinations were used to categorize CYP2D6 metabolism phenotypes as PM, IM, and EM (Table 1). Additionally, the frequency of the homozygous (T/T) variant of CYP2D6*10 was significantly high in the Lure (χ2=8.162, P<0.017) and low in the Kurd (χ2=12.25, P<0.002) ethnicities. However, the frequency of the homozygous (T/T) variant of CYP2D6*10 in ethnic Fars was similar to that of ethnic Mazani (Table 1). The frequency of the heterozygous (C/T) variant of CYP2D6*10 was significantly high in Mazani (χ2=10.857, P<0.004) and low in Kurds (χ2=12.25, P<0.002). Moreover, the frequency of the wild-type (C/C) variant of CYP2D6*10 was highest in Kurds, yet lowest in Lures (Table 1). The results indicate that the frequency of the homozygous (T/T) variant of CYP2D6*10 in central Iran was significantly the highest (χ2=15.354, P<0.001), while the south of Iran had the lowest frequency (χ2=23.164, P<0.001). The frequency of the heterozygous (C/T) variant of the CYP2D6*10 allele in the east of Iran had the highest frequency, while the west had the lowest frequency (Figure 4; Table 1). The frequency of the C/T variant of the CYP2D6*10 allele was significantly a bit higher (P<0.001) in females compared to males, while the frequencies of the T/T variant in females was similar to males, which are 24.4% and 24.3%, respectively (Table 1). However, the CYP2D6*4 (G1846A) and *14 (G1758A) allelic frequencies were not detected in different ethnicities, demonstrating the absence of a significant contribution of these alleles in Iranian populations. Additionally, the CYP2D6*10 allele was directly detected using the PCR-RFLP and -SSCP. The PCR-RFLP analysis of the CYP2D6*10 allele using the HphI enzyme indicates the heterozygous C/T, homozygous T/T, and wild-type C/C genotypes (Figure 1). The PCR-SSCP analysis demonstrates the heterozygous C/T, homozygous T/T, and wild-type C/C genotypes with four, three, and two fragments, respectively (Figure 2).
Table 1.
Statistical analysis of the CYP2D6*10 genotypes in different ethnicities and different geographical regions of Iran
| Genotype | C/C | C/T | T/T | χ2 | P-value |
|---|---|---|---|---|---|
|
| |||||
| Metabolic phenotype | EM | IM | PM | ||
| Frequency (overall) | 60.6% (55.0–66.1) | 15% (11.0–19.0) | 24.4% (19.5–29.0) | 90.02 | <0.01* |
| Ethnicities | |||||
| Fars | 61% (52.2–69.8) | 15.3% (8.8–21.7) | 23.7% (1.61–31.4) | 41.96 | <0.001* |
| Kurd | 66.7% (47.8–85.5) | 12.5% (0.7–25.7) | 20.8% (4.53–37) | 12.25 | 0.002* |
| Lure | 54.1% (38.4–70.1) | 16.2% (4.3–28.1) | 29.7% (15.0–44.4) | 8.162 | 0.017* |
| Mazani | 57.1% (41.1–72.1) | 19% (7.1–30.1) | 23.8% (11–36.7) | 10.857 | 0.004* |
| Turk | 59.5% (48.3–70.3) | 16.5% (8.3–24.7) | 24.1% (14.7–33.5) | 25.013 | <0.001* |
| Geographic | |||||
| Central | 55.4% (43.4–67.5) | 16.9% (7.8–26.0) | 27.7% (16.8–38.6) | 15.354 | <0.001* |
| East | 54.7% (41.9–67.9) | 20.8% (9.9–31.8) | 24.5% (12.9–36.1) | 11.019 | <0.001* |
| North | 56.7% (44.8–68.5) | 17.9% (8.7–27.1) | 25.4% (15–35.8) | 17.045 | <0.001* |
| South | 63.6% (50.8–76.3) | 14.5% (5.2–23.8) | 21.8% (10.9–32.7) | 23.164 | <0.001* |
| West | 63.3% (51.1–75.5) | 11.7% (3.5–19.8) | 25% (14.0–35.0) | 25.90 | <0.001* |
| Sex | |||||
| Female | 60.5% (51.7–69.3) | 15.1% (8.7–21.6) | 24.4% (16.7–32.1) | 41.059 | <0.001* |
| Male | 60.8% (53.7–67.9) | 14.9% (9.7–20.1) | 24.3% (18.1–30.6) | 63.725 | <0.001* |
Notes:
Statistically significant (P<0.01). Values shown in brackets represent 95% CI.
Abbreviations: CI, confidence interval; EM, extensive metabolizer; IM, intermediate metabolizer; PM, poor metabolizer.
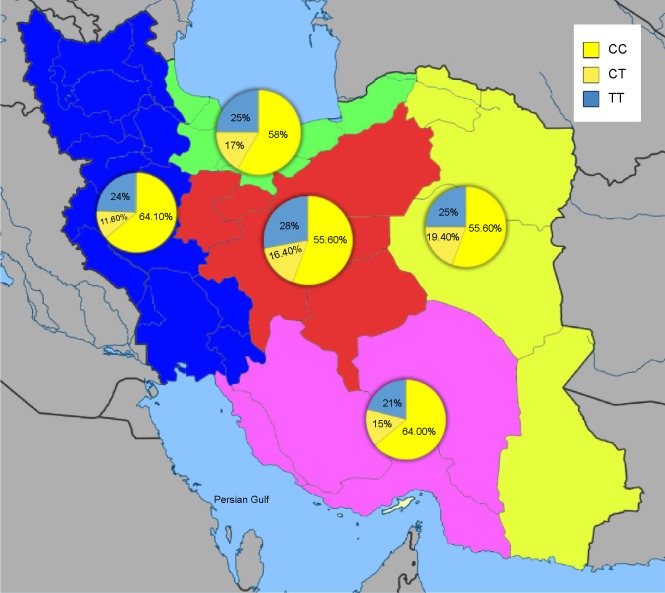
Figure 4.
Geographical distribution of the CYP2D6*10 genotypes among 300 normal Iranian individuals of different ethnicities.
Notes: The T/T, C/T, and C/C genotype frequencies of CYP2D6*10 were significantly different (χ2=90.02, P<0.01): 24.3%, 15%, and 60.7%, respectively; The results indicate that the frequency of the homozygous T/T variant of CYP2D6*10 in the central Iran was the highest (χ2=15.354, P<0.001), while the south of Iran had the lowest frequency (χ2=23.164, P<0.001); The frequency of the heterozygous C/T variant of CYP2D6*10 allele is highest in the east and lowest in the west; The green, pink, yellow, blue, and red colors indicate the north, south, east, west, and center of Iran, respectively.
Discussion
CYP2D6 is a clinically important enzyme that metabolizes numerous therapeutic drugs.16 Among the alleles of CYP2D6, CYP2D6*10 (C100T) is the most common allele in the Asian population, with a frequency of between 30% and 69.2%, where 13%–49% of the aforementioned population are homozygous for this allele.5,6 Accordingly, the majority of the Asian population is at risk of either harmful effects or adverse reactions towards medication.6 Thus, considering the effects of the CYP2D6*10 allele on drug metabolism in Iran, along with allele genotyping, including that of CYP2D6*wt/*wt, CYP2D6*wt/*10, and CYP2D6*10/*10, may prevent the harmful effects of drugs and drug interactions, thereby reducing mortality risk in Iranian populations.
The CYP2D6*10 “C100T” mutation in patients leads to reduced enzyme activity and decrease of appropriate response to drug treatments, such as tamoxifen, codeine, antidepressants, and antipsychotics.4,22–24 Additionally, there has been some research linking the CYP2D6 alleles and drug metabolism in the Iranian population,19,25 but study of the CYP2D6*10 allele regarding its significant role in drug metabolism in Asian populations26 have been neglected. Therefore, genotyping of the CYP2D6*10 allele, including CYP2D6*wt/*wt, CYP2D6*wt/*10, and CYP2D6*10/*10, in different ethnicities including Lure, Turk, Fars, Kurd, and Mazani is considered as an important determinant for the prevention of the harmful effects of drugs and drug interactions, resulting in better survival and reduced mortality risk.
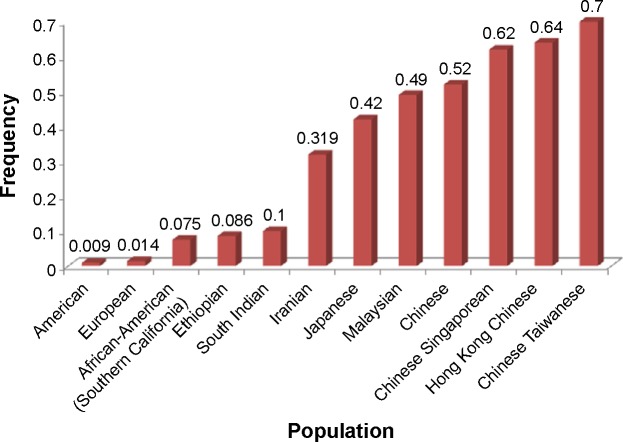
The CYP2D6 allele frequency in our Iranian population with different ethnicities including Lure, Turk, Mazani, Fars, and Kurd were compared with the frequencies seen in other ethnic populations. The study by Kouhi et al19 showed that the prevalence of CYP2D6*10 was significant only in the eastern part (Tabriz) of Azerbaijan (Turk). However, in our study, the samples were selected from both the western and eastern parts of Azerbaijan (Turk), indicating no significant difference in Turk ethnicity. The CYP2D6*10 allele was found to have a frequency of 0.319 in the Iranian population (Figure 5), which is higher than that of South Indian27 and Ethiopian populations,28 but not as high as what was reported in other populations such as the Japanese,29 Malaysian,8 and Chinese.30
Figure 5.
Frequency of the CYP2D6*10 allele in different populations.
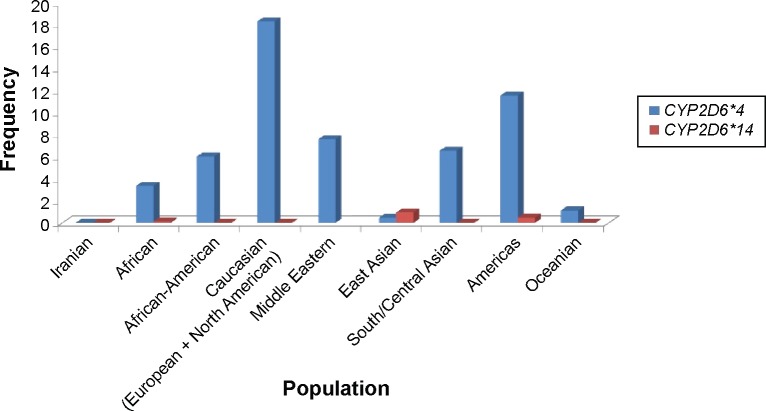
There are four variations including C100T, G1758A, C2850T, and G4180C in the CYP2D6*14 allele, in which identification of the G1758A polymorphism is exclusively used for the CYP2D6*14 genotype determination.17,18,20 In our study, the CYP2D6*4 (G1846A) and *14 (G1758A) allelic frequencies were not identified in different ethnicities including Fars, Turk, Lure, Kurd, and Mazani, demonstrating the absence of a significant contribution of these alleles in Iranian populations compare to other populations (Figure 6).
Figure 6.
Frequency of the CYP2D6*4 and CYP2D6*14 alleles in different populations.
Notes: The CYP2D6*14 allele frequency was not determined in the Middle Eastern population. Americas is a study from a white North American population. Permission to reprint data from Clinical Pharmacogenetics Implementa tion Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Crews KR, Gaedigk A, Dunnenberger HM, et al; Clinical Pharmacogenetics Implementation Consortium. Clin Pharmacol Ther. 91(2). Copyright © (2012).31
In conclusion, this study demonstrates the absence of the CYP2D6*4 (G1846A) and *14 (G1758A) alleles, whereas approximately 24% of the Iranian population has the homozygous T/T CYP2D6*10, suggesting susceptibility to adverse or poor drug responses, which may lead to increase in mortality risk. It is clinically important to identify those individuals who probably have altered pharmacokinetics for CYP2D6 substrates in order to avoid adverse drug reactions. CYP2D6 polymorphism analysis in different ethnicities indicates a very useful forensic tool toward personalized medicine. These data also allow comparison of pharmacogenetic variation between populations with different ethnicities. Therefore, the determination of the presence of this allele in individuals via genetic tests is a basic step to prevent the harmful effects of drugs.
Acknowledgments
We would like to thank all the participants for their blood donation to the Medical Genetics Department at the Special Medical Center, Tehran, Iran. This work was supported by Dr M Houshmand, to whom we give our grateful appreciation. The authors would like to express their utmost gratitude and appreciation to the University of Malaya Research Grant (RG084-13BIO), IPPP grant (PG082-2013B), and BKP grant (BK020-2012) for providing partial financial support to conduct this study.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Meisel C, Roots I, Cascorbi I, Brinkmann U, Brockmöller J. How to manage individualized drug therapy: application of pharmacogenetic knowledge of drug metabolism and transport. Clin Chem Lab Med. 2000;38(9):869–876. doi: 10.1515/CCLM.2000.126. [DOI] [PubMed] [Google Scholar]
- 2.De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46(4):594–598. [PubMed] [Google Scholar]
- 3.Gurwitz D, Motulsky AG. ‘Drug reactions, enzymes, and biochemical genetics’: 50 years later. Pharmacogenomics. 2007;8(11):1479–1484. doi: 10.2217/14622416.8.11.1479. [DOI] [PubMed] [Google Scholar]
- 4.Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006;58(3):521–590. doi: 10.1124/pr.58.3.6. [DOI] [PubMed] [Google Scholar]
- 5.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116(3):496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Kwong EH. A Novel Genotyping Algorithm for the CYP2D6* 10 Allele in Asians Using Real-Time, Rapid-Cycle PCR and Multiplex PCR [master’s thesis] Vancouver: University of British Columbia; 2004. Available from: https://circle.ubc.ca/bitstream/2429/15602/1/ubc_2004-0519.pdf. [Google Scholar]
- 7.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48(12):761–804. doi: 10.2165/11318070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 8.Teh LK, Ismail R, Yusoff R, Hussein A, Isa MN, Rahman AR. Heterogeneity of the CYP2D6 gene among Malays in Malaysia. J Clin Pharm Ther. 2001;26(3):205–211. doi: 10.1046/j.1365-2710.2001.00347.x. [DOI] [PubMed] [Google Scholar]
- 9.Ishiguro A, Kubota T, Sasaki H, Yamada Y, Iga T. Common mutant alleles of CYP2D6 causing the defect of CYP2D6 enzyme activity in a Japanese population. Br J Clin Pharmacol. 2003;55(4):414–415. doi: 10.1046/j.1365-2125.2003.01782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishida Y, Fukuda T, Yamamoto I, Azuma J. CYP2D6 genotypes in a Japanese population: low frequencies of CYP2D6 gene duplication but high frequency of CYP2D6*10. Pharmacogenetics. 2000;10(6):567–570. doi: 10.1097/00008571-200008000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Dahl ML, Yue QY, Roh HK, et al. Genetic analysis of the CYP2D locus in relation to debrisoquine hydroxylation capacity in Korean, Japanese and Chinese subjects. Pharmacogenetics. 1995;5(3):159–164. doi: 10.1097/00008571-199506000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjöqvist F, Ingelman-Sundberg M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994;46(3):452–459. [PubMed] [Google Scholar]
- 13.Hicks JK, J Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab. 2014;15(2):218–232. doi: 10.2174/1389200215666140202215316. [DOI] [PubMed] [Google Scholar]
- 14.Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):89–104. doi: 10.1007/s00210-003-0819-z. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev. 2009;41(4):573–643. doi: 10.1080/03602530903118729. [DOI] [PubMed] [Google Scholar]
- 16.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 17.Kubota T, Yamaura Y, Ohkawa N, Hara H, Chiba K. Frequencies of CYP2D6 mutant alleles in a normal Japanese population and metabolic activity of dextromethorphan O-demethylation in different CYP2D6 genotypes. Br J Clin Pharmacol. 2000;50(1):31–34. doi: 10.1046/j.1365-2125.2000.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji L, Pan S, Marti-Jaun J, Hänseler E, Rentsch K, Hersberger M. Single-step assays to analyze CYP2D6 gene polymorphisms in Asians: allele frequencies and a novel *14B allele in mainland Chinese. Clin Chem. 2002;48(7):983–988. [PubMed] [Google Scholar]
- 19.Kouhi H, Hamzeiy H, Barar J, Asadi M, Omidi Y. Frequency of five important CYP2D6 alleles within an Iranian population (Eastern Azerbaijan) Genet Test Mol Biomarkers. 2009;13(5):665–670. doi: 10.1089/gtmb.2009.0009. [DOI] [PubMed] [Google Scholar]
- 20.Wang SL, Lai MD, Huang JD. G169R mutation diminishes the metabolic activity of CYP2D6 in Chinese. Drug Metab Dispos. 1999;27(3):385–388. [PubMed] [Google Scholar]
- 21.de Leon J, Arranz MJ, Ruaño G. Pharmacogenetic testing in psychiatry: a review of features and clinical realities. Clin Lab Med. 2008;28(4):599–617. doi: 10.1016/j.cll.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Ramón y Cajal T, Altés A, Paré L, et al. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat. 2010;119(1):33–38. doi: 10.1007/s10549-009-0328-y. [DOI] [PubMed] [Google Scholar]
- 23.Punglia RS, Burstein HJ, Winer EP, Weeks JC. Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst. 2008;100(9):642–648. doi: 10.1093/jnci/djn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 phar-macogenetics and cancer. Oncogene. 2006;25(11):1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 25.Afshar M, Rouini M, Ala S. Dextromethorphan metabolic phenotyping in an Iranian population. Eur J Clin Pharmacol. 2005;60(12):849–854. doi: 10.1007/s00228-004-0859-4. [DOI] [PubMed] [Google Scholar]
- 26.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3(2):229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 27.Naveen AT, Adithan C, Soya SS, Gerard N, Krishnamoorthy R. CYP2D6 genetic polymorphism in South Indian populations. Biol Pharm Bull. 2006;29(8):1655–1658. doi: 10.1248/bpb.29.1655. [DOI] [PubMed] [Google Scholar]
- 28.Aklillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in an ethiopian population carrying duplicated and mul-tiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278(1):441–446. [PubMed] [Google Scholar]
- 29.Tateishi T, Chida M, Ariyoshi N, Mizorogi Y, Kamataki T, Kobayashi S. Analysis of the CYP2D6 gene in relation to dextro methorphan O-demethylation capacity in a Japanese population. Clin Pharmacol Ther. 1999;65(5):570–575. doi: 10.1016/S0009-9236(99)70077-9. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Zhang H, He F, Fang X. Effect of the CYP2D6*10 C188T polymorphism on postoperative tramadol analgesia in a Chinese population. Eur J Clin Pharmacol. 2006;62(11):927–931. doi: 10.1007/s00228-006-0191-2. [DOI] [PubMed] [Google Scholar]
- 31.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91(2):321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]