Abstract
The mouse limb deformity (ld) mutations cause limb malformations by disrupting epithelial–mesenchymal signaling between the polarizing region and the apical ectodermal ridge. Formin was proposed as the relevant gene because three of the five ld alleles disrupt its C-terminal domain. In contrast, our studies establish that the two other ld alleles directly disrupt the neighboring Gremlin gene, corroborating the requirement of this BMP antagonist for limb morphogenesis. Further doubts concerning an involvement of Formin in the ld limb phenotype are cast, as a targeted mutation removing the C-terminal Formin domain by frame shift does not affect embryogenesis. In contrast, the deletion of the corresponding genomic region reproduces the ld limb phenotype and is allelic to mutations in Gremlin. We resolve these conflicting results by identifying a cis-regulatory region within the deletion that is required for Gremlin activation in the limb bud mesenchyme. This distant cis-regulatory region within Formin is also altered by three of the ld mutations. Therefore, the ld limb bud patterning defects are not caused by disruption of Formin, but by alteration of a global control region (GCR) required for Gremlin transcription. Our studies reveal the large genomic landscape harboring this GCR, which is required for tissue-specific coexpression of two structurally and functionally unrelated genes.
Keywords: cis regulation, Formin, global control region, Gremlin, limb development, regulatory landscape
The mouse is the genetic model of choice to study mammalian development and disease. In addition to alteration of specific genes by gene targeting and transgenesis, mutant mouse strains identified by phenotypic screens are commonly used to analyze developmental and disease processes (for reviews, see Justice 2000; Perkins 2002). A significant fraction of spontaneous mutations in mice (and humans) cause congenital limb malformations and have proven crucial to unravel the molecular mechanisms regulating vertebrate limb bud morphogenesis (for review, see Gurrieri et al. 2002). In particular, several alleles of the recessive mouse limb deformity (ld) mutation disrupt patterning of the distal limb skeleton. Over the years, a total of five ld alleles have been identified by phenotypic and genetic complementation analysis. All ld homozygous newborn mice display limb patterning defects characterized by synostosis of the zeugopod in combination with oligo- and syndactyly of metacarpal bones and digits (for review, see Zeller et al. 1999). In addition, ld homozygous newborn mice display varying degrees of uni- and bilateral renal aplasias depending on allele “strength” (Maas et al. 1994). Molecular analysis showed that the two ld alleles (ldTgHd, ldTgBri) that arose by chance insertional mutagenesis disrupt the C-terminal region of the Formin gene (for details, see Wang et al. 1997). The ldIn2 allele arose by an ∼40-Mb inversion with breakpoints in the C-terminal region of Formin and the Agouti locus (Maas et al. 1990; Woychik et al. 1990). To date the molecular lesions in the two first identified ld alleles, ldOR and ldJ (for review, see Zeller et al. 1999), remained obscure as there are no alterations in the Formin open reading frame (ORF; Wynshaw-Boris et al. 1997). Formin (or Formin-1) is the founding member of a multigene family that mediates cytoskeletal rearrangements in response to signals that induce, for example, cell polarization (for review, see Evangelista et al. 2003). The Formin proteins are encoded by at least 24 exons spread over ∼400 kb, and alternative splicing gives rise to several protein isoforms of ∼180 kDa. All Formin isoforms share a proline-rich FH1 domain (interacting with SH3 domains and Profilin) and a highly conserved C-terminal FH2 domain, which is required to stimulate polymerization of linear actin filaments (for reviews, see Zeller et al. 1999; Kobielak et al. 2004). However, inactivation of specific Formin isoforms by gene targeting in the mouse resulted in partial renal agenesis phenotypes, but failed to reproduce the ld limb phenotype as limb morphogenesis was normal (Wynshaw-Boris et al. 1997; Chao et al. 1998).
In ld homozygous embryos, the epithelial–mesenchymal signaling interactions regulating limb bud development are disrupted (for review, see Panman and Zeller 2003). Limb bud growth and patterning are coordinately controlled by two main signaling centers, the Sonic hedgehog (Shh)-expressing polarizing region, located in the posterior limb bud mesenchyme and the apical ectodermal ridge (AER), which expresses several types of signals including Fibroblast growths factors (Fgfs) and Bone morphogenetic proteins (Bmps). Molecular analysis of ld mutant limb buds revealed that activation of Fgf4 expression in the posterior AER, establishment of the SHH/FGF4 feedback loop, and thereby up-regulation of Shh expression by the polarizing region are disrupted (for review, see Panman and Zeller 2003). A functional screen for mesenchymal signals able to relay SHH to the AER resulted in identification of the BMP antagonist Gremlin as the signal lacking from ld mutant limb bud mesenchyme. Grafts of Gremlin-expressing cells into ld mutant limb buds restore Fgf4 expression and the SHH/FGF4 feedback loop (Zuniga et al. 1999). Analysis of Gremlin-deficient mouse embryos generated by gene targeting has confirmed its essential functions during limb bud development (Khokha et al. 2003; Michos et al. 2004). Induction of Fgf8-expressing AER cells occurs normally, but a morphologically distinct and functional AER fails to form in Gremlin-deficient embryos. Gremlin-mediated BMP antagonism is required in the limb bud mesenchyme to enable expression of various types of AER signals such as Fgfs and Bmps and for survival of core mesenchymal cells (Michos et al. 2004). The general disruption of AER function in Gremlin-deficient embryos in turn blocks propagation of Shh expression by the polarizing region as is also observed in ld mutant limb buds. In addition, the induction of metanephric kidney organogenesis and complete differentiation of lung airway epithelia fail to occur, which causes neonatal lethality. The studies by Michos et al. (2004) reveal a more general role of Gremlin-mediated BMP antagonism in epithelial–mesenchymal signaling during vertebrate organogenesis. Interestingly, the Gremlin transcription unit maps only ∼40 kb downstream from the Formin gene on mouse chromosome 2 and is transcribed in opposite orientation (University of California at Santa Cruz Genome Browser, http://genome.ucsc.edu). Finally, Khokha et al. (2003) established allelism between one of the ld alleles (ldJ) and a Gremlin null allele generated by gene targeting, but did not analyze the genetic and molecular basis for these rather unexpected findings.
In the present study we establish that the ldJ mutation is a point mutation affecting splicing of Gremlin transcripts that results in truncation of the 5′ part of the Gremlin ORF. Furthermore, the complete Gremlin ORF encoded by exon 2 is deleted by the ldOR mutation. Therefore, these two ld alleles are spontaneous Gremlin loss-of-function alleles, which also establishes that the ld complementation group encompasses both the Gremlin and Formin loci. Using gene targeting we show that disruption of the Formin FH2 domain by deleting coding exon 10 (FmnΔ10 mutation) does not reproduce the ld phenotype. In contrast, deletion of the genomic region encompassing exons 10–24 (FmnΔ10.24 mutation) results in the characteristic ld limb phenotype. Interestingly, the FmnΔ10.24 deletion, but not the FmnΔ10 mutation, is allelic to Gremlin loss-of-function mutations. Further analysis establishes that the deleted region encodes regulatory elements required to activate both Formin and Gremlin expression in the posterior limb bud mesenchyme and mediate responsiveness to SHH, but not to FGF signaling. BAC transgenic analysis positively identifies this cis-regulatory region and shows that it is required to activate Gremlin transcription in the posterior–distal limb bud mesenchyme. The features of this large cis-regulatory landscape are reminiscent of a recently discovered global control region, a novel type of chromosomal regulatory element that controls expression of 5′Hoxd genes in the distal limb bud mesenchyme (Spitz et al. 2003). Taken together, our studies establish that the ld limb phenotype is a direct consequence of losing Gremlin expression in the limb bud mesenchyme and not due to disrupting Formin functions. We also discuss how such large regulatory landscapes that control tissue-specific coexpression of functionally unrelated genes may have arisen.
Results
The ldJ and ldOR alleles are loss-of-function mutations directly disrupting the Gremlin gene products
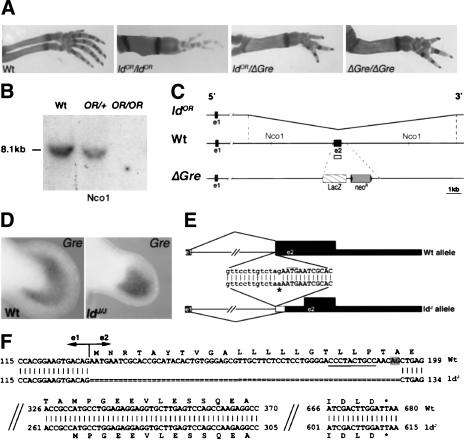
In contrast to the three independent alleles of the mouse ld mutation that truncate Formin (see the introduction), no molecular alterations of the Formin ORF were identified for the ldOR and ldJ alleles (Wynshaw-Boris et al. 1997). However, both these ld mutations are allelic to null mutations in the neighboring Gremlin gene (generated by gene targeting; Michos et al. 2004) as compound heterozygous mice display the characteristic ld limb phenotype (Fig. 1A; Khokha et al. 2003). Furthermore, newborn ldOR homozygous mice lack kidneys and ureters (Supplementary Fig. 1) similar to Gremlin-deficient and ldJ homozygous mice (Maas et al. 1994). This renal agenesis in combination with lung defects (Supplementary Fig. 1) causes death shortly after birth identical to Gremlin null mutant mice (Michos et al. 2004). Molecular analysis of the ldOR allele indeed reveals a 12.7-kb genomic deletion removing the complete Gremlin ORF (Fig. 1B,C), which establishes the ldOR mutation as a spontaneous Gremlin null allele. In sharp contrast, Gremlin transcripts remain expressed in embryos homozygous for the ldJ allele (Fig. 1D), despite the fact that activation of Fgf4 in the posterior AER is disrupted and the Shh expressing polarizing region is not maintained (Haramis et al. 1995). Sequence analysis of the Gremlin locus in the ldJ allele reveals a specific G-to-A base change at the first intron–exon 2 boundary (asterisk in Fig. 1E). This point mutation eliminates the intronic AG motif at the 3′ splice site, which is essential for correct pre-mRNA splicing (Faustino and Cooper 2003). Indeed, a single aberrantly spliced transcript with a 65-base deletion removing the 5′ part of exon 2 was identified in ldJ homozygous embryos (Fig. 1F; data not shown). This aberrant splice makes use of a downstream AG dinucleotide within exon 2 that is preceded by a pyrimidinerich stretch required for splicing (Fig. 1F; Faustino and Cooper 2003). This deletion within the Gremlin transcript removes the AUG translational start codon, thereby abolishing translation of full-length secreted Gremlin protein (Fig. 1F).
Figure 1.
The ldOR and ldJ mutations are Gremlin loss-of-function alleles. (A) The ldOR mutation is allelic to a Gremlin null allele (ΔGre) generated by gene targeting (Michos et al. 2004). (B) Southern blot analysis reveals that the Gremlin ORF encoded by exon 2 is deleted in the ldOR mutation. Genomic DNA isolated from embryos was digested by Nco1 and probed with a Gremlin exon 2 probe (open box in scheme, C). (Wt) Wild-type littermate; (OR/+) heterozygous embryo; (OR/OR) homozygous embryo. (C) Schematic representation of the Gremlin locus on chromosome 2 in the ldOR allele, wild-type, and ΔGre mutation. (ldOR) Kinked line indicates the 12.7-kb region deleted in the ldOR mutation. (Wt) Open box indicates the probe used for the Southern blot analysis shown in B. (ΔGre) LacZ and the NeoR replace coding exon 2 in the Gremlin null allele generated by gene targeting. (e1) Exon 1; (e2) exon 2. (D) Gremlin remains expressed in ldJ homozygous embryos. Shown are limb buds of a wild-type (Wt) and ldJ homozygous (ldJ/J) mouse embryo at E11.5. (E) The G-to-A mutation at the intron–exon 2 junction of the Gremlin gene in the ldJ allele (indicated by an asterisk) disrupts splicing. Lowercase indicate intronic, uppercase indicate exonic sequences. Thick black boxes indicate the Gremlin ORF, thin boxes indicate the 5′ and 3′ noncoding regions. The thin white box indicates the deletion of mRNA due to aberrant splicing. Thin black lines indicate intronic sequences and splices. (F) Aberrant pre-mRNA splicing deletes the first 65 bases of the Gremlin ORF. Shown is an alignment of the cDNA sequences of wild-type (Wt) and ldJ alleles with the respective ORFs. The truncated ldJ Gremlin transcript could potentially encode a Gremlin protein of 117 amino acids (instead of 184) lacking the signal peptide (Avsian-Kretchmer and Hsueh 2003). The AG dinucleotide used for splicing in the ldJ allele is shaded gray. The required upstream poly-pyrimidine tract is underlined (Faustino and Cooper 2003).
Not the disruption of the Formin FH2 domain, but deletion of the corresponding genomic region causes the ld limb phenotype
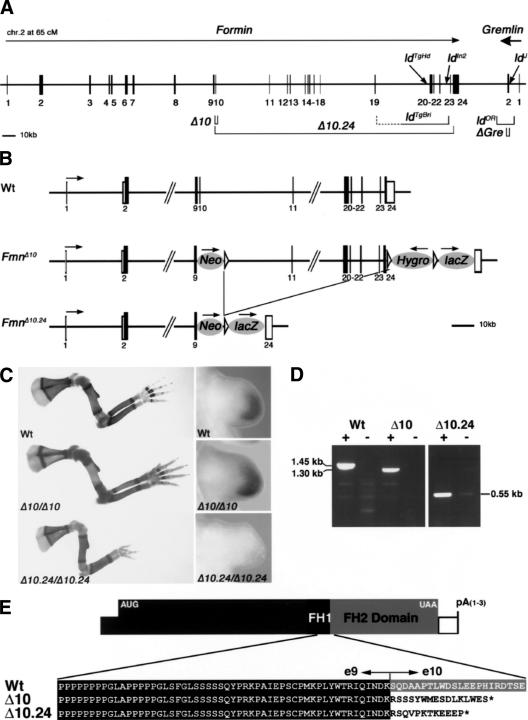
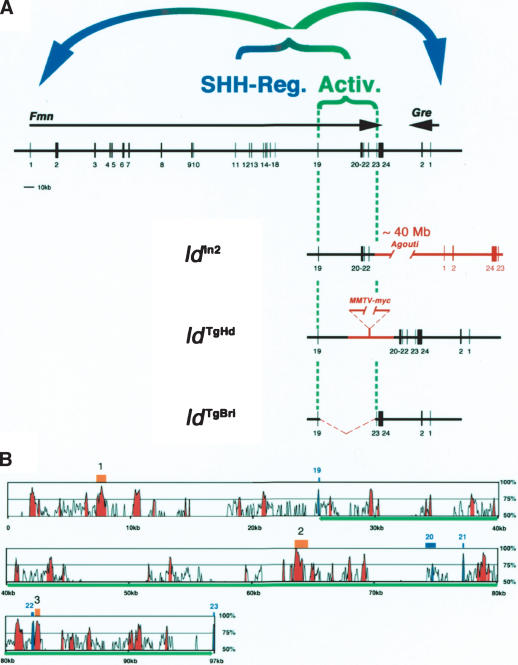
The fact that the ldOR and ldJ alleles are Gremlin loss-of-function mutations (Fig. 1) reiterates the question of whether the disruption of Formin functions is indeed the primary cause of the ld phenotype. In particular, the Gremlin gene is located ∼40 kb downstream from Formin and transcribed in opposite orientation, extending the ld complementation group to ∼450–500 kb in size (Fig. 2A). Previous analysis showed that the other three ld alleles (ldIn2, ldTgBri, and ldTgHd; for details, see Wang et al. 1997) disrupt the genomic region encoding the C-terminal part of Formin (encoded by exons 10–24; Fig. 2A). In an attempt to reproduce the ld phenotype and possibly generate a null allele by reverse genetics, Formin exon 10 was deleted (FmnΔ10 allele; Fig. 2B; for details, see Materials and Methods). Exon 10 was chosen as its deletion results in a frame shift that disrupts translation of the C-terminal protein domain completely (Fig. 2E). Rather unexpectedly, FmnΔ10 homozygous mice are phenotypically wild type (Fig. 2C; data not shown), thereby establishing that deletion of exon 10 is not sufficient to reproduce the ld phenotype. Next, we deleted the complete genomic region spanning exons 10–24 (Fig. 2A; region 10.24), which results in the FmnΔ10.24 allele (Fig. 2B; for details, see Materials and Methods). Mice homozygous for the FmnΔ10.24 allele indeed display the characteristic ld limb phenotype (Fig. 2C). However, FmnΔ10.24 homozygous newborn mice display neither renal agenesis nor lung patterning phenotypes and survive to adulthood (data not shown). Molecular analysis of FmnΔ10.24 homozygous embryos reveals that Gremlin expression is lost specifically from the limb bud mesenchyme, whereas it is normal in FmnΔ10 homozygous embryos (Fig. 2C; data not shown). In agreement with limb bud specific loss of Gremlin expression, activation of Fgfs in the posterior AER and SHH–GRE/AER feedback signaling are disrupted in FmnΔ10.24 homozygous embryos (Supplementary Fig. 2), but not in FmnΔ10 homozygous embryos (data not shown). One possible explanation for the lack of an ld limb phenotype in FmnΔ10 homozygous embryos could be the rescue of Formin function due to aberrant splicing. However, thorough analysis of Formin transcripts extending 3′ to exon 9 by RT–PCR provided no evidence for aberrant splicing (Fig. 2D; data not shown). Furthermore, sequence analysis of the altered Formin transcripts in FmnΔ10 and FmnΔ10.24 homozygous embryos establishes that the Formin ORF is truncated in both alleles at the level of the exon 9/10 boundary (Fig. 2E). These results show that the ld limb phenotype in the FmnΔ10.24 mutation is not caused by disruption of the C-terminal Formin protein domain as previously concluded (for review, see Zeller et al. 1999), but by the deletion of other essential elements located in the genomic region 10.24.
Figure 2.
Not disruption of the Formin FH2 domain, but deletion of the corresponding genomic region causes the ld limb phenotype. (A) Schematic representation of the ld complementation group consisting of Formin and Gremlin loci. The Formin gene is encoded by at least 24 exons (transcriptional direction indicated by arrow; Wang et al. 1997), whereas the Gre gene is transcribed in reverse orientation (bold arrow) and contains only two exons. The intergenic region separating the two genes is ∼38 kb. The Formin FH2 domain is encoded by exons 10–24 and is present in all Formin protein isoforms (Wang et al. 1997). The following genetically engineered mutations are indicated: (Δ10) FmnΔ10 allele; (Δ10.24) FmnΔ10.24 allele; (ΔGre) GreΔORF null allele (Michos et al. 2004). The spontaneous ld alleles are indicated: (ldTgBri) transgene induced deletion of genomic region between exons 19 and 23 (Vogt et al. 1992); (ldTgHd) transgene insertional mutagenesis (Woychik et al. 1985); (ldIn2) 40-Mb inversion involving Formin and Agouti loci (Woychik et al. 1990); (ldOR) deletion of the Gre ORF; (ldJ) point mutation disrupting Gre pre-mRNA splicing. (B) Schematic representation of the genetically engineered FmnΔ10 and FmnΔ10.24 alleles. (Neo) PGK-NeoR gene used to select ES-cell clones (first round of gene targeting); (Hygro) PGK-HygroR gene used to select ES-cell clones (second round of gene targeting); (lacZ) IRES-LacZ gene used to tag Fmn transcripts. Arrows indicate direction of transcription. Formin exons are numbered as in A. (C, left panels) Limb skeletal phenotypes of wild-type and homozygous mice. Genotypes are indicated in the panels. (Right panels) Gremlin expression in limb buds of wild-type and homozygous embryos (E10.75). For nomenclature see the legend for A. (D) RT–PCR of Formin transcripts isolated from wild-type (Wt), FmnΔ10 (Δ10) and FmnΔ10.24 (Δ10.24) homozygous embryos. Wild-type and FmnΔ10 mRNAs extending downstream from exon 9 were detected using primers in exons 9 and 23, FmnΔ10.24 mRNAs extending downstream from exon 9 were detected using primers in exon 9 and the IRES-LacZ tag (see Materials and Methods). (+) Reverse transcriptase included; (-) reverse transcriptase omitted (control). Note that the difference in size between wild-type (Wt) and FmnΔ10 transcripts is 150 bases, as expected. (E) Amino acid sequence deduced from the sequences of the Formin transcripts arising from wild-type, FmnΔ10 and FmnΔ10.24 alleles.
The genomic region 10.24 exerts cis-effects on Gremlin expression in the limb bud mesenchyme
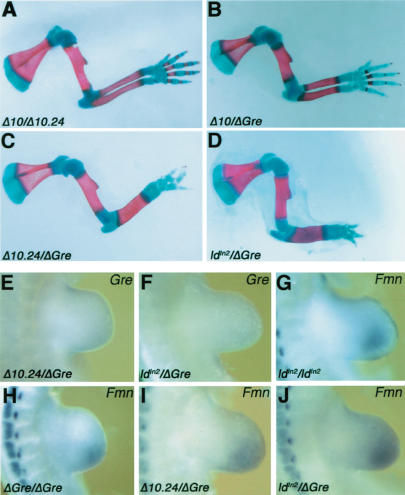
The phenotypic analysis indicated that all ld mutations belong to the same complementation group in spite of their disrupting either the Formin or Gremlin loci (Fig. 2A); therefore, allelism between the FmnΔ10, FmnΔ10.24, and GreΔORF mutations was assessed. Limbs of FmnΔ10/Δ10.24 and FmnΔ10; GreΔORF compound heterozygous mice are normal (Fig. 3A,B), confirming the wild-type phenotypic nature of the FmnΔ10 mutation. In contrast, the FmnΔ10.24 mutation is a hypomorphic allele of the GreΔORF null mutation as FmnΔ10.24/+; GreΔORF/+ mice display a fully penetrant ld limb phenotype (Fig. 3C), but not the phenotypes causing neonatal lethality (data not shown). The ldIn2 allele is of particular interest, as an inversion between Formin and Agouti (Woychik et al. 1990) relocates the Gremlin gene ∼40 Mb away on mouse chromosome 2. Compound ldIn2/+; GreΔORF/+ heterozygous mice also display the ld limb phenotype (Fig. 3D), which indicates that integrity of the genomic region encoding Formin and Gremlin (Fig. 2A) is required in cis for normal limb bud development. Therefore, the loss of Gremlin expression in FmnΔ10.24 (Fig. 2E) and ldIn2 homozygous limb buds seems to be a consequence of either deleting (FmnΔ10.24) or disrupting (ldIn2) a distant cis-regulatory element of Gremlin expression in limb buds rather than disrupting Formin functions (Zuniga et al. 1999). Indeed, Gremlin transcription is lost from the limb bud mesenchyme of FmnΔ10.24; GreΔORF and ldIn2; GreΔORF compound heterozygous embryos (Fig. 3E,F). Conversely, Formin remains expressed in limb buds of ldln2 and GreΔORF homozygous (Fig. 3G,H) and compound heterozygous embryos (Fig. 3I,J). These results indicate that the relevant elements in question are located upstream of Formin coding exon 23. As no additional genes have been found in the genomic region 10.24 (data not shown), these results indicate that this region is required for cis regulation of Gremlin expression in the limb bud mesenchyme.
Figure 3.
The FmnΔ10.24 and ldIn2 mutations are allelic to GreΔORF by disrupting cis regulation of Gremlin in the limb bud mesenchyme. (A–D) Forelimb skeletal phenotypes of compound heterozygous mice. (E,F) Gremlin is no longer expressed in the limb bud mesenchyme of FmnΔ10.24/+; GreΔORF/+ (E) and ldIn2/+; GreΔORF/+ (F) compound heterozygous embryos. (G–J) Formin remains expressed in the limb bud mesenchyme of ldIn2/In2 (G), GreΔORF/ΔORF (H), FmnΔ10.24/+; GreΔORF/+ (I), and ldIn2/+; GreΔORF/+ (J) compound heterozygous embryos. Note: Samples G–J were pretreated prior to whole mount in situ hybridization for optimal detection of Formin transcripts in the mesenchyme. Such pretreatment results in loss of the AER. Genotypes are indicated as defined in the legend for Figure 2A.
Cis-regulatory elements in region 10.24 mediate activation and SHH responsiveness in the limb bud mesenchyme
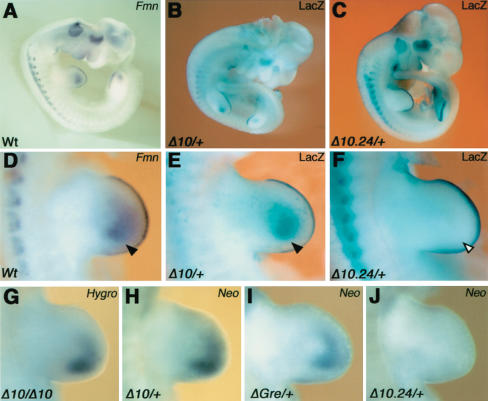
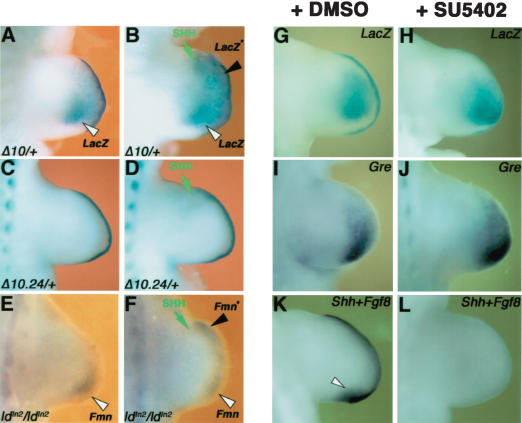
To explore the molecular mechanism for cis regulation of Gremlin, we analyzed the expression of exogenous genes inserted into the ld locus (Fig. 2A,B). In the FmnΔ10 and FmnΔ10.24 alleles, expression of the LacZ reporter gene is controlled by the endogenous Fmn promoters (Fig. 2B). As a consequence, the LacZ distribution recapitulates Formin expression perfectly in FmnΔ10 heterozygous embryos (Fig. 4, cf. A and B). In FmnΔ10.24 heterozygous embryos, LacZ activity (Fig. 4C) is specifically lost from the limb bud mesenchyme (arrowheads in Fig. 4, cf. D,E and F). These results reveal that the genomic region 10.24 is required for limb bud mesenchymal expression of both Gremlin and Formin (Figs. 3E, 4C,F). Furthermore, insertion of PGK promoters driving expression of NeoR and HygroR genes at various positions (Fig. 2B) results in these exogenous transcripts being expressed like Formin and Gremlin in the limb bud mesenchyme (Fig. 4G–I), irrespective of transgene insertion site and orientation (Fig. 2B). In contrast, expression of the NeoR transgene is lost in limb buds heterozygous for the FmnΔ10.24 mutation (Fig. 4J). These results reveal the presence of cis-regulatory elements within Fmn genomic region 10.24 able to drive expression of exogenous genes in the limb bud mesenchyme. As both Gremlin and Formin expression are positively regulated by SHH signaling in the limb bud mesenchyme (Zuniga et al. 1999), a potential role of region 10.24 in mediating this SHH responsiveness was assessed. Indeed, anterior grafts of SHH expressing cells induce ectopic LacZ expression in cultured limb buds of FmnΔ10 heterozygous embryos (Fig. 5, cf. A and B). In contrast, SHH is unable to induce LacZ expression in limb buds of FmnΔ10.24 heterozygous embryos (Fig. 5, cf. C and D), which indicates that this region participates in mediating SHH responsiveness of Formin and Gremlin in the limb bud mesenchyme. In contrast to the FmnΔ10.24 allele (Fig. 5D), Formin but not Gremlin (Zuniga et al. 1999) expression can be ectopically induced by SHH in ldIn2 homozygous limb buds (Fig. 5, cf. E and F; data not shown). These results show that the inversion affecting the ldIn2 allele separates the Gremlin, but not Formin transcription unit from the SHH response elements (see also Fig. 7A, below).
Figure 4.
The Formin genomic region 10.24 regulates expression of endogenous and exogenous transcription units inserted into the ld locus. (A) Formin transcript distribution in a wild-type embryo around gestational day 10.5 (hemisection). (B) LacZ recapitulates the Formin transcript distribution in the FmnΔ10 allele. (C) Limb bud mesenchymal LacZ is specifically lost in the FmnΔ10.24 allele. (D–F) Forelimb buds of the embryos shown in A–C. Black arrowheads indicate Fmn/LacZ distribution in the mesenchyme and open arrowhead indicates the loss of LacZ in the FmnΔ10.24 allele. (G–J) Limb bud mesenchyme-specific expression of the HygroR (G) and NeoR (H–J) genes inserted into the ld locus. Genotypes are indicated as defined in the legend for Figure 2A.
Figure 5.
The limb bud regulatory region 10.24 is responsive to SHH signaling. SHH-expressing cells were grafted to the anterior limb bud mesenchyme (E10.25, 32–34 somites) and trunks cultured for 16–20 h prior to analysis. (A) Nongrafted limb bud of an FmnΔ10 heterozygous embryo (control). (B) Ectopic LacZ in the contralateral limb bud having received an anterior graft of SHH expressing cells. (C) Control FmnΔ10.24 heterozygous limb bud. (D) Failure to induce LacZ expression in response to SHH-expressing cells in an FmnΔ10.24 heterozygous limb bud. (E) Control limb bud of an ldIn2 homozygous embryo. (F) Induction of Fmn expression in response to ectopic SHH signaling in an ldIn2 homozygous embryo. In A–F, a green arrow indicates the position of SHH expressing cells, a black arrowhead and an asterisk indicate ectopic gene expression, and an open arrowhead indicates endogenous expression. (G–L) Gremlin and Formin expression are maintained in limb buds in which FGF signaling transduction has been blocked by the inhibitor SU5402. Forelimb buds of FmnΔ10/Δ10 embryos (E10.0, 29–32 somites) were cultured in the presence of 10 μM SU5402 (+SU5402; stock dissolved in DMSO) for 14–16 h prior to analysis. Controls were cultured in the presence of an equal concentration of DMSO (+DMSO; 0.03% final concentration in medium). (G) LacZ detection in an untreated limb bud. (H) LacZ remains in a limb bud cultured in the presence of SU5402. Note the down-regulation of LacZ in the AER due to flattening in the absence of FGF signal transduction. (I) Gremlin expression in an untreated limb bud. (J) Gremlin remains in a limb bud cultured in the presence of SU5402. (K) Detection of Shh (arrowhead) and Fgf8 transcripts (AER) in an untreated limb bud. (L) Loss of both Shh and Fgf8 expression in a limb bud cultured in the presence of SU5402.
Figure 7.
The large regulatory landscape required for activation of Gremlin expression in the limb bud mesenchyme. (A) Region 19.23 is disrupted by all relevant ld alleles. (Activ.) A global control region (GCR) located in the genomic region encompassing Formin exons 19–23 is required in cis for activation of Gremlin and Formin expression in the posterior limb bud mesenchyme. This region is disrupted in the ldIn2, ldTgHd, and ldTgBri alleles. (SHH-Reg.) The region necessary for SHH-mediated regulation of Gremlin and Formin (see Fig. 5A–F) is most likely located upstream of Formin exon 19. Schemes show how the ldIn2, ldTgHd, and ldTgBri mutations disrupt the activator GCR. (B) Alignment of the orthologous region 19.23 from the mouse and human genome using the mVISTA program (window size, 100 bases; homology threshold, 65%; Mayor et al. 2000). This alignment reveals multiple blocks of intronic sequences highly conserved between the two species. Exons 19–23 are indicated in blue and the parts of intronic sequences conserved more than 75% are marked as red peaks. The green line indicates the genomic sequences driving LacZ expression in BAC construct C (see Fig. 6C). The chicken genomic region 19.23 is only partially available, with some gaps in the regions containing the blocks of sequence conserved between mouse and human genome that precluded complete analysis. However, three regions (indicated in orange) highly conserved among all three species have been identified. Region 1 (upstream of exon 19): 77.4% identity over 243 bases. Region 2 (upstream of exon 20): 85.1% over 329 bases. Region 3 (just downstream from exon 22): 81% over 352 bases. The human, mouse, and chicken genomic sequences were obtained from the Ensembl Genome Browser (http://www.ensembl.org) using genome assembly releases v19.34b, v19.32.2, and prerelease1, respectively.
Analysis of Gremlin expression in chicken embryos suggested that its expression in the limb bud mesenchyme may depend on FGF signaling by the AER (Merino et al. 1999). However, limb buds of FmnΔ10/Δ10 embryos cultured in the presence the FGF signaling inhibitor SU5402 (Mohammadi et al. 1997; for details, see Materials and Methods) continue to express LacZ and Gremlin (Fig. 5, cf. G,I and H,J). As expected, inhibition of FGF signaling causes flattening of the AER (data not shown) and subsequent down-regulation of Shh and Fgf expression due to disrupting feedback signaling (Fig. 5K,L; data not shown). The results shown in Figure 5G–J indicate that mesenchymal Formin and Gremlin expression does not depend significantly on FGF signaling. In agreement, genetic analysis reveals that Gremlin, but not Shh, remains expressed in mouse limb buds lacking both Fgf8 and Fgf4 (Sun et al. 2002).
The genomic region 19.23 is sufficient to activate gene expression in the limb bud mesenchyme
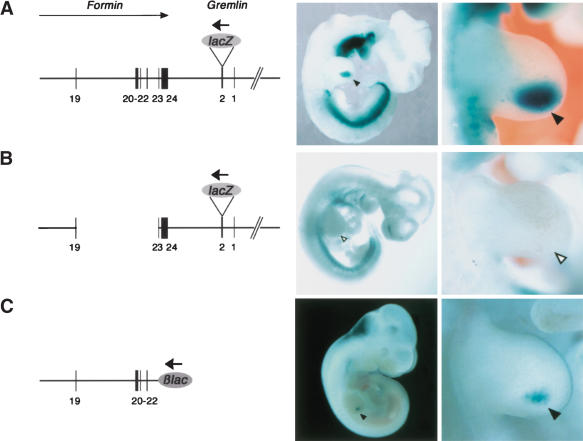
Using a BAC-based strategy to generate transient transgenic mouse embryos, we have positively identified the relevant cis-regulatory region (Fig. 6). Initially, a BAC containing the mouse Fmn genomic region spanning exons 19–24, the intergenic region, and the complete Gremlin transcription unit (tagged by LacZ in exon 2) was injected into fertilized oocytes and embryos stained for LacZ activity during gestational days 10.5 (Fig. 6A). This transgene (BAC construct A) is expressed in the posterior limb bud mesenchyme (arrowheads in Fig. 6A, n = 5/6), which indicates that all the required cis-regulatory elements are present. In contrast, LacZ activity is specifically lost from the limb bud mesenchyme of embryos harboring BAC construct B, which lacks region 19.23 (Fig. 6B, n = 7/7). The potential autonomy of region 19.23 (Fig. 6C) was assessed by inserting it downstream from a LacZ gene under control of a minimal β-globin promoter (Morgan et al. 1996). Indeed, BAC construct C (Fig. 6C) and a shorter construct containing region 20.23 (data not shown) are sufficient to drive LacZ expression into the posterior limb bud mesenchyme (Fig. 6C, arrowheads, n = 2/6). These results establish that region 19.23 encodes cis-regulatory elements sufficient to activate gene expression in combination with either the endogenous Gremlin or an exogenous minimal promoter (albeit with lower efficiency; Fig. 6, cf. A and C). Furthermore, region 19.23 is sufficient to activate LacZ expression in both dorsal and ventral posterior limb bud mesenchyme similar to Formin and Gremlin (Supplementary Fig. 3). Reduction of this genomic region reduces expression further, indicating that the required regulatory elements are spread over a larger region (see also Fig. 7; data not shown).
Figure 6.
The Fmn locus encodes a regulatory region sufficient to activate Gremlin transcription in the limb bud mesenchyme. (A) Construct A was generated by in frame insertion of a LacZ gene 30 bases downstream from the Gremlin ATG (exon 2) into BAC #113H17. This BAC encodes Fmn exons 19–24, the intergenic region, entire Gremlin gene and extends ∼150 kb upstream of Gremlin exon 1. Note that Gremlin (bold arrow) is transcribed in reverse orientation to Formin (arrow). (B) Construct B was generated by deleting the genomic region delimited by exons 19–23 from construct A. (C) Construct C was generated by replacing Fmn exon 23 and all downstream sequences with the β-lac reporter gene in BAC #113H17. Note that the β-lac reporter gene inserted in construct C is transcribed like Gremlin, that is, in reverse orientation (bold arrow) to Formin; exons are numbered as in Figure 2A. For all constructs, the LacZ distribution is shown in founder embryos around gestational day 10.5. Left panels show whole embryo views and right panels show forelimb buds. Note that different embryos are shown in the left and right panels. Black arrowheads point to the LacZ expression domains in the limb bud. Open arrowheads indicate the lack of LacZ expression in the limb bud of an embryo transgenic for construct B.
Discussion
We establish that the ld phenotype is caused by disrupting either the regulatory landscape controlling transcriptional activation of the BMP antagonist Gremlin in the limb bud mesenchyme or directly the Gremlin transcription unit. Therefore, the ld phenotype has been wrongly attributed to disruption of Formin functions. We now show that all ld alleles together with the FmnΔ10.24 and GreΔORF mutations define one allelic series of variable phenotypic strength. The ldOR, ldJ, and GreΔORF alleles are the strongest alleles as the Gremlin gene products are either deleted or truncated, which causes a pleiotropic loss-of-function phenotype. The complete renal agenesis and lung septation defects in ldOR homozygous newborn mice result in fully penetrant neonatal lethality identical to the GreΔORF null allele generated by gene targeting (Michos et al. 2004). A second class of ld alleles is hypomorphic (FmnΔ10.24, IdIn2, ldTgBri, ldTgHd). These ld alleles display the characteristic and fully penetrant ld limb phenotype, but either lack or only show low frequencies of renal abnormalities and thereby generally survive to adulthood (see also Maas et al. 1994). A cis-regulatory region located within Formin that is required for Gremlin activation in the limb bud mesenchyme is either deleted or disrupted by all these hypomorphic ld alleles (Fig. 7A). Therefore, these mutations induce limb bud specific loss of Gremlin expression in cis and not in trans as a consequence of disrupting Formin (Zuniga et al. 1999). Despite the fact that FmnΔ10 homozygous mice (lacking the complete C-terminal Formin domain) are normal, it cannot be excluded that other Formin protein domains function during embryonic development in pathways other than the ones disrupted due to the lack of the BMP antagonist Gremlin. In particular, specific inactivation of Formin isoform IV results in low penetrance and mostly partial renal agenesis phenotypes, whereas limbs are phenotypically wild type (Wynshaw-Boris et al. 1997; Chao et al. 1998). The partial disruption of kidney development in these mutations and some of the hypomorphic ld alleles (see before) correlate well with abundant Formin expression during kidney morphogenesis (for review, see Zeller et al. 1999).
Our studies reveal the molecular disruption of Gremlin in the ldOR and ldJ alleles and positively identify the shared limb bud cis-regulatory elements in Formin genomic region 19.23. In particular, these latter studies explain satisfactorily why disruption of Gremlin functions is the primary cause of the limb phenotypes observed in ld homozygous mice. In the ldIn2 allele, the inversion between Formin and Agouti (Woychik et al. 1990) relocates the Gremlin gene ∼40 Mb away from the genomic region 19.23 (Fig. 7A). The ldTgHd allele arose by insertion of multiple copies of a MMTV–myc transgene in combination with an ∼1-kb deletion between Formin exons 19 and 20 (Woychik et al. 1985). The transgene insertion site disrupts the genomic region 19.23 and is likely to tether long-range-enhancing activity due to insertion of several strong exogenous promoters (Fig. 7A). Last but not least, the genomic region 19.23 is deleted in the ldTgBri allele (Vogt et al. 1992). We establish that such a deletion completely abolishes Gremlin activation in the posterior limb bud mesenchyme (Fig. 7A). Our results also reveal the need to reexamine the human congenital malformations mapping to the orthologous locus on chromosome 15q13-14 (Maas et al. 1991) with phenotypes similar to limb deformity. Despite the fact that mutations in the Gremlin and Formin ORF have been excluded (Bacchelli et al. 2001; Morgan et al. 2003), mutations in the homologous cis-regulatory elements may cause these malformations.
The positively identified cis-regulatory region 19.23 (Fig. 7A) activates transcription of unrelated genes in a promoter- and orientation-independent manner at large distances. These features are strikingly similar to the ones of a recently identified global control region (GCR), which regulates limb bud specific expression of 5′Hoxd and the unrelated Evx2 and Lunapark (Lnp) genes (Spitz et al. 2003). Such GCRs seem to consist of multiple regulatory regions and/or enhancers that form a chromosomal regulatory landscape and act at a distance to coactivate sets of neighboring genes in specific tissues. Comparison of the orthologous mouse and human genomic regions 19.23 reveals highly conserved sequences within introns (at least 100 bases with more than 75% identity) that are spread over the whole region (Fig. 7B). However, no obvious consensus binding sites for, for instance, GLI transcription factors have been found within these conserved blocks (F. Spitz, unpubl.).
Interestingly, neither Lnp nor Formin are essential for limb development in spite of these loci harboring the essential GCR and being expressed during limb bud development. Cis regulation of the further downstream 5′Hoxd and Gremlin genes by the GCR is however essential for distal limb bud morphogenesis (Zákány and Duboule 1996; Khokha et al. 2003; Michos et al. 2004). Experimental evidence indicates that the Formin landscape region 10.24 not only contains the regulatory elements necessary for activation, but also the ones mediating response to SHH signaling (Fig. 7A). Regulation of these two genes in the limb bud mesenchyme is likely regulated by interaction of several control regions scattered over the genomic landscape (Fig. 7A,B). Our studies also reveal for the first time that such landscapes and GCRs are not a peculiar feature of regulating functionally related and clustered genes such as, for example, 5′Hoxd and globin genes (for review, see Zeller and Deschamps 2002). Rather, they seem to represent a novel mechanism by which tissue-specific coexpression of neighboring genes is orchestrated, even if they are structurally and functionally not related. These studies are likely to reveal the tip of the iceberg and further exploration of these regulatory landscapes will be necessary to understand if GCRs are composed of novel types of tissue-specific activator/enhancer elements or if they harbor elements enabling the known activators and enhancers to act over greater than usual distances (Fig. 7; Spitz et al. 2003). Last but not least, Shh expression in the limb bud mesenchyme is itself controlled by a regulatory region located ∼800 kb upstream within the unrelated Lmbr1 gene (Lettice et al. 2002).
It could well be that during evolution of vertebrates an initial selective constraint resulted in Formin and Gremlin being kept neighboring genes and their expression became coregulated as part of a larger regulatory landscape. In fact, both genes are also expressed in similar but not identical patterns during kidney organogenesis, and genetic analysis has revealed essential functions for both genes during kidney development (Wynshaw-Boris et al. 1997; Michos et al. 2004). Initial intertwining of their regulation could have resulted in them becoming inseparably linked or trapped into this regulatory landscape, in spite of eventual diversification of their functions during vertebrate evolution. The present study establishes that Gremlin and Formin are neither part of the same pathway nor a common synexpression group (Niehrs and Pollet 1999), in spite of these two genes being coexpressed in various embryonic tissues. Much of the gene diversity during vertebrate evolution is thought to have arisen following gene and chromosomal duplications. Interestingly, the arrangement of Formin-2 (Leader and Leder 2000) and Gremlin-2 (or Prdc; Minabe-Saegusa et al. 1998) on mouse chromosome 1 is identical to chromosome 2, as the two genes are also located next to one another and transcribed in reverse orientation (see also UCSC Genome Browser, http://genome.ucsc.edu). During organogenesis, both Formin-2 and Gremlin-2 are expressed in the developing neural tube (Minabe-Saegusa et al. 1998; Leader and Leder 2000), suggesting that they could also be part of a common regulatory landscape.
Large-scale phenotypic screens have become a renewed and major effort to genetically analyze vertebrate model organisms such as zebrafish and mouse (Justice 2000). Such screens provide powerful tools to identify the gene cascades controlling development, physiology, and disease by scoring for the relevant phenotypes. However, the present study reveals that in specific cases, the identification and analysis of the essential gene and/or cascades can be rather tedious. The molecular alterations in the Formin locus together with the Formin transcript and protein distribution established this gene as the obvious candidate, whose disruption causes the ld phenotype (for reviews, see Zeller et al. 1999; Panman and Zeller 2003). Only the combination of experimental embryology with advanced reverse genetics and transgenesis has finally revealed the true nature of the ld limb phenotype and established Gremlin as the essential one of the two disrupted genes. As many vertebrate genes contain rather large intronic and noncoding regions, such large regulatory landscapes may be rather common and more surprises with respect to assigning phenotypes to alterations of particular genes and/or pathways may well be in store.
Materials and methods
Mapping and identification of the ldOR and ldJ mutations
The deletion of the Gremlin ORF in the ldOR allele was initially detected by Southern blotting (Fig. 1B) and mapped using a combination of Southern blot and long-range PCR analysis (Jansen et al. 1997). A PCR fragment spanning the deletion breakpoints in the ldOR allele was isolated. This fragment was sequenced and the extent of the deletion identified by sequence comparison to the wild-type Gremlin locus. The ldOR allele was crossed into 129S3/SvImJ, C57BL/6, and CD1 strains, as the penetrance of the kidney phenotype depends on genetic background. In the 129S and CD1 backgrounds, the kidney phenotype is fully penetrant (Supplementary Fig. 1). The point mutation affecting splicing of Gremlin in the ldJ allele was identified as follows. Total RNAs were isolated from wild-type and ldJ/J mouse embryos (embryonic day 12 [E12]) using the RNeasy kit (Qiagen) and cDNAs synthesized using standard procedures. Gremlin transcripts were amplified using specific primers in exons 1 and 2. The Gremlin locus was analyzed comparing genomic DNA from wild-type and ldJ/J embryos. The genomic region containing the intron–exon 2 boundary was amplified by PCR and PCR products separated on a 1.0% agarose gel, and in all cases (cDNAs and genomic DNAs) only one specific fragment was amplified (data not shown). These amplified DNA bands were cloned and their sequences analyzed using the ClustalW-X program (http://www.ebi.ac.uk/clustalw). Sequences of all primers used for these studies are available upon request.
Generation of Fmn mutant alleles
The FmnΔ10 allele was obtained by successive targeting in R1 ES cells (Nagy et al. 1993). Initially, Fmn exon 10 (151 bases) was completely replaced by a PGK-NeoR expression cassette with an SV40 polyadenylation site and a LoxP site was inserted further downstream in the intron (Fig. 2B). One of three correctly recombined ES-cell clones was selected for additional gene targeting. A PGK-HygroR cassette flanked by two loxP sites and an En-2 splice acceptor-IRES-LacZ expression cassette (Mountford et al. 1994) was inserted into Fmn exon 24 using a Spe1 site 86 bases downstream of the translational stop codon. Fifty-three correctly targeted ES-cell clones were obtained and characterized extensively by genomic Southern blot analysis to confirm correct alterations of both targeted genomic sites. To identify clones carrying both targeting sites in cis, 25 ES-cell clones were electroporated with a Cre expression vector. For 14 ES-cell clones, the correct Cre-mediated excision patterns were obtained for all possible combinations between the three LoxP sites (Fig. 2B). ES-cell clones carrying the FmnΔ10 allele were injected into C57BL/6 blastocysts and germ-line transmission was obtained. FmnΔ10 mice were maintained by breeding a mixed 129xC57/BL6 background. The FmnΔ10.24 allele was generated by intercrossing FmnΔ10/+ mice with the Cre deleter mouse strain (Schwenk et al. 1995). PCR analysis was used to show that excision had occurred between the two most distant LoxP sites, causing deletion of the genomic region spanning Fmn exons 10–24 (170 kb).
RT–PCR analysis of Formin expression
Total RNA from wild-type, FmnΔ10, and FmnΔ10.24 homozygous embryos was isolated using the RNeasy extraction kit (Qiagen). First strand cDNAs were synthesized according to standard procedures by using 17 μg of total RNA. Subsequently, PCR was performed to detect Formin transcripts. The following primer pairs were used: forward primer in exon 9 (e9, 5′-GCTCTTCCTAACAGTGGAGGTCC-3′) and reverse primer in exon 15 (e15, 5′-CACACTCTTCATGTGCAACAA-3′) or exon 23 (e23L1, 5′-CTTTGTCTCCACTTTCTTCTCTGATG TC-3′) for wild-type and FmnΔ10/Δ10 cDNAs, forward primer in exon 9 and reverse primer in IRES (IRES-1, 5′-GCTTCCTTC ACGACATTCAACAGACC-3′) for FmnΔ10.24/Δ10.24 cDNAs. PCR products were separated on a 1.0% agarose gel and cloned for sequence analysis and alignment using the ClustalW-X program (http://www.ebi.ac.uk/clustalw).
Culture of mouse limb buds (trunk cultures) and inhibition of FGF signaling
Shh expressing cells were grafted into mouse limb buds (30–33 somites, E10.0–E10.25) cultured as described (Zuniga et al. 1999). Alternatively, FGF signaling was blocked by supplementing the culture medium with 10 μM SU5402 (final concentration), an efficient inhibitor of FGF signal transduction (Mohammadi et al. 1997). SU5402 (Calbiochem) was dissolved in 100% DMSO at 10 mg/mL (stock solution). Experimental controls were treated with an equal concentration of DMSO in culture medium (0.03% final). The SU5402 concentration blocking FGF signaling efficiently was established in pilot experiments and is well within the commonly used range of concentrations. Both grafted and SU5402-treated limb buds were cultured for 14–16 h prior to analysis by in situ hybridization.
Skeletal preparations, whole mount in situ hybridization and LacZ staining
Skeletal preparations and whole mount in situ hybridization assays were carried out as previously described (Zuniga and Zeller 1999). β-Galactosidase (LacZ) activity was detected in whole mounts (Knittel et al. 1995) with the following modification: Embryos were stained in the dark at 37°C in 1 mg/mL X-Gal, 0.25 mM K3Fe(CN)6, 0.25 mM K4Fe(CN)6, 0.01% NP40, 0.4 mM MgCl2 in 1× PBS.
BAC constructs and generation of transient transgenic mouse embryos
The genomic organization of the Gremlin and Formin loci and the appropriate BACs were identified using sequences from the Mouse Genome Sequencing Consortium (Waterston et al. 2002) and analyzed with the UCSC Genome Browser (http://genome.ucsc.edu). Mouse BAC clones were obtained from BacPac Resources (Children's Hospital Oakland, USA) and modified by ET recombination as described (Spitz et al. 2003). Construct A was engineered by inserting a LacZ reporter gene in frame into the Gremlin ORF encoded by BAC RP23-113H17 (Fig. 6A) using a Zeocin resistance cassette (Invitrogen). Construct B (Fig. 6B) was obtained by deleting the region between exons 19 and 23 from construct A using a Kanamycin resistance cassette. Construct C (Fig. 6C) was generated by targeted replacement of Fmn exon 23 and all downstream 3′ sequences from BAC 113H17 by a LacZ reporter gene driven by a β-globin minimal promoter (Spitz et al. 2003). This cassette is flanked by 50 bases of BAC DNA sequence, which borders the region to be deleted by homologous recombination. All BAC constructs were injected into the pronucleus of fertilized mouse eggs according to standard procedures (Spitz et al. 2001; Nagy et al. 2002) and embryos were collected during gestational day 10 (E10.5) and analyzed by LacZ staining.
Acknowledgments
We are grateful to H. Goedemans, N. Lagarde, and C. Lehmann for technical assistance and mouse husbandry, and to K. O'Leary for help in preparation of the manuscript. We are grateful to T. Kondo, M. Kmita, S.L. Ang, and F. Guillemot for providing targeting vectors, probes, and advice, and to A.M. van der Linden for advice in mapping and isolating the deletion breakpoints in the ldOR mutation. We thank J. Deschamps, F. Meijlink, I. Mattaj, D. Sussman, and C. Torres de los Reyes for helpful discussions and comments on the manuscript. This research was supported by the Faculty of Biology at Utrecht University, grants from the Dutch KNAW (to A.Z.) and NWO (to R.Z.), the Swiss National Science Foundation (to R.Z. and D.D.) and the two cantons of Basel (to R.Z).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.299904.
References
- Avsian-Kretchmer O. and Hsueh, A.J. 2003. Comparative genomic analysis of the eight-membered-ring cystine-knot-containing bone morphogenetic protein (BMP) antagonists. Mol. Endocrinol. 18: 1-12. [DOI] [PubMed] [Google Scholar]
- Bacchelli C., Goodman, F.R., Scambler, P.J., and Winter, R.M. 2001. Cenani-Lenz syndrome with renal hypoplasia is not linked to FORMIN or GREMLIN. Clin. Genet. 59: 203-205. [DOI] [PubMed] [Google Scholar]
- Chao C.W., Chan, D.C., Kuo, A., and Leder, P. 1998. The mouse formin (Fmn) gene: Abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 4: 614-628. [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Zigmond, S., and Boone, C. 2003. Formins: Signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116: 2603-2611. [DOI] [PubMed] [Google Scholar]
- Faustino N.A. and Cooper, T.A. 2003. Pre-mRNA splicing and human disease. Genes & Dev. 17: 419-437. [DOI] [PubMed] [Google Scholar]
- Gurrieri F., Kjaer, K.W., Sangiorgi, E., and Neri, G. 2002. Limb anomalies: Developmental and evolutionary aspects. Am. J. Med. Genet. 115: 231-244. [DOI] [PubMed] [Google Scholar]
- Haramis A.G., Brown, J.M., and Zeller, R. 1995. The limb deformity mutation disrupts the SHH/FGF-4 feedback loop and regulation of 5′HoxD genes during limb pattern formation. Development 121: 4237-4245. [DOI] [PubMed] [Google Scholar]
- Jansen G., Hazendonk, E., Thijssen, K.L., and Plasterk, R.H. 1997. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17: 119-121. [DOI] [PubMed] [Google Scholar]
- Justice M.J. 2000. Capitalizing on large-scale mouse mutagenesis screens. Nat. Rev. Genet. 1: 109-115. [DOI] [PubMed] [Google Scholar]
- Khokha M.K., Hsu, D., Brunet, L.J., Dionne, M.S., and Harland, R.M. 2003. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 34: 303-307. [DOI] [PubMed] [Google Scholar]
- Knittel T., Kessel, M., Kim, M.H., and Gruss, P. 1995. A conserved enhancer of the human and murine Hoxa-7 gene specifies the anterior boundary of expression during embryonal development. Development 121: 1077-1088. [DOI] [PubMed] [Google Scholar]
- Kobielak A., Pasolli, H.A., and Fuchs, E. 2004. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6: 21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader B. and Leder, P. 2000. Formin-2, a novel formin homology protein of the cappuccino subfamily, is highly expressed in the developing and adult central nervous system. Mech. Dev. 93: 221-231. [DOI] [PubMed] [Google Scholar]
- Lettice L.A., Horikoshi, T., Heaney, S.J., Van Baren, M.J., Van Der Linde, H.C., Breedveld, G.J., Joosse, M., Akarsu, N., Oostra, B.A., Endo, N., et al. 2002. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl. Acad. Sci. 99: 7548-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas R.L., Zeller, R., Woychik, R.P., Vogt, T.F., and Leder, P. 1990. Disruption of formin-encoding transcripts in two mutant limb deformity alleles. Nature 346: 853-855. [DOI] [PubMed] [Google Scholar]
- Maas R.L., Jepeal, L.I., Elfering, S.L., Holcombe, R.F., Morton, C.C., Eddy, R.L., Byers, M.G., Shows, T.B., and Leder, P. 1991. A human gene homologous to the formin gene residing at the murine limb deformity locus: Chromosomal location and RFLPs. Am. J. Hum. Genet. 48: 687-695. [PMC free article] [PubMed] [Google Scholar]
- Maas R., Elfering, S., Glaser, T., and Jepeal, L. 1994. Deficient outgrowth of the ureteric bud underlies the renal agenesis phenotype in mice manifesting the limb deformity (ld) mutation. Dev. Dyn. 199: 214-228. [DOI] [PubMed] [Google Scholar]
- Mayor C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046-1047. [DOI] [PubMed] [Google Scholar]
- Merino R., Rodriguez-Leon, J., Macias, D., Ganan, Y., Economides, A.N., and Hurle, J.M. 1999. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development 126: 5515-5522. [DOI] [PubMed] [Google Scholar]
- Michos O., Panman, L., Vintersten, K., Beier, K., Zeller, R., and Zuniga, A. 2004. Gremlin mediated BMP antagonism induces the epithelial–mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development (in press). [DOI] [PubMed]
- Minabe-Saegusa C., Saegusa, H., Tsukahara, M., and Noguchi, S. 1998. Sequence and expression of a novel mouse gene PRDC (protein related to DAN and cerberus) identified by a gene trap approach. Dev. Growth Differ. 40: 343-353. [DOI] [PubMed] [Google Scholar]
- Mohammadi M., McMahon, G., Sun, L., Tang, C., Hirth, P., Yeh, B.K., Hubbard, S.R., and Schlessinger, J. 1997. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276: 955-960. [DOI] [PubMed] [Google Scholar]
- Morgan B.A., Conlon, F.L., Manzanares, M., Millar, J.B., Kanuga, N., Sharpe, J., Krumlauf, R., Smith, J.C., and Sedgwick, S.G. 1996. Transposon tools for recombinant DNA manipulation: Characterization of transcriptional regulators from yeast, Xenopus, and mouse. Proc. Natl. Acad. Sci. 93: 2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan N.V., Bacchelli, C., Gissen, P., Morton, J., Ferrero, G.B., Silengo, M., Labrune, P., Casteels, I., Hall, C., Cox, P., et al. 2003. A locus for asphyxiating thoracic dystrophy, ATD, maps to chromosome 15q13. J. Med. Genet. 40: 431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountford P., Zevnik, B., Duwel, A., Nichols, J., Li, M., Dani, C., Robertson, M., Chambers, I., and Smith, A. 1994. Dicistronic targeting constructs: Reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. 91: 4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Rossant, J., Nagy, R., Abramow-Newerly, W., and Roder, J.C. 1993. Viable cell culture-derived mice from early passage embryonic stem cells. Proc. Natl. Acad. Sci. 90: 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsensten, M., Vintersten, K., and Behringer, R. 2002. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, New York.
- Niehrs C. and Pollet, N. 1999. Synexpression groups in eukaryotes. Nature 402: 483-487. [DOI] [PubMed] [Google Scholar]
- Panman L. and Zeller, R. 2003. Patterning the limb before and after SHH signalling. J. Anat. 202: 3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A.S. 2002. Functional genomics in the mouse. Funct. Integr. Genomics 2: 81-91. [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron, U., and Rajewsky, K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23: 5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Gonzalez, F., Peichel, C., Vogt, T.F., Duboule, D., and Zakany, J. 2001. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes & Dev. 15: 2209-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Gonzalez, F., and Duboule, D. 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113: 405-417. [DOI] [PubMed] [Google Scholar]
- Sun X., Mariani, F.V., and Martin, G.R. 2002. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418: 501-508. [DOI] [PubMed] [Google Scholar]
- Vogt T.F., Jackson Grusby, L., Wynshaw Boris, A.J., Chan, D.C., and Leder, P. 1992. The same genomic region is disrupted in two transgene-induced limb deformity alleles. Mamm. Genome 3: 431-437. [DOI] [PubMed] [Google Scholar]
- Wang C.C., Chan, D.C., and Leder, P. 1997. The mouse formin (Fmn) gene: Genomic structure, novel exons, and genetic mapping. Genomics 39: 303-311. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Woychik R.P., Stewart, T.A., Davis, L.G., D'Eustachio, P. and Leder, P. 1985. An inherited limb deformity created by insertional mutagenesis in a transgenic mouse. Nature 318: 36-40. [DOI] [PubMed] [Google Scholar]
- Woychik R.P., Generoso, W.M., Russell, L.B., Cain, K.T., Cacheiro, N.L.A., Bultman, S.J., Selby, P.B., Dickinson, M.E., Hogan, B.L.M., and Rutledge, J.C. 1990. Molecular and genetic characterisation of a radiation-induced structural rearrangement in mouse chromosome 2 causing mutations at the limb deformity and agouti loci. Proc. Natl. Acad. Sci. 87: 2588-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A., Ryan, G., Deng, C.X., Chan, D.C., Jackson-Grusby, L., Larson, D., Dunmore, J.H., and Leder, P. 1997. The role of a single formin isoform in the limb and renal phenotypes of limb deformity. Mol. Med. 3: 372-384. [PMC free article] [PubMed] [Google Scholar]
- Zákány J. and Duboule, D. 1996. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 384: 69-71. [DOI] [PubMed] [Google Scholar]
- Zeller R. and Deschamps, J. 2002. Developmental biology: First come, first served. Nature 420: 138-139. [DOI] [PubMed] [Google Scholar]
- Zeller R., Haramis, A., Zuniga, A., McGuigan, C., Dono, R., Davidson, G., Chabanis, S., and Gibson, T. 1999. Formin defines a large family of morphoregulatory genes and functions in establishment of the polarising region. Cell Tissue Res. 296: 85-93. [DOI] [PubMed] [Google Scholar]
- Zuniga A. and Zeller, R. 1999. Gli3 (Xt) and formin (ld) participate in the positioning of the polarising region and control of posterior limb-bud identity. Development 126: 13-21. [DOI] [PubMed] [Google Scholar]
- Zuniga A., Haramis, A.P., McMahon, A.P., and Zeller, R. 1999. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401: 598-602. [DOI] [PubMed] [Google Scholar]