Abstract
Jasmonates (JA) are important regulators of plant defense responses that activate expression of many wound-induced genes including the tomato proteinase inhibitor II (pin2) and leucine aminopeptidase (LAP) genes. Elements required for JA induction of the LAP gene are all present in the -317 to -78 proximal promoter region. Using yeast one-hybrid screening, we have identified the bHLH-leu zipper JAMYC2 and JAMYC10 proteins, specifically recognizing a T/G-box AACGTG motif in this promoter fragment. Mutation of the G-box element decreases JA-responsive LAP promoter expression. Expression of JAMYC2 and JAMYC10 is induced by JA, with a kinetics that precedes that of the LAP or pin2 transcripts. JAMYC overexpression enhanced JA-induced expression of these defense genes in potato, but did not result in constitutive transcript accumulation. Using footprinting assays, an additional protected element was identified, located directly adjacent to the T/G-box motif. Mutation of this element abolishes JA response, showing that recognition of this duplicated element is also required for gene expression. Knockout mutants in the AtMYC2 homolog gene of Arabidopsis are insensitive to JA and exhibit a decreased activation of the JA-responsive genes AtVSP and JR1. Activation of the PDF1.2 and b-CHI, ethylene/JA-responsive genes, is, however, increased in these mutants. These results show that the JAMYC/AtMYC2 transcription factors function as members of a MYC-based regulatory system conserved in dicotyledonous plants with a key role in JA-induced defense gene activation.
Keywords: pin2, LAP, wounding, herbivore attack, JA cross-talk
Plants respond to insect attack and wounding by activating a set of genes involved in herbivore deterrence, wound-healing, and defense against pathogen infection (Ryan 2000; León et al. 2001; Li et al. 2002; Turner et al. 2002). Activation of wound-induced defense genes involves signal transduction pathways that operate both locally at the site of wounding and systemically in undamaged leaves (Ryan 2000). Mechanical wounding or herbivore attack to tomato or potato leaflets results in systemic accumulation of multiple defense-related proteins, including proteinase inhibitors (PI), exopeptidases like leucine aminopeptidase (LAP), and components of the wound signal pathway (Ryan 2000). Wound-induced expression of these defense genes is controlled by the jasmonate family of signaling molecules (Ryan 2000; Turner et al. 2002). Genetic analysis in tomato has indicated that the 18-amino-acid peptide systemin and its precursor protein, prosystemin, are upstream components of the signaling cascade involving systemic induction of jasmonates (Howe and Ryan 1999; Li et al. 2001, 2002). Recognition of systemin by its 160-kDa receptor LRR-receptor-like kinase (Scheer and Ryan 2002) activates a cascade of intracellular signaling events that leads to the release of linolenic acid from membrane lipids and the synthesis of jasmonates via the octadecanoid pathway (Schaller 2001; Howe and Schilmiller 2002).
A role for jasmonates in intercellular signaling is supported by the fact that application of JA to one leaf induces PI expression in distal untreated leaves (Ryan 2000). Evidence for a function of JA as a long-distance signal for systemic wound-activation has also been obtained from grafting experiments using mutants deficient in JA biosynthesis or JA perception. These studies indicated that the peptide systemin might not function as a long-distance signal, although it is required for production or translocation of the systemic signal to unwounded leaves (Li et al. 2002; Lee and Howe 2003).
In Arabidopsis, wounding activates independent signaling pathways regulating different sets of target genes either at the wound site or in distal leaves (Rojo et al. 1999). JA induces systemic expression of wound-responsive genes such as VSP, JR1, or Thi1.2 (Berger et al. 1995; Titarenko et al. 1997; Bohlmann et al. 1998), but induction of these genes in the wounded leaves is negatively regulated by local synthesis of ethylene (Rojo et al. 1999). JA and ethylene, however, synergistically cooperate to activate expression of basic PR proteins such as b-CHI, PR3, and PDF1.2 (Xu et al. 1994; Penninckx et al. 1998). Pathogen-induced expression of b-CHI and PDF1.2 is blocked in mutants affected in their response to JA (coi1-1 and jar1) or ethylene (ein2-1 and etr1-1), these mutants being more susceptible to the attack by different fungal pathogens (Staswick et al. 1998; Thomma et al. 1999). These differences in signaling may reflect different mechanisms that evolved in each plant to optimize spatial and temporal defense-gene expression, although a better characterization of the mechanisms integrating JA and ethylene signaling is still needed.
Despite the vast amount of information available for the primary signalling components (for review, see Schaller 2001; Turner et al. 2002), limited data exist on the latter signaling steps leading to defense gene activation in response to JA. Mutant screens in Arabidopsis for insensitivity to JA and coronatine or altered JA-regulated gene expression have identified only a few genes, including coi1 (Feys et al. 1994), jar1 (Staswick et al. 2002), jin1 and jin4 (allelic to jar1; Berger et al. 1996), cev1 (Ellis and Turner 2001), and the jasmonate underexpressing mutations jue1, jue2, and jue3 (Jensen et al. 2002).
Upstream regions required for JA-induced gene expression have been identified in the promoters of several JA-regulated genes (Kim et al. 1992; Ishikawa et al. 1994; Samach et al. 1995; Ruíz-Rivero and Prat 1998), but the transcription factors binding to these cis-elements have not yet been isolated. JA-responsive elements have also been identified in the promoter regions of the barley LOX1 (Rouster et al. 1997) and the soybean VspB genes (Mason et al. 1993), although a consensus element could not be deduced from the identified sequences. Whereas a G-box element mediates JA-regulated expression of the potato pin2 and soybean VspB genes, a TGACG-related motif was identified in the barley LOX1 gene (Rouster et al. 1997) or the nos/minimal 35S promoters (Xiang et al. 1996).
In Arabidopsis, constitutive expression of the downstream component of the ethylene/JA-signaling pathway ethylene response factor 1 (ERF1; Solano et al. 1998) increases resistance to several necrotrophic pathogens (Berrocal-Lobo et al. 2002). Overexpression of the AP2 domain ERF1 factor rescues the defense response defects of the coi1 and ein2 mutations and leads to constitutive activation of a large number of ethylene/JA-induced genes, including the PDF1.2 and b-CHI genes (Lorenzo et al. 2003). Interestingly, a GCC-box-like element like that recognized by ERF1 (Fujimoto et al. 2000) was also identified as a JA- and elicitor-responsive element involved in regulation of the terpenoid indole alkaloid (TIA) biosynthetic genes in Catharantus roseus (Menke et al. 1999). This element is bound by the elicitor- and JA-induced AP2 transcription factors ORCA2 and ORCA3 (Menke et al. 1999; van der Fits and Memelink 2000). Overexpression of ORCA3 leads to increased accumulation of terpenoid indole alkaloids (van der Fits and Memelink 2000, 2001), therefore suggesting a conserved role of these AP2/ERF-transcription factors in ethylene (elicitor)/JA-dependent gene activation.
Here we describe the isolation of two MYC regulatory proteins that bind to JA-responsive elements in the tomato LAP and pin2 promoters. Elements required for JA induction of the LAP gene are present in the -317 to -78 proximal promoter region (Ruíz-Rivero and Prat 1998). Using yeast one-hybrid interaction assays, we have isolated two cDNA clones, JAMYC2 and JAMYC10, encoding bHLH-Leu zipper DNA-binding proteins that specifically recognize a T/G-box motif in this promoter region. We have used transgenic potatoes to analyze the function of the T/G-box cis-acting element and the trans-activation ability of these JAMYC transcription factors, because of the faster generation of transformants in this plant species. Potato plants are, indeed, suitable for these studies, as they are genetically very close to tomato, and systemic wound- and JA-induced expression of the LAP, pin2, as well as other defense-response genes is highly conserved in these two species. Arabidopsis lines with T-DNA insertions in the AtMYC2 gene, encoding the closest homolog to the tomato JAMYC regulatory proteins, are insensitive to JA and exhibit an altered pattern of activation of the VSP, PDF1.2, and b-CHI genes. These findings demonstrate a role of JAMYC/AtMYC2 in JA-induced gene activation, these proteins functioning as conserved master switch regulators that activate expression of differentially JA-regulated genes while repressing expression of genes regulated by the ethylene/JA pathway.
Results
Isolation of cDNAs encoding LAP promoter DNA-binding proteins
Tomato LAP is induced in response to wounding and JA treatment and is thought to play a role in protein turnover during defense gene activation (Hildmann et al. 1992; Pautot et al. 1993). This exopeptidase is encoded by a gene family comprised of at least three genes (Ruíz-Rivero and Prat 1998). Previous work in our laboratory has led to the isolation of two genomic clones encoding LAP that share nearly identical promoter sequences up to position -317 relative to the ATG. Transcriptional promoter fusions to the GUS reporter gene showed that these were active gene copies, driving constitutive GUS expression in flowers and wound- or MeJA-induced expression in leaves (Ruíz-Rivero and Prat 1998). A promoter deletion down to this conserved region (-317LAP) was still able to direct MeJA-induced GUS expression, indicating that elements required for JA-responsive gene expression are present in this region. A -317 to -78 fragment fused to the minimal CaMV 35S promoter was also able to confer JA responsiveness to this minimal promoter (Ruíz-Rivero and Prat 1998), this region therefore used as bait in a yeast one-hybrid screening to search for transcription factors that mediate JA response of the LAP gene. A yeast YM4271 strain containing this promoter fragment fused to the HIS3 and β-GAL selection markers was used to screen for specific DNA-binding proteins in a tomato JA-induced leaf cDNA library fused to the GAL4 activation domain. Screening of 2 million transformants yielded 18 positive clones. Of these, 17 encoded proteins with a basic-helix–loop–helix (bHLH) cMYC DNA-binding domain and one encoded a transcription factor of the WRKY family, with a zinc finger DNA-binding domain exclusive to plants (Eulgem et al. 2000). Alignment of the MYC-deduced amino sequences revealed that these clones correspond to two related genes, designated as JAMYC2 and JAMYC10. The deduced JAMYC amino acid sequences shared highest homology with LEJA3 of tomato (AF011557), a partial cDNA isolated in one-hybrid screening studies using the threonine deaminase (Tda) promoter; with the PG1 and PG2 (Phaseolin G-box-binding proteins) regulatory factors from pea (Kawagoe and Murai 1996), and with the AtMYC2 gene from Arabidopsis, reported to function in drought and ABA signaling (Abe et al. 2003).
Full-length clones (Fig. 1A) were isolated by screening a λ-ZAP library. JAMYC2 and JAMYC10 encode proteins with a molecular mass of 75 kDa and 70 kDa, respectively, and a DNA-binding/dimerization domain consisting of a bHLH region followed by a Leu-zipper protein–protein interaction domain located at the C-terminal part of the protein. A conserved acidic region postulated to function as the transcriptional activation domain (Abe et al. 1997) is located in the N-terminal region.
Figure 1.
Amino acid sequences and patterns of expression of the JAMYC bHLH-Zip transcription factors. (A) JAMYC2 and JAMYC10 deduced amino acid sequences. The bHLH-Leu Zip domain is shown as a gray box, and the acidic region is underlined. The start point of the partial clones isolated by one hybrid screening is indicated as an arrowhead. (B) Northern analysis of tomato leaves induced by wounding or application of MeJA. (C) Pattern of tissue-specific expression at 30 min of MeJA treatment. Each lane was loaded with 30 μg of total RNA. Blots were hybridized with probes corresponding to the 3′-noncoding regions of JAMYC2 or JAMYC10, or to the LAP cDNA clone as indicated. Equal loading was verified by EtBr staining of the gel.
JAMYC2 and JAMYC10 expression is induced by wounding and JA treatment
JAMYC mRNA levels were analyzed after wound induction and JA application. As shown in Figure 1B, basal levels of JAMYC2 and JAMYC10 mRNAs were detected in control untreated leaves, but a rapid induction of these transcripts was observed after wounding or JA treatment. High levels of expression of these transcripts were observed as early as 30 min after JA addition, indicating that these transcription factors correspond to early jasmonate-responsive genes. A biphasic induction pattern in response to both wounding or JA application (less mRNA is detected at 2 h as compared with 30 min or 4 h) was consistently detected for transcript JAMYC10, with levels of this transcript returned to control levels by 24 h after induction. Hybridization of the same blots with an LAP cDNA probe showed that induction of the LAP transcript starts by 2 h of treatment, with maximal levels of mRNA observed after 12–24 h of exposure to JA (Fig. 1B), peaks of JAMYC2 and JAMYC10 expression therefore preceding maximal induction of the LAP gene. These results are consistent with a possible function of these transcription factors in LAP gene regulation.
Analysis of the tissue-specific pattern of expression of these genes showed elevated levels of mRNA in flowers. JA application induced accumulation of these transcripts in leaves and flowers and to a lower extent in stem, but did not modify basal levels of JAMYC mRNAs in fruit (Fig. 1C).
The JAMYC proteins recognize a T/G-box motif in the LAP promoter and a G-box motif required for JA induction of the pin2 promoter
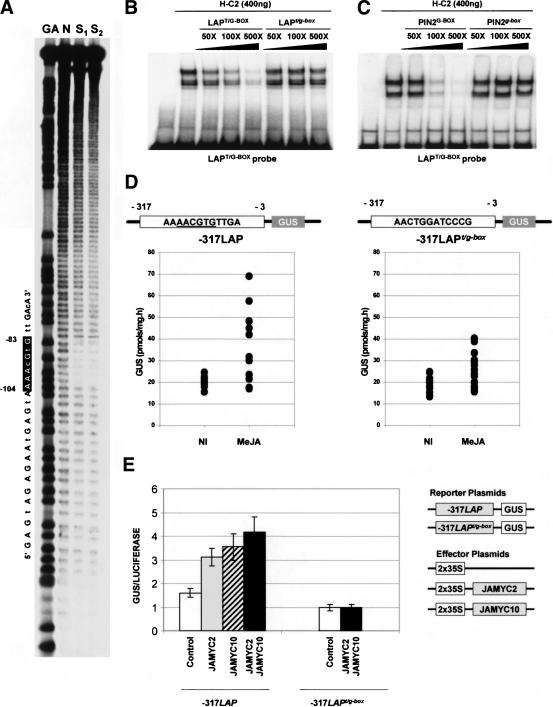
To map the recognition site for these transcription factors, hydroxyl radical interference experiments (Hayes and Tullius 1989) of the -125 LAP fragment were performed with His-tagged fusions of the JAMYC2 and JAMYC10 proteins. As shown in Figure 2A, seven nucleotides were found to be protected from hydroxylation in these assays, with identical protected windows observed for both slower and faster migrating complexes. This footprinted region identifies an AAACGTG element (T/G-box motif) as the binding site for the JAMYC transcription factors. Noteworthy, G-box (CACGTG) sequence motifs are preferential targets for cMYC bHLH DNA-binding proteins (Toledo-Ortiz et al. 2003). These results were further confirmed by mobility shift (EMSA) studies using oligonucleotide probes with intact or mutagenized AAACGTG motifs. As shown in Figure 2B, oligonucleotides containing an intact AACGTG element (LAPT/G-BOX) efficiently competed for complex formation. Competition was totally abolished by replacement of the C and T residues within the AACGTG motif (LAPt/g-box), indicating that the AAACGTG element is critical for JAMYC recognition.
Figure 2.
Characterization of the JAMYC DNA recognition motif. (A) Hydroxyl radical interference assays identify a T/G-box element in the -125LAP promoter fragment as the recognition site for the His-C2 protein. The -125LAP probe was asymmetrically labeled with 32P-dCTP and treated with hydroxyl radical before incubation with the purified protein. Free probe (N) and retarded bands (S1 and S2) were purified by PAGE and separated on a sequencing gel. A G + A Maxam reaction (GA) on the same fragment was loaded as marker. Protected residues are shown as a black box on the nucleotide sequence on the left. Identical results were obtained for both His-C2 and His-C10 proteins. (B) Binding activity of His-C2 protein in gel retardation assays. End-labeled LAPT/G-BOX double-stranded oligonucleotides containing an intact T/G-box motif were used as probe. The oligonucleotide pairs LAPT/G-BOX, with an intact motif, or LAPt/g-box, in which the T/G-box motif was mutagenized, were used as competitors for binding. Oligonucleotides were added in 50- to 500-fold excess for competition. (C) JAMYC2 recognizes a G-box motif required for JA induction of the pin2 promoter. Binding of the His-C2 protein to the LAPT/G-BOX probe was competed by incubation with a 50- to 500-fold excess of the unlabeled PIN2G-BOX (intact G-box) or PIN2g-box (mutated G-box) oligonucleotide pairs. (D) Site-directed mutagenesis of the LAP promoter T/G-box motif. Transgenic potato plants carrying either the -317LAP or the -317LAPt/g-box constructs were grown on soil for 4 wk, and GUS activity was measured in noninduced leaves or in leaves treated for 24 h with 50 μM MeJA. Dots represent the values of GUS activity measured in each individual transformant. (E) DNA binding and trans-activation activity of the JAMYC2 and JAMYC10 transcription factors in BY2 cells. The reporter constructs -317LAP:GUS and -317LAPt/g-box:GUS, including intact or mutagenized copies of the T/G-box motif, were cotransfected with the effector plasmids 2x35S:JAMYC2 (JAMYC2) and 2X35S:JAMYC10 (JAMYC10), expressing the JAMYC proteins under control of the 2x35S promoter. The same vector without insert (2x35S) was used as a control. A 35S:Luciferase plasmid was also used as internal control to normalize cotransfection efficiency. Histograms represent the mean GUS/LUC values for each set of replicates.
JAMYC recognition of the T/G-box motif prompted us to investigate whether these transcription factors would also bind a G-box (CACGTG) element present in the pin2 promoter and required for JA-induced expression of this gene (Kim et al. 1992). EMSA assays were performed with pairs of oligonucleotides complementary to this promoter segment, containing an intact (PIN2G-BOX) or mutagenized (PIN2g-box) copy of the G-box element. Binding of the JAMYC factors to the AACGTG LAP motif (LAPT/G-BOX) was strongly competed by incubation with the wild-type pin2 promoter segment (PIN2G-BOX), and this competition was completely abolished by mutation of the G-box element (PIN2g-box; Fig. 2C). Identical results were obtained when these oligonucleotide pairs were used as probes (data not shown), demonstrating a specific recognition of the tomato pin2 promoter G-box element by these transcription factors.
Site-directed mutagenesis of the LAP promoter T/G-box motif results in reduced JA activation
To determine whether the T/G-box motif functions as a JA-responsive element, transgenic potato lines were obtained bearing a mutated version of the -317 to -3 LAP promoter region fused to the GUS reporter gene (Fig. 2D). Mutation of the T/G-box element caused a strong decrease in the levels of JA-induced activation of this promoter (cf. levels of GUS activity measured in the leaves of the -317LAP or the -317LAPt/g-box lines treated with JA). These results demonstrate a role of the AACGTG motif in JA-induced activation of the LAP promoter, although an additional element appears to be present in this proximal promoter region, this motif being responsible for the residual inducible activity detected in the -317LAPt/g-box transformants.
T/G-Box dependent activation of the LAP promoter by the JAMYC transcription factors was further investigated by transient overexpression of these proteins in BY2 cells. Effector constructs expressing the JAMYC2/JAMYC10 transcription factors under control of the 35S promoter were cotransfected with the -317LAP or the -317LAPt/g-box–GUS reporter fusions (Fig. 2E). Expression of JAMYC2, JAMYC10, or both factors together was able to enhance expression of the -317LAP reporter fusion but did not result in any activation of the -317LAPt/g-box construct. Basal levels of GUS activity obtained for this latter construct were lower than those obtained for -317LAP, consistent with a function of the T/G-box motif in JA-induced promoter expression (wound signaling is activated in bombarded cells). Overexpression of both transcription factors led to higher levels of activation than expression of each protein alone (Fig. 2E). This observation indicates that these factors can heterodimerize, the heterodimer exhibiting higher transactivation ability than the respective homodimers.
Overexpression of the JAMYC transcription factors enhances JA-induced expression of the pin2 and LAP genes
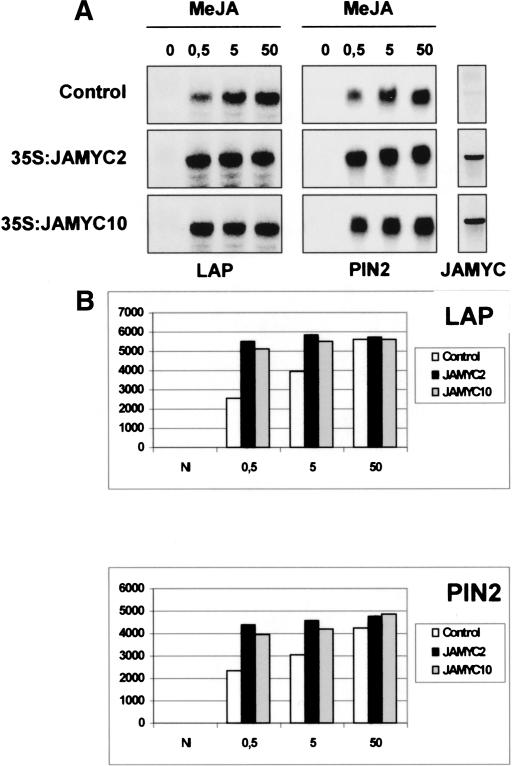
The ability of the JAMYC regulatory proteins to activate expression of the LAP and pin2 genes was also examined in transgenic potato plants that constitutively expressed the JAMYC2 or JAMYC10 transcription factors under control of the 35S promoter (35S:JAMYC2 and 35S: JAMYC10 lines). Individual transformants were analyzed for high levels of expression of the JAMYC transcripts, and the two lines exhibiting a higher level of expression of the transgene were selected for further analysis. Plants were treated with increasing doses of JA, and expression of the LAP and pin2 genes was analyzed after 6 h of treatment. Similar steady-state levels of pin2 and LAP mRNAs were detected in the noninduced transformants and wild-type controls (Fig. 3). A marked increase in the levels of accumulation of these mRNAs was, however, observed in the JAMYC transformants after treatment with subsaturating doses of JA (cf. the levels of pin2 and LAP transcripts detected in the 35S:JAMYC2 or 35S: JAMYC10 lines and in the controls, treated with 0.5 μM or 5 μM JA). At higher JA concentrations, differences between overexpresser lines and the controls were less evident, likely because of response saturation.
Figure 3.
Potato JAMYC2 and JAMYC10 overexpressers show an enhanced induction of the LAP and pin2 genes upon MeJA treatment. (A) LAP and pin2 gene expression in wild-type controls (control) and the 35S:JAMYC2 and 35S:JAMYC10 transgenic potato plants after 6 h of treatment with increasing concentrations of MeJA. Each lane was loaded with 20 μg of total RNA. Blots were hybridized with the LAP, pin2, and JAMYC2 or JAMYC10 probes as indicated. Equal loading was verified by EtBr staining of the gel. (B) Histograms represent the levels of expression of LAP and pin2 as quantified by densitometric scanning of the blot.
These results denote an enhanced JA-induced activation of the pin2 and LAP transcripts in the overexpresser lines and demonstrate a role of both JAMYC2 and JAMYC10 tomato factors in regulated expression of these target genes. Activation of these genes is only observed after JA application, suggesting that JA-induced posttranscriptional modification of these transcription factors, or further induction of an additional regulatory activity, is required for gene expression.
In vivo footprinting of the LAP promoter region
To identify other motifs involved in JA-regulated expression of the -317 to -78 LAP promoter fragment, in vivo footprinting analysis was performed (Busk et al. 1997). Two G residues (positions -96 and -107) were identified on the top DNA strand that were partially protected from methylation in JA-treated leaves (Fig. 4A). Upstream of these two protected nucleotides, an additional G residue (position -118) exhibited enhanced reactivity to DMS treatment (see Fig. 4A). A correlation was observed between the inducible footprint and the time course of LAP mRNA accumulation. No further significant differences in the pattern of methylation were detected within the 300-bp promoter. Differences in methylation were also not observed on the lower DNA strand, owing to the lack of G residues within the protected region. A CGCGG sequence is identified by the hypersensitive G residue (Fig. 4C), whereas a GAGTA duplicated element, between positions -109 and -94, is identified by the protected G residues. A duplicated GAGTA motif similar to the one identified here is present in the cathepsin D inhibitor (CDI) promoter, in a region that has been reported to be required for JA response of this gene (Ishikawa et al. 1994). These findings suggest a function of this duplicated motif in JA-regulated expression of these genes.
Figure 4.
DMS in vivo footprinting of the proximal LAP promoter region identified two G residues in a GAGTA repeated motif that are protected from methylation and shown to be required for JA response. (A) DMS in vivo footprinting of the -270 to -3 LAP promoter region. Control tomato leaves (control) and leaves of plants treated with 50 μM MeJA (8, 16, and 24 h) were directly incubated with DMS. DNA was extracted from these leaves, cleaved with piperidine, and subjected to LmPCR. Amplification of in vitro methylated DNA (naked) is also included as control. Protected G residues are indicated by filled circles and residues with enhanced reactivity (hypersensitivity) by open circles. The position of both protected and hyperreactive residues is indicated on the left. (B) Deletion analysis and site-directed mutagenesis of the proximal LAP promoter region. Transgenic potato plants carrying the -317LAP, -125LAP, -78LAP, and -317LAPgagta constructs fused to the uidA gene were grown on soil for 4 wk, and GUS activity was measured in noninduced leaves or in leaves treated for 24 h with 50 μM MeJA. The histogram shows the average values of GUS activity detected in nontreated and MeJA-treated plants. (C) Nucleotide sequence of the proximal LAP promoter. Hypermethylated G residues, GAGTA repeats, and the T/G-box motif are shown in the -125LAP promoter context. Mutations introduced to yield construct -317LAPgagta are indicated.
Protection of the AACGTG JAMYC target motif was not detected in these assays. Transcripts encoding these regulatory proteins are relatively abundant in noninduced plants. Therefore, it is possible that in noninduced conditions, this element is partially occupied by JAMYC in a transcriptionally inactive state.
Mutations of the protected G residues in the LAP promoter abolish the JA response
To determine whether these footprinted elements are required for JA induction, two progressive 5′-promoter deletions down to position -125 (-125LAP) and to position -78 (-78LAP) were obtained and introduced into transgenic potato plants. Both CGCGG sequence and GAGTA repeats are present in the -125LAP deletion, whereas these elements were removed in the -78LAP construct. Incubation with JA induced a substantial increase in the levels of GUS activity in the plants transformed with the -125LAP construct (Fig. 4B). In contrast, a complete loss in JA responsiveness was observed in the -78LAP transgenic lines. Contribution of the repeated GAGTA element to JA-induced expression was further analyzed by mutation of this duplicated motif in the context of the -317 to -3 LAP promoter–GUS fusion (-317LAPgagta construct; Fig. 4C). As shown in Figure 4B, mutation of the GAGTA element completely abolished JA response of the -317 promoter deletion. An intact GAGTA repeat is, therefore, required for JA response of the LAP promoter, indicating that this duplicated motif along with the JAMYC binding T/G-box element form a bifactorial JA-responsive (JARE) element that mediates JA-induced expression of the LAP genes.
The Arabidopsis AtMYC2 gene shares strong homology to JAMYC2 and JAMYC10 and is induced by JA treatment
To further assess the function of JAMYC2/JAMYC10 in regulated expression of the pin2 and LAP genes, transgenic lines were generated with down-regulated levels of expression of these transcripts by antisense inhibition. However, only a small reduction in the levels of expression of the JAMYC genes was observed in some antisense lines. As an alternative to the inefficient down-regulation results obtained in these transformants, we searched for gene orthologs in Arabidopsis, to identify loss-of-function mutations in these genes. Comparison of the JAMYC2 and JAMYC10 coding regions to the complete Arabidopsis sequence identified three genes with significant similarity (60% identity at the amino acid level) to the tomato regulatory proteins. Phylogenetic analysis revealed that gene RD22BP1 or AtMYC2 (Abe et al. 1997) encodes the closest homolog to the tomato JAMYC proteins (Fig. 5A). This gene had been reported to be induced by drought and ABA, and to play a role in transcriptional activation of the rd22 promoter through cooperative interaction with the MYB-related protein AtMYB2 (Urao et al. 1993; Abe et al. 1997). RNA profiling analysis of AtMYC2/AtMYB2 overexpressers shows a constitutive activation of several drought and osmotic stress response genes, including the rd22 target, but also increased levels of some JA- and pathogen-induced transcripts, like VSP2 (Abe et al. 2003).
Figure 5.
The Arabidopsis AtMYC2 gene shares strong homology with JAMYC2 and JAMYC10 and is rapidly induced by JA. (A) Phylogenetic tree generated by CLUSTALW alignment of the tomato JAMYC2 and JAMYC10 proteins and related bHLH proteins from Arabidopsis. (B) RNA blot analysis of the induction of AtMYC2 by MeJA. Total RNA was isolated from 3-week-old Col-0 plants grown in soil after different times of MeJA application, and 30 μg was loaded per line. Equal loading was verified by rRNA visualization after EtBr staining.
As a first approach to investigate whether AtMYC2 corresponds to a functional homolog of the JAMYC2/JAMYC10 factors, we investigated if expression of AtMYC2 is induced in response to JA treatment. JA application resulted in rapid activation of the AtMYC2 gene, with high levels of transcript observed as early as 30 min after JA treatment (Fig. 5B). AtMYC2 thus corresponds to an early JA-responsive gene, which agrees with a role of this transcription factor in JA-regulated expression.
Arabidopsis lines carrying an insertion within the AtMYC2 gene are insensitive to JA
A search for AtMYC2 insertion mutants in the T-DNA SALK collection (Alonso et al. 2003) retrieved mutations SALK_040500 (atmyc2-1) and SALK_083483 (atmyc2-2) with insertions at nucleotides 57 (Asn 18) and 1237 (Asp 411), respectively, within the protein coding region. RNA blot analysis of homozygous plants for these insertions revealed transcripts of either smaller or larger sizes, indicative of a loss of gene function (Fig. 6A). In Arabidopsis, JA induces strong inhibition of root growth (Staswick et al. 1992; Feys et al. 1994; Berger et al. 1996). Thus, we first investigated whether JA regulation was affected in these mutants by analyzing JA-induced root growth inhibition. Seeds of the atmyc2-1 and atmyc2-2 mutants or wild-type plants were germinated on JA plates, and roots were scored 2–3 wk later for growth. Normal growth and similar root lengths were observed for wild-type controls and atmyc2 mutants in plates without JA (Fig. 6B). Increasing concentrations of JA resulted in severe root growth inhibition in wild-type plants, but did not affect root growth in the atmyc2-1 and atmyc2-2 mutants (Fig. 6B). Only a small reduction in root length was observed in these insertion lines at high (100 μM) JA concentrations, these mutants therefore being insensitive to JA.
Figure 6.
Arabidopsis lines carrying an insertion within the AtMYC2 gene are insensitive to JA and exhibit altered JA-regulated gene expression. (A) Northern analysis of the atmyc2-1 (SALK_040500) and atmyc2-2 (SALK_083483) mutants. Fifteen micrograms of total RNA from Col-0 and atmyc2 mutant seedlings noninduced or treated for 8 h with 50 μM MeJA was loaded per lane and hybridized with the AtMYC2 probe. (B) MeJA inhibition of root growth in the atmyc2 mutants and Col-0 seedlings grown for 10 d on MS plates, or MS plates with 10, 50, and 100 μM MeJA. (C) MeJA induction of the VSP, JR1, PDF1.2, and b-CHI transcripts in Col-0 and the atmyc2 mutants. Fifteen micrograms of total RNA from noninduced seedlings (NI) or seedlings treated for 8 h with 50 μM MeJA (JA) was loaded per lane and hybridized with the indicated probes. (D) Complementation of the atmyc2-2 JA-insensitive root growth phenotype by overexpression of the JAMYC2 and JAMYC10 proteins (JAMYC/atmyc2-2 lines). Seeds were grown for 10 d on MS medium containing 50 μM MeJA. (E) Molecular complementation of the atmyc2-2 phenotype by expression of the Arabidopsis AtMYC2 gene or the tomato JAMYC factors. Fifteen micrograms of total RNA from seedlings treated for 8 h with 50 μM MeJA was loaded per lane and hybridized with the VSP and PDF1.2 probes.
Ratios of germination and growth retardation of the atmyc2-1 and atmyc2-2 mutants germinated on ABA plates were comparable to those of the controls (data not shown). Therefore, these lines apparently do not exhibit a reduced sensitivity to ABA like that reported for a Ds-insertion mutant in the AtMYC2 gene between residues 107 and 108 of the protein (Abe et al. 2003).
The AtMYC2 insertion lines exhibit altered JA-regulated gene expression
To further study the effects of these mutations on JA-regulated gene expression, we analyzed steady-state levels of the JA-regulated transcripts VSP and JR1 (Berger et al. 1995; Titarenko et al. 1997) and the JA/ethylene (ET)-regulated transcripts PDF1.2 and b-CHI (Penninckx et al. 1998; Lorenzo et al. 2003) in the atmyc2-1 and atmyc2-2 insertion lines. RNA blot analysis showed low levels of mRNA for these transcripts in untreated wild-type controls or the atmyc2 mutants (Fig. 6C). However, whereas JA application induced normal levels of accumulation of these genes in wild-type seedlings, induction of the JA/wound-responsive transcripts VSP and JR1 was strongly reduced in the atmyc2 mutants (reduction was more evident for the VSP transcript). JA treatment, in contrast, induced much greater levels of expression of the pathogen defense-related transcripts PDF1.2 and b-CHI in the atmyc2 mutants than in wild-type seedlings, suggesting a role of AtMYC2 in repression of these JA/ET-responsive genes. Together, these findings indicate a function of AtMYC2 in JA-induced gene expression, this transcription factor having a role in activation of the JA/wound-responsive genes VSP and JR1, but also in repression of the JA/ET-responsive genes PDF1.2 or b-CHI.
Expression of tomato JAMYC2 and JAMYC10 complements the JA-related phenotype of the AtMYC2 mutants
Evidence for a functional homolog activity of the tomato JAMYC and the Arabidopsis AtMYC2 transcription factors was further obtained by complementation of the atmyc2-2 mutation with the tomato proteins. Mutant plants expressing the JAMYC overexpression constructs were generated and examined for JA-induced inhibition of root growth. Overexpression of the tomato JAMYC transcription factors was able to recover the root growth phenotype of the atmyc2-2 mutation as a severe root growth inhibition was observed on JA plates in transgenic JAMYC/atmyc2-2 seedlings compared with the atmyc2-2 mutant controls (Fig. 6D). Such root growth differences were not observed in plates without JA (data not shown), thus evidencing JA mediation of this growth effect. JA-induced expression of the VSP and PDF1.2 genes was also restored in these transgenic lines (Fig. 6E). JA induction of the VSP transcript was increased to levels comparable to those of wild-type plants in the JAMYC overexpressers. JA-induced expression of the PDF1.2 gene was also sensibly reduced in these lines, to levels similar to those observed in transgenic atmyc2-2 mutants complemented with the endogenous Arabidopsis gene (see Fig. 6E). These results demonstrate that overexpression of the tomato JAMYC transcription factors complements the JA-insensitive phenotype of the atmyc2-2 mutation, these transcription factors therefore corresponding to the functional homologs of AtMYC2.
Discussion
Isolation of cDNA clones encoding the JAMYC factors
Using yeast one-hybrid screening, we have identified two transcription factors binding the cis-elements in the proximal promoter region of the tomato LAP genes. A -317 to -78 promoter region shown to be sufficient for JA-regulated gene expression was used as bait for screening, to yield clones JAMYC2 and JAMYC10 with >80% overall identity, encoding proteins with a basic-helix–loop–helix leucine zipper (bHLH-ZIP) DNA-recognition domain. JAMYC2 and JAMYC10 transcripts are rapidly induced in leaves by wounding and JA treatment, with a kinetics that precedes that of the LAP gene, supporting a function of these transcription factors in JA-induced LAP gene expression.
Hydroxyl radical in vitro footprinting studies identified a 7-bp AAACGTG motif as the binding site for the JAMYC factors. Notably, members of the bHLH-ZIP (MYC-like) family of transcription factors were found to bind preferentially to canonical G-box (CACGTG) elements, although they can also recognize T/G-box sequences like the AACGTG motif identified here (Blackwell et al. 1993). EMSA studies showed that a G-box element required for JA-induced expression of the pin2 gene (Kim et al. 1992) is able to compete efficiently for JAMYC binding to the AAACGTG motif. These proteins can bind a pin2 promoter fragment including this G-box motif, indicating that pin2 is a likely target for JAMYC-regulated transcriptional activation. JAMYC2, on the other hand, shares near-complete nucleotide sequence identity with LEJA3, a partial tomato cDNA clone identified in one-hybrid screening studies with the promoter region of the tomato threonine deaminase (Tda) gene (L. Broday and E. Lifschitz, pers. comm.). Notably, pin2, Tda, and LAP are coordinately induced in response to wounding and JA application (Hildmann et al. 1992; Samach et al. 1995; Ryan 2000). Consistent with this coordinated pattern of expression, in vitro studies showed that the JAMYC factors bind to these promoters and suggest a role for these proteins in JA-regulated transcriptional activation.
JAMYC-regulated gene expression
Mutation of the AAACGTG element in the -317 to -3 LAP promoter resulted in reduced JA activation of the -317LAPt/g-box construct. A residual level of activity was, however, observed in the transformants containing this construct, suggesting the presence of additional JA-responsive motifs in this promoter region. In line with this observation, overexpression of the JAMYC2 and JAMYC10 proteins did not lead to constitutive activation of the target pin2 or LAP genes, although enhanced expression of the pin2 and LAP transcripts was observed in the JAMYC-OE lines after treatment with JA. Therefore, it is possible that JA is required for activation of the JAMYC factors through phosphorylation or other types of posttranslational modification of these proteins or, alternatively, that JA induces expression of other rate-limiting transcription factors required for defense gene activation. Consistent with this latter hypothesis, animal and plant MYC-like proteins were often found to require interaction with partner proteins to activate target gene expression (Roth et al. 1991; Kretzner et al. 1992; Payne et al. 2000). Supporting evidence for interaction of additional regulatory proteins has, indeed, been obtained from in vivo footprinting analyses, where a GAGTA repeat situated directly adjacent to the T/G-box motif was found to be protected from methylation in induced leaves. Mutation of this duplicated sequence (-317LAPgagta construct) abolishes JA response of the -317 promoter deletion, indicating a requirement of this repeated element for JA-induced expression. No apparent sequence homology was observed between this duplicated motif and other JA-responsive cis-elements, with the exception of the potato CDI promoter (Ishikawa et al. 1994), which also has a duplicated GAGTA motif in a region required for JA-regulated expression of the gene.
Conservation of MYC-based JA signaling in Arabidopsis
Amino acid sequence comparisons showed elevated overall identity of the tomato JAMYC proteins with the Arabidopsis AtMYC2 gene, with a reported function in ABA-regulated responses to drought stress (Abe et al. 2003). In RNA blot analysis, we observed that JA induces a rapid accumulation of this transcript to even greater levels than those obtained in response to ABA application (data not shown), thus suggesting a role of this gene as functional homolog of the tomato JAMYC factors. Consistent with this observation, knockout mutations in this gene (atmyc2-1 and atmyc2-2 mutants) rendered plants insensitive to root growth inhibition by JA, indicating an impaired JA response in these plants. These insertion lines, on the other hand, were male fertile, which indicates that AtMYC2 is not required for normal stamen and pollen development.
JA-induced expression of VSP and JR1 is blocked in the atmyc2 lines, these mutants exhibiting in response to JA treatment much greater levels of activation of the PDF1.2 and b-CHI genes. These results are indicative of a function of AtMYC2 in transcriptional activation of JA-regulated genes but also in repression of JA/ET-regulated gene expression.
Expression of the tomato transcription factors rescued the root elongation phenotype of the Arabidopsis atmyc2 mutants and recovered JA-induced expression of both JA- and JA/ET-responsive genes, thus confirming that AtMYC2 and the JAMYC genes are functional homologs. Overexpression of the JAMYC or AtMYC2 factors did not lead to constitutive activation of the VSP gene, indicating that, as observed in tomato, activation of an as-yet-unidentified regulatory protein is also required for transcriptional activation of the JA-responsive genes.
Cross-talk regulation of the JA- and JA/ET-signaling pathways
In Arabidopsis, JA regulates defense responses against both herbivore (McConn et al. 1997) and necrotrophic pathogen attack (Pieterse et al. 1998; Thomma et al. 1999). Activation of these two responses involves interaction of both JA- and ethylene (ET)-signaling pathways. A synergistic interaction of these pathways is involved in activation of pathogen-related defense genes like PR5, PDF1.2, and b-CHI (Xu et al. 1994; Penninckx et al. 1998; Ellis and Turner 2001), whereas a negative interaction would repress expression in the directly damaged tissues of JA/wound-related genes like VSP or Thi2.1 (Berger et al. 1995; Vignutelli et al. 1998; Rojo et al. 1999).
JA/ET-dependent induction of pathogen-related genes is regulated by the ethylene response factor (ERF1) regulatory protein (Lorenzo et al. 2003). Constitutive expression of ERF1 rescues the defense response defects of the coi1 and ein2 mutants (Berrocal-Lobo et al. 2002) and leads to constitutive activation of JA/ET-regulated genes (Lorenzo et al. 2003). Interestingly, a down-regulated expression of the VSP and Thi2.1 genes was observed in RNA profiling analysis of ERF1 plants (Lorenzo et al. 2003), indicative of a negative regulatory function of ERF1 on JA-responsive gene expression.
In this work, we have shown that knockout mutations in the AtMYC2 gene have much reduced levels of expression of VSP and JR1 in response to JA treatment, but accumulate higher levels of the JA/ET-regulated transcripts PDF1.2 and b-CHI. A model of our current view of the mechanism of action of these regulatory proteins is shown in Figure 7. Herbivore or pathogen attack induces both systemic accumulation of JA and local synthesis of ET (Penninckx et al. 1998; Rojo et al. 1999). JA activates expression of VSP, JR1, and Thi2.1, which accumulate systemically in response to mechanical wounding or herbivore attack. Expression of these genes is repressed at the wound site by the local synthesis of ET. A positive interaction between JA and ET, in turn, is responsible for induction of PDF1.2 and b-CHI in response to necrotrophic pathogen attack. Activation of these genes is mediated by ERF1 (Lorenzo et al. 2003) and AtMYC2 (this work). ERF1 activates JA/ET-regulated gene expression and possibly plays a role in repression of genes differentially regulated by JA (Lorenzo et al. 2003). AtMYC2, in turn, is involved in JA-regulated gene expression and repression of the JA/ET pathway. Interaction between these two signaling pathways therefore occurs downstream in the signaling process, likely at the level of gene-specific promoters.
Figure 7.
Model for the proposed function of AtMYC2 and ERF1 in cross-talk regulation of the JA and JA/ET pathways in Arabidopsis. Herbivore and pathogenic attack promote a transient increase of both JA and ethylene. JA is involved in systemic activation of wound-responsive genes, and ethylene prevents local expression of these genes. A positive interaction between JA and ethylene is involved in activation of the JA/ET-pathway regulated genes in response to necrotrophic pathogen attack. Whereas ERF1 activates JA/ET-regulated gene expression (Lorenzo et al. 2003), AtMYC2 is involved in activation of JA-regulated genes and also in repression of JA/ET-induced gene expression. Arrows and bars indicate positive and negative interactions, respectively. A possible function of ERF1 in repression of the JA-regulated pathway is indicated by a question mark. (OGAs) Oligogalacturonides; (JA) jasmonic acid; (ET) ethylene.
ERF transcription factors bind a GCCGCC (GCC-box) element present in the promoter region of several PR genes (Fujimoto et al. 2000; Ohme-Takagi et al. 2000; Gu et al. 2002). Deletion analysis of the PDF1.2 promoter has recently identified a GCC-box element associated with JA/ET-responsive expression of this gene (Brown et al. 2003). This GCC-box element is directly followed by a G-box motif (-255 to -250), which might be a functional binding site for AtMYC2. Noteworthy, close proximity of G-box and GCC-box elements is a common feature of many tobacco PR genes (Sessa et al. 1995; Buttner and Singh 1997), binding of AtMYC2 to the G-box motif being likely to compete for binding of ERF1 to the GCC-box element. Hence, it is possible that competitive binding of these TFs mediates negative cross-talk regulation of the JA and JA/ET pathways.
A JA-responsive element in the AtVSP1 promoter has also been recently identified (Guerineau et al. 2003). This regulatory element is comprised of an inverted repeat containing a G-box-like element, located 150 bp upstream of the TATA-box. Mutation of the G-box-like element or the left part of the inverted repeat both have a strong effect on JA-regulated gene expression (Guerineau et al. 2003). GCC-box elements indicative of a direct binding of ERF1, to repress expression of this gene, are not observed in the AtVSP1 promoter, suggesting that negative cross-talk regulation by ERF1 might involve a different regulatory mechanism.
JA-pathway regulation in tomato
Important differences were observed in JA signaling between Arabidopsis and tomato. Arabidopsis mutants defective in JA synthesis or perception are male sterile (Feys et al. 1994), whereas tomato mutants were reported to be male fertile (Howe et al. 1996). Systemic induction of JA responses in tomato requires the 18-amino-acid peptide systemin (Ryan 2000), but evidence for a similar pathway in Arabidopsis has not been observed. Also, ethylene signaling is required for wound induction of pin2 in tomato (O'Donnell et al. 1996), whereas in Arabidopsis it seems to suppress JA-dependent gene expression in the wounded tissues (Rojo et al. 1999). Recent advances, however, suggest that these differences are mainly due to gaps in knowledge, rather than actual discrepancies in the signaling pathways in these two species. Grafting experiments using mutants deficient in either JA biosynthesis or JA perception, for example, have questioned the role of systemin as a systemic wound signal and pointed to JA as the long-distance moving signal (Li et al. 2002). Pollen examination of the tomato jasmonic acid insensitive-1 (jai1) mutant showed that it has reduced pollen viability and germination and that JA is also required for viable pollen formation in tomato (Li et al. 2004). Defects in this mutant are caused by a lesion in the LeCOI1 gene, encoding the tomato homolog of COI1 (Li et al. 2004). Like the Arabidopsis coi1 mutant, jai1 plants are insensitive to coronatine and highly resistant to coronatine-producing strains of Pseudomonas syringae (Zhao et al. 2003). Microarray analysis has shown that expression of all JA up-regulated genes is blocked in the jai-1 plants, thus demonstrating a similar function of COI1 in Arabidopsis and tomato.
The tomato ERF1-related factors Pti4, Pti5, and Pti6 were identified in a yeast two-hybrid screen with the Pto resistance gene (Zhou et al. 1997; Kim et al. 2002). A role of these tomato regulatory factors in plant defense activation was proved by expression in Arabidopsis. Expression of Pti4 causes the activation of several Arabidopsis pathogenesis-related genes, including PDF1.2, b-CHI, and PR4, and results in resistance to fungal pathogens and increased tolerance to the bacterial pathogen P. syringae (Gu et al. 2002; Wu et al. 2002), indicating that Pti4 might correspond to a tomato homolog of ERF1.
In this report, we have presented evidence for a functional homolog function of the tomato JAMYC and Arabidopsis AtMYC2 proteins. These factors have a main function in JA-regulated gene expression and cross-talk repression of the JA/ET-signaling pathway. Conservation of all these signaling components (COI1, Pti4/ERF1, and JAMYC/AtMYC2) thus suggests a similar regulatory network in these plants, although target genes are different in these two species (i.e., genes that encode PIs are not present in Arabidopsis). Noteworthy, wound/JA-regulated gene expression has been largely studied in tomato, whereas identification of the regulatory mechanisms underlying JA/ET-regulated gene activation derive mainly from studies in Arabidopsis. Therefore, further study of the complementary pathways will be required to confirm identical pathways for defense gene regulation in these species.
In conclusion, we have identified a conserved component of the JA-signaling pathway with a key function in JA-regulated expression and cross-talk inhibition of JA/ET signaling. Besides a function in JA-regulated transcriptional activation, JAMYC/AtMYC2 serve as cross-talk or integration points of both the JA- and JA/ET-signal outputs, concerted action between these MYC proteins and the ERF1/Pti4 playing a key role in orchestrating activation of different sets of defense-related genes in response to both herbivore or necrotrophic pathogen challenges.
Materials and methods
Plant materials and treatments
Tomato (Lycopersicum esculentum cv Moneymaker) and potato (Solanum tuberosum var Désirée) plants were grown in soil in the greenhouse under a 16 h light/8 h dark regime. For in vitro conditions, plants were grown in MS media supplemented with 2% sucrose. For Northern blot experiments, plants were grown in soil for 4–5 wk and sprayed with a 50 μM MeJA solution or wounded by midrib cutting of the leaves. Leaves (second to fourth from the apex) were collected at different times after treatment. JAMYC2 and JAMYC10 overexpressers were obtained by transformation of Solanum tuberosum var. Désirée with the 35S:JAMYC2 or 35S:JAMYC10 constructs in pBin19.
Arabidopsis thaliana (ecotype Columbia-0) and mutants SALK_040500 (atmyc2-1) and SALK_083483 (atmyc2-2) were grown in soil in the greenhouse under a 12 h light/12 h dark regime. For in vitro conditions, seeds were surface-sterilized and sown in MS supplemented with 1% sucrose. For Northern blot experiments, seedlings were grown on plates for 2 wk and incubated in a 50 μM MeJA solution for 8 h during the light period. For JA-induced root growth inhibition, seeds were sown on MS plates containing 0, 10, 50, or 100 μM MeJA, and cultured in a vertical position.
atmyc2-2/JAMYC2 and atmyc2-2/JAMYC10 plants were obtained by transformation of the atmyc2-2 homozygous mutants with the 35S:JAMYC2 or 35:JAMYC10 constructs.
Yeast one-hybrid screening
The -308/-78 wild-type EcoRI/HindII fragment of the LAP17.1 promoter was inserted into the EcoRI/filled XbaI site of the pHISi vector (-308LAP:HIS3) or the EcoRI/SmaI site of the pLacZi vector (-308LAP:LacZ). Plasmid -308LAP:LacZ was linearized with NcoI and transformed into the yeast strain YM4271 (Clontech). Recombinants were selected on SD-Ura medium and retransformed with the -308LAP:HIS3 plasmid linearized with XhoI to obtain the double reporter yeast strain -308LAP:HIS3/-308LAP:LacZ. Recombinants were selected on SD-His-Ura and single recombination events were verified by Southern blot. Poly(A)+ RNA was prepared from tomato leaves treated with 50 μM MeJA and used for cDNA synthesis using the Stratagene cDNA synthesis kit. This cDNA was directionally cloned into the EcoRI/XhoI sites of the HybriZap vector and packaged to obtain a primary library into this vector. This library was amplified and in vivo excised to a pAD-GAL4 plasmid vector as described by the manufacturer (Stratagene). The cDNA library was screened with the double reporter yeast strain in the presence of 30 mM 3-AT, as described by the manufacturers (Matchmaker One-Hybrid system; Clontech). A λ-Zap cDNA library obtained from leaves was screened with the partial cDNA clones C2 and C10 identified in the one-hybrid screening, to isolate the full-length clones JAMYC2 and JAMYC10.
Hydroxyl Radical Interference
Hydroxyl radical interference experiments were performed as described by Hayes and Tullius (1989). A -125LAP fragment, asymmetrically labelled with 32P-dCTP by fill-in with the Klenow polymerase fragment, was used as probe for JAMYC binding and subsequent hydroxyl reaction.
Electrophoretic mobility shift assays (EMSA)
The double-stranded oligonucleotides LAPT/G-BOX (5′-GGG AATGACATAGACGCGGTTAAAGAGTAGAGAATGAGTA AAACGTGTTGACATGC-3′; 5′-TTGCATGTCAACACGTTT TACTCATTCTCTACTCTTTAACCGCGTCTATGTCAC-3′), LAPt/gbox (5′-GGGAATGACATAGACGCGGTTAAAGAGTA GAGAATGAGTAAATCGGGTTGACATGC-3′; 5′-TTGCAT GTCAACCCGATTTACTCATTCTCTACTCTTTAACCGCG TCTATGTCATTC-3′), PIN2G-BOX (5′-GGGAGTTCCAACT TAATTATCACGTGGACTTATAAGAAACCGA-3′; 5′-GGT CGGTTTCTTATAAGTCCACGTGATAATTAAGTTGGAA CTC-3′), and PIN2g-box (5′-GGGAGTTCCAACTTAATTATC TCGGGGACTTATAAGAAACCGA-3′; 5′-GGTCGGTTTCT TATAAGTCCCCGAGATAATTAAGTTGGAACTC-3′) were used as probes and competitors. The double-stranded oligonucleotides were end-labeled with α-32P-dATP/dCTP by fill-in with the Klenow polymerase fragment, and purified on NAP5 columns (Pharmacia) according to the manufacturer's instructions.
The EcoRI/SalI fragment corresponding to C2 was cloned into the pET28a vector (Novagen) and transformed into Escherichia coli BL21 cells. Production and purification of the His-Tag fusion protein (His-C2) was performed according to the manufacturer's instructions. The radioactive probe was incubated with 300 ng of the purified protein in 20 μL of 1× binding buffer (20 mM HEPES at pH 7.8, 40 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM DTT, 1 μg/μL BSA, 0.01% Triton X-100, 10% glycerol) and 100 ng poly(dI-dC), in the presence or absence of nonradiolabeled competitor for 30 min on ice, before loading on a 1× TBE, 5% polyacrylamide gel.
DNA constructs and plant transformation
The JAMYC2 and JAMYC10 coding regions were amplified by PCR using synthetic primers (J2D: 5′-GTGTTTATGGAATG AC-3′; J2R: 5′-GACGATTTCTATCTAC-3′ for JAMYC2 and J10D: 5′-GATTGAATGACGGAC-3′; J10R: 5′-GATAATTTCA TCGCG-3′ for JAMYC10) and cloned into the SmaI site of pUC18. Constructs 35S:JAMYC2 and 35S:JAMYC10 were obtained by excising the XbaI/SacI JAMYC2 and JAMYC10 fragments from pUC18 and cloning into the pBin19–35S:nosT vector. Constructs -317LAPt/g-box:GUS and -317LAPgagta:GUS were obtained by inverse PCR reactions on the -317LAP–pGUS plasmid (Ruíz-Rivero and Prat 1998), using synthetic primers with a 10-bp sequence including a BamHI site in their 5′-ends that replaces the original T/G-box site (LS8A: 5′-CTGGATCC CGCATGCAAATATCTTGTTC-3′; LS8B: 5′-CGGGATCCAG TTACTCATTCTCTACTC-3′) for the first construct, or using the oligonucleotides FPLA: 5′-CAATCATCAAAACGTGTTG ACATGC-3′ and FPLB: 5′-TATGAACTTTAATTGCGTCTAT GTCATTCA-3′ for the second one. Constructs -317LAP:GUS, -317LAPt/g-box:GUS, and -317LAPgagta:GUS were obtained by EcoRI/HindIII digestion of the corresponding pGUS plasmid constructs and cloning into the pBin19 vector.
Transformation of S. tuberosum var Désirée plants was performed as previously described (Carrera et al. 2000). Transformation of A. thaliana Col-0 or the atmyc2-2 mutant was performed as described by Bechtold and Pelletier (1998).
Transient expression assays
Tobacco BY2 cells freshly subcultured for 5 d were spread on filter paper 1 d before bombardment and incubated on NT medium (5 g/L Murashige and Skoog salts, 1 mg/L thiamine, 200 mg/L KH2PO4, 0.2 mg/L 2,4-D, 100 mg/L myo-inositol, 30 g/L sucrose, 2 g/L gelrite) overnight at 26°C in the dark. Four hours before bombardment, cells were moved to NT medium with 200 mM mannitol. DNA absorption to gold particles and bombardment using a helium-driven particle accelerator (PDS-1000; Bio-Rad) was performed according to the manufacturer's recommendations. Cells were transformed with 1 μg of the pUC18 -317LAP:GUS or -317LAPt/g-box:GUS reporter plasmids, 1 μg of the pUC18 35S:LUC plasmid as internal standard, and up to 2 μg of either the pUC18 35S:JAMYC2 or 35S:JAMYC10 constructs or the 35S:nosT vector without insert (control). After bombardment, cells were moved to fresh NT medium and incubated for 22 h at 26°C in the dark, before freezing in liquid nitrogen.
For activity assays, samples were thawed and homogenized on ice in a buffer containing 25 mM Tris (pH 7.8), 2 mM CDTA, 2 mM DTT, 10% glycerol, and 1% Triton X-100, and cleared by centrifugation at 12,000g for 5 min. For luciferase (LUC) and GUS assays, 60–100 μg of protein extract was used. LUC activity was determined using the Luciferase Assay System kit (Promega), according to the manufacturer's instructions. GUS activity was measured by fluorometric assay as described by Jefferson (1987).
Analysis of gene expression
Total RNA was prepared as described by Logemann et al. (1987). Hybridizations were performed at 42°C with washes at 65°C as described by Amasino (1986). 3′-Noncoding ends corresponding to C2 and C10 were used as probes for hybridizations in tomato. For analysis of the potato transgenic lines, total cDNAs corresponding to JAMYC2, JAMYC10, LAP, and pin2 were used as probes. Total cDNAs corresponding to AtMYC2, VSP, JR1, PDF1.2, and b-CHI were used as probes for Northern blot analysis in Arabidopsis. GUS activity of leaf protein extracts from the -317LAP:GUS, -317LAPt/gbox:GUS, and -317LAPgagta:GUS transgenic plants was measured by fluorometric assay as previously described (Jefferson 1987).
LmPCR in vivo footprinting
In vivo footprinting was performed from leaves of Lycopersicon esculentum cv Moneymaker untreated or treated with MeJA, and directly incubated with dimethyl sulphate (DMS). DMS treatment and DNA purification and cleavage at modified guanines were performed as described previously (Busk et al. 1997).
Ligation-mediated polymerase chain reaction (LmPCR) conditions were as previously described (Busk et al. 1997). Oligonucleotides used for LmPCR amplification of the sense strand were: Lap12: 5′-AGAATTGAAGTGTCTAAGCGAAATTT CG-3′; Lap13: 5′-GCGAAATTTCGTGCCAACTTTAGGTGG-3′; and Lap14: 5′-CGTGCCAACTTTAGGTGGTTACCGAT AGTTAGACC-3′; and those for the antisense strand: Lap16: 5′-CTGAAGGGTTAGAATGCAAAGATG-3′; Lap17: 5′-GAT GAAGAAGAAGCAAACAATGAAGAAACTC-3′; and Lap18: 5′-GAAGAAGAAGCAAACAATGAAGAAACTCTTAGTGT TGCCATTG-3′.
Acknowledgments
We thank Javier Paz-Ares, José Sánchez-Serrano, and David Hannapel for critical reading of the manuscript. We also thank Pilar Fontanet and Carme Badía for excellent plant care. This research was supported by grants BIO99-0961 and BIO2002-02045 from the Spanish MCyT and by predoctoral grants from the Catalan CERBA and Spanish AECI, respectively, to M.B. and A.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.297704.
References
- Abe H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. 1997. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Amasino R.M. 1986. Acceleration of nucleic acid hybridization rate by polyethyleneglycol. Anal. Biochem. 152: 304-307. [DOI] [PubMed] [Google Scholar]
- Bechtold N. and Pelletier, G. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259-266. [DOI] [PubMed] [Google Scholar]
- Berger S., Bell, E., Sadka, A., and Mullet, J.E. 1995. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 27: 933-942. [DOI] [PubMed] [Google Scholar]
- Berger S., Bell, E., and Mullet, J.E. 1996. Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111: 525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina, A., and Solano, R. 2002. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23-32. [DOI] [PubMed] [Google Scholar]
- Blackwell T.K., Huang, J., Ma, A., Kretzner, L., Alt, F.W., Eisenman, R.N., and Weintraub, H. 1993. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell Biol. 13: 5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann H., Vignutelli, A., Hilpert, B., Miersch, O., Wasternack, C., and Apel, K. 1998. Wounding and chemicals induce expression of the Arabidopsis thaliana gene Thi2.1, encoding a fungal defense thionin, via the octadecanoid pathway. FEBS Lett. 437: 281-286. [DOI] [PubMed] [Google Scholar]
- Brown R.L., Kazan, K., McGrath, K.C., Maclean, D.J., and Manners, J.M. 2003. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132: 1020-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk P.K., Jensen, A.B., and Pagés, M. 1997. Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J. 11: 1285-1295. [DOI] [PubMed] [Google Scholar]
- Buttner M. and Singh, K.B. 1997. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. 94: 5961-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E., Bou, J., García-Martinez, J.L., and Prat, S. 2000. Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J. 22: 247-256. [DOI] [PubMed] [Google Scholar]
- Ellis C. and Turner, J.G. 2001. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton, P.J., Robatzek, S., and Somssich, I.E. 2000. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199-206. [DOI] [PubMed] [Google Scholar]
- Feys B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. 1994. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate and resistant to a bacterial pathogen. Plant Cell 6: 751-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. 2000. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y.Q., Wildermuth, M.C., Chakravarthy, S., Loh, Y.T., Yang, C., He, X., Han, Y., and Martin, G.B. 2002. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14: 817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau F., Benjdia, M., and Zhou, D.X. 2003. A jasmonate-responsive element within the A. thaliana vsp1 promoter. J. Exp. Bot. 54: 1153-1162. [DOI] [PubMed] [Google Scholar]
- Hayes J.J. and Tullius, T.D. 1989. The missing nucleoside experiment: A new technique to study recognition of DNA by protein. Biochemistry 28: 9521-9527. [DOI] [PubMed] [Google Scholar]
- Hildmann T., Ebneth, M., Peña-Cortés, H., Sánchez-Serrano, J.J, Willmitzer, L., and Prat, S. 1992. General roles of abscisic and jasmonic acid in gene activation as a result of mechanical wounding. Plant Cell 4: 1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A. and Ryan, C.A. 1999. Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153: 1411-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A. and Schilmiller, A.L. 2002. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 5: 230-236. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Lightner, J., Browse, J., and Ryan CA. 1996. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defence against insect attack. Plant Cell 8: 2067-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A., Yoshihara, T., and Nakamura K. 1994. Jasmonate-inducible expression of a potato cathepsin D inhibitor-GUS gene fusion in tobacco cells. Plant Mol. Biol. 26: 403-414. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. 1987. Assaying chimeric genes in plants: The GUS–gene fusion system. Plant Mol. Biol. Rep. 5: 387-405. [Google Scholar]
- Jensen A.B., Raventós, D., and Mundy, J. 2002. Fusion genetic analysis of jasmonate-signalling mutants in Arabidopsis. Plant J. 29: 595-606. [DOI] [PubMed] [Google Scholar]
- Kawagoe Y. and Murai, N. 1996. A novel basic region/helix-loop-helix protein binds to the G-box motif of the bean β-phaseolin gene. Plant Sci. 116: 47-57. [Google Scholar]
- Kim S.R., Choi, J.L., Costa, M.A., and An, G. 1992. Identification of G-box sequence as an essential element for Methyl Jasmonate response of potato Proteinase inhibitor II promoter. Plant Physiol. 99: 627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Lin, N.C., and Martin, G.B. 2002. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589-598. [DOI] [PubMed] [Google Scholar]
- Kretzner L., Blackwood, E.M., and Eisenman, R.N. 1992. Myc and Max proteins possess distinct transcriptional activities. Nature 359: 426-429. [DOI] [PubMed] [Google Scholar]
- Lee G.I. and Howe, G.A. 2003. The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33: 567-576. [DOI] [PubMed] [Google Scholar]
- León J., Rojo, E., and Sánchez-Serrano, J.J. 2001. Wound signalling in plants. J. Exp. Bot. 52: 1-9. [DOI] [PubMed] [Google Scholar]
- Li L., Li, C.Y., and Howe, G.A. 2001. Genetic analysis of wound signaling in tomato. Evidence for a dual role of jasmonic acid in defense and female fertility. Plant Physiol. 127: 1414-1417. [PMC free article] [PubMed] [Google Scholar]
- Li L., Li, C., Lee, G.I., and Howe, G.A. 2002. Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. 99: 6416-6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao, Y., McCaig, B.C., Wingerd, B.A., Wang, J., Whalon, M.E., Pichersky, E., and Howe, G.A. 2004. The tomato homolog of CORONATINEINSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J., Schell, J., and Willmitzer, L. 1987. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163: 16-20. [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason H.S., De Wald, D.B., and Mullet, J.E. 1993. Identification of a methyl jasmonate-responsive domain in the soybean vspB promoter. Plant Cell 5: 241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. 1997. Jasmonate is essential for insect defence in Arabidopsis. Proc. Natl. Acad. Sci. 94: 5473-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke F.L., Champion, A., Kijne, J.W., and Memelink, J. 1999. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18: 4455-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. 1996. Ethylene as a signal mediating the wound response of tomato plants. Science 274: 1914-1917. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M., Suzuki, K., and Shinshi, H. 2000. Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 41: 1187-1192. [DOI] [PubMed] [Google Scholar]
- Pautot V., Holzer, F.M., Reisch, B., and Walling, L.L. 1993. Leucine aminopeptidase: An inducible component of the defense response in Lycopersicon esculentum (tomato). Proc. Natl. Acad. Sci. 90: 9906-9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang, F., and Lloyd, A.M. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.P., and Broekaert, W.F. 1998. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M., van Wees, S.C., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and van Loon, L.C. 1998. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., León, J., and Sánchez-Serrano, J.J. 1999. Cross-talk between wound-signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20: 135-142. [DOI] [PubMed] [Google Scholar]
- Roth B.A., Goff, S.A., Klein, T.M., and Fromm, M.E. 1991. C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell 3: 317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouster J., Leah, R., Mundy, J., and Cameron-Mills, V. 1997. Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 11: 513-523. [DOI] [PubMed] [Google Scholar]
- Ruíz-Rivero O.J. and Prat, S. 1998. A -308 deletion of the tomato LAP promoters is able to direct flower-specific and MeJA-induced expression in transgenic plants. Plant Mol. Biol. 36: 639-648. [DOI] [PubMed] [Google Scholar]
- Ryan C.A. 2000. The systemin signalling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477: 112-121. [DOI] [PubMed] [Google Scholar]
- Samach A., Broday, L., Hareven, D., and Lifschitz, E. 1995. Expression of an amino acid biosynthesis gene in tomato flowers: Developmental upregulation and MeJA response are parenchyma-specific and mutually compatible. Plant J. 8: 391-406. [DOI] [PubMed] [Google Scholar]
- Schaller F. 2001. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J. Exp. Bot. 52: 11-23. [PubMed] [Google Scholar]
- Scheer J.M. and Ryan Jr., C.A. 2002. The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. 99: 9585-9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Meller, Y., and Fluhr, R. 1995. A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol. Biol. 28: 145-153. [DOI] [PubMed] [Google Scholar]
- Solano R., Stepanova, A., Chao, Q., and Ecker, J.R. 1998. Nuclear events in ethylene signaling: A transcriptional cascade mediated ETHYLENEINSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Dev. 12: 3703-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su, W.P., and Howell, S.H. 1992. Methyl jasmonate inhibition of root-growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. 89: 6837-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Yuen, G.Y., and Lehman, C.C. 1998. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15: 747-754. [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki, I., and Rowe, M.L. 2002. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. 1999. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121: 1093-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E., Rojo, E., León, J., and Sánchez-Serrano, J.J. 1997. JA-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 115: 817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq, E., and Quail, P.H. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.G., Ellis, C., and Devoto, A. 2002. The jasmonate signal pathway. Plant Cell 14: S153-S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. 1993. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L. and Memelink, J. 2000. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295-297. [DOI] [PubMed] [Google Scholar]
- ____. 2001. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25: 43-53. [DOI] [PubMed] [Google Scholar]
- Vignutelli A., Wasternack, C., Apel, K., and Bohlmann, H. 1998. Systemic and local induction of an Arabidopsis thionin gene by wounding and pathogens. Plant J. 14: 285-295. [DOI] [PubMed] [Google Scholar]
- Wu K., Tian, L., Hollingworth, J., Brown, D.C., and Miki, B. 2002. Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128: 30-37. [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Miao, Z.H., and Lam, E. 1996. Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol. Biol. 32: 415-426. [DOI] [PubMed] [Google Scholar]
- Xu Y., Chang, P.F.L., Liu, D., Narasimhan, M.L., Raghptanma, K.G., Gasegawa, P.M., and Bressan, R.A. 1994. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6: 1077-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Thilmony, R., Bender, C.L., Schaller, A., He, S.Y., and Howe, G.A. 2003. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36: 485-499. [DOI] [PubMed] [Google Scholar]
- Zhou J., Tang, X., and Martin, G.B. 1997. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16: 3207-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]