Abstract
The core promoter is the ultimate target of the vast network of regulatory factors that contribute to the initiation of transcription by RNA polymerase II. Here we describe the MTE (motif ten element), a new core promoter element that appears to be conserved from Drosophila to humans. The MTE promotes transcription by RNA polymerase II when it is located precisely at positions +18 to +27 relative to A+1 in the initiator (Inr) element. MTE sequences from +18 to +22 relative to A+1 are important for basal transcription, and a region from +18 to +27 is sufficient to confer MTE activity to heterologous core promoters. The MTE requires the Inr, but functions independently of the TATA-box and DPE. Notably, the loss of transcriptional activity upon mutation of a TATA-box or DPE can be compensated by the addition of an MTE. In addition, the MTE exhibits strong synergism with the TATA-box as well as the DPE. These findings indicate that the MTE is a novel downstream core promoter element that is important for transcription by RNA polymerase II.
Keywords: RNA polymerase II, core promoter, DPE, Inr, TATA-box
Transcription is a critical control point in the regulation of gene expression. In eukaryotes, the transcription of protein-coding genes by RNA polymerase II is mediated by a complex network of factors that include sequence-specific DNA-binding proteins, transcriptional coregulators, chromatin-remodeling factors, enzymes that covalently modify histones and other proteins, and the basal transcriptional machinery (for reviews, see Struhl 1999; Hirose and Manley 2000; Lee and Young 2000; Myers and Kornberg 2000; Näär et al. 2001; Brivanlou and Darnell 2002; Maniatis and Reed 2002; Orphanides and Reinberg 2002; Levine and Tjian 2003; Kadonaga 2004).
A significant proportion of the regulatory information that specifies the transcriptional program of each gene is encoded in the DNA sequence, such as in promoters and enhancers. However, the ultimate target sequence of the vast array of factors that control the initiation of transcription is the core promoter (for reviews, see Smale 1997, 2001; Butler and Kadonaga 2002; Kadonaga 2002; Hochheimer and Tjian 2003; Smale and Kadonaga 2003). The core promoter encompasses the RNA start site and directs the accurate initiation of transcription. Core promoters are typically ∼50 nt in length, and consist of functional subregions termed core promoter elements. These core promoter elements are not universally present in all core promoters. Rather, core promoter elements confer the specific properties of each core promoter, such as the interactions of core promoters with enhancer elements (for reviews, see Smale 2001; Butler and Kadonaga 2002; Smale and Kadonaga 2003). Therefore, in the analysis of the regulation of a gene, it is essential to identify the particular core promoter elements that direct the initiation process.
Some of the known core promoter elements are the TATA-box, initiator (Inr), transcription factor IIB (TFIIB) recognition element (BRE), and the downstream core promoter element (DPE). The TATA-box (Goldberg 1979) is located ∼26–31 nt upstream of the start site and has a consensus sequence of TATAWAAR. (Degenerate nucleotides are designated according to the IUPAC code; http://www.chem.qmul.ac.uk/iubmb/misc/naseq.html.) The TATA-box is a recognition site for the binding of the TATA-box-binding protein (TBP) subunit of the transcription factor IID (TFIID) complex. The BRE, which interacts with TFIIB, has a consensus of SSRCGCC and is located immediately upstream of ∼12% of TATA-boxes (Lagrange et al. 1998). The Inr (Smale and Baltimore 1989) encompasses the transcription start site and has a consensus sequence of YYANWYY in humans and TCAKTY in Drosophila, where the A nucleotide is typically designated as the +1 position of the core promoter. The Inr is recognized by the TAF1 (TAFII250) and TAF2 (TAFII150) subunits of the TFIID complex (e.g., see Kaufmann and Smale 1994; Purnell et al. 1994; Verrijzer et al. 1994; Chalkley and Verrijzer 1999) as well as several other factors (discussed in Smale and Kadonaga 2003). The DPE (Burke and Kadonaga 1996) is located from +28 to +32 (+33) and has a consensus of RGWYV(T). TFIID binds cooperatively to the Inr and DPE motifs (Burke and Kadonaga 1996). Photocross-linking analyses indicated that the TAF6 (TAFII60) and TAF9 (TAFII40) subunits of Drosophila TFIID are in close proximity to the DPE (Burke and Kadonaga 1997). In addition, studies of TAF9 in vivo indicated that it associates preferentially with the DPE-containing human IRF-1 promoter relative to a DPE-mutant version of the promoter (Chen and Manley 2003).
It is likely that there are many core promoter elements that remain to be discovered. For instance, a statistical analysis of 205 Drosophila core promoters suggested that ∼31% of the core promoters contained neither TATA-box nor DPE motifs (Kutach and Kadonaga 2000). More recently, a potential core promoter element was identified as an overrepresented sequence motif in a computational analysis of nearly 2000 Drosophila core promoters (Ohler et al. 2002). This sequence, motif 10, is located ∼20–30 nt downstream of the transcription start site (Ohler et al. 2002). In this work, we sought to determine whether the motif 10 sequence affects transcription as well as if it is related to the DPE. These studies have revealed that the motif 10 sequence contains a new core promoter element termed the MTE, for motif ten element. The MTE can promote transcription by RNA polymerase II in conjunction with the Inr but independently of the TATA-box or DPE.
Results
The motif 10 sequence is located at positions +18 to +29 in the core promoter
A computational analysis of core promoter sequences in Drosophila (Ohler et al. 2002) identified four overrepresented sequence motifs that are located at a distinct position relative to the transcription start site. These four motifs are the TATA-box (“motif 3”), Inr (“motif 4”), DPE (“motif 9”), and a new sequence termed motif 10. Motif 10 was found to be located downstream from the transcription start site in the vicinity of +20 to +30.
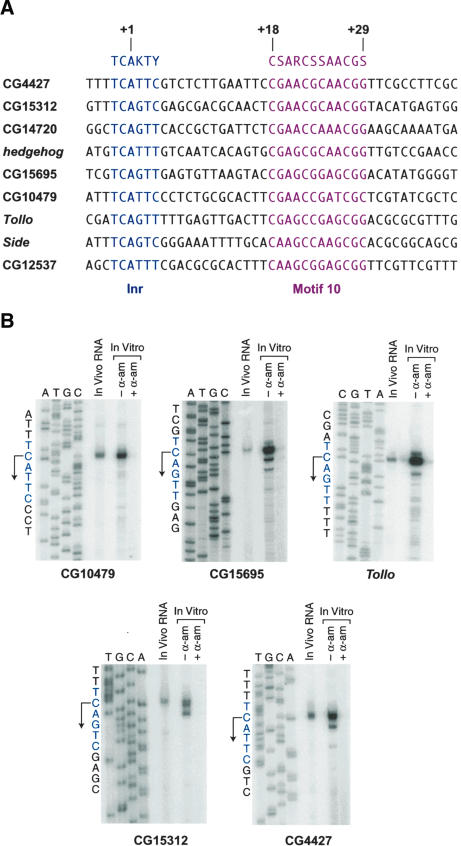
Based on these findings, we sought to determine whether motif 10 is involved in transcription by RNA polymerase II. Given the downstream location of motif 10, it also seemed possible that it could be a variant of the DPE. To investigate the function of motif 10, we initially examined some motif 10-containing core promoter sequences that were identified in Ohler et al. (2002). This analysis suggested that the motif 10 consensus sequence (CSARCSSAACGS) is located from +18 to +29 relative to the A+1 position in the Inr. Then, by using the JDSA search program (http://www-biology.ucsd.edu/labs/kadonaga/JDSA.html), we surveyed the Drosophila genome database for sequences (which are upstream of an open reading frame) that contain a motif 10 consensus from +18 to +29 relative to A+1 of a consensus Inr sequence. Some of these putative motif 10-containing core promoters are shown in Figure 1A. None of these sequences appears to contain a TATA-box in the -20 to -30 region upstream of A+1.
Figure 1.
Identification of core promoters that contain a motif 10 sequence. (A) Putative core promoters that contain a motif 10 consensus sequence were identified in the Drosophila genome database by using the JDSA search program. In these putative core promoters, the motif 10 consensus (in red) is located from +18 to +29 relative to the A+1 position in the initiator (Inr) consensus sequence (in blue). (B) Identification of five motif 10-containing core promoters. The in vivo start sites were mapped by primer extension analysis with poly(A)+ RNA (15 μg) from Drosophila embryos that were collected from 0 to 12 h after egg deposition. The in vitro start sites were mapped with RNA that was synthesized in vitro with a nuclear extract derived from Drosophila embryos. Where indicated, α-amanitin (α-am; 4 μg/mL) was included in the in vitro transcription reactions. For each promoter, the primer extension products were subjected to denaturing polyacrylamide gel electrophoresis in parallel with DNA sequencing ladders that were generated from the same primers that were used in the primer extension analyses of the RNA. Transcription from each of these promoters appears to start from the C nucleotide that is immediately upstream of the “A+1” position of the Inr consensus. For consistency of nomenclature, however, we will continue to refer to “A+1” as the “+1” position of these promoters.
To determine whether these motif 10-containing sequences function as core promoters in vivo, we carried out primer extension analyses of poly(A)+ RNA that was isolated from Drosophila embryos. We found that the nine sequences listed in Figure 1A are indeed the sites of transcription initiation in vivo (Fig. 1B; data not shown). In addition, in vitro transcription reactions revealed an excellent correlation between the in vivo and in vitro start sites (Fig. 1B). Transcription from these motif 10-containing promoters appears to initiate at the “C-1” position of the Inr consensus sequence. For consistency of nomenclature, however, we will continue to refer to “A+1” as the “+1” position. We also observed that transcription in vitro is inhibited by 4 μg/mL α-amanitin (α-am; Fig. 1B), which indicates that the transcription is mediated by RNA polymerase II.
Motif 10 contains a promoter element termed the MTE
To test whether motif 10 contains a promoter element, we initially carried out a scanning mutational analysis of the downstream region of two motif 10-containing promoters, CG4427 and CG15312. In these experiments, we generated a series of triple substitution mutations that spanned from positions +11 to +37, and then subjected the wild-type and mutant promoters to in vitro transcription analysis. With the CG4427 promoter, mutation of +17–19 as well as +29–31 resulted in a strong decrease in transcription, whereas mutations within the +20–27 region of motif 10 had little or no effect on transcription levels (Fig. 2). Similarly, we found that mutations in the +17–19 and +29–31 positions in the CG15312 promoter led to a reduction in transcriptional activity, whereas mutations in the +20–27 region did not result in a significant loss of activity (data not shown). These results suggest that the +17–19 and +29–31 regions are important for transcription from these two motif 10-containing core promoters.
Figure 2.
Scanning mutational analysis of the motif 10 sequence. A series of mutant CG4427 core promoters was constructed in which triple nucleotide substitutions were introduced in the downstream promoter region that encompasses the motif 10 sequence and the DPE. Outside the motif 10 sequence, A, T, and G nucleotides were mutated to C, and C nucleotides were mutated to A. Within the motif 10 sequence, the substitution mutations were designed to minimize the similarity of the sequence to the motif 10 consensus. The wild-type and mutant promoters were subjected to in vitro transcription analysis with a Drosophila nuclear extract. The transcriptional activity of each mutant promoter is reported relative to that of the wild-type promoter.
It is important to note, however, that the motif 10 sequence (+18 to +29) overlaps with the DPE consensus (+28 to +33). Hence, the sensitivity of the +29–31 mutants in the CG4427 and CG15312 promoters could be due to the presence of DPE activity in those promoters. Thus, it was important to determine whether there are distinct functions of the motif 10 sequence and the DPE. To this end, we sought to create mutations that specifically inactivate the DPE but not a motif 10-specific promoter activity, and vice versa.
We therefore used our knowledge of the DPE consensus (Kutach and Kadonaga 2000; A. Kutach, S. Iyama, and J.T. Kadonaga, unpubl.) to design a mutation that inactivates the DPE but does not alter the region of the motif 10 consensus sequence (+18 through +29). This analysis indicated that the CATA nucleotides are disfavored and/or underrepresented at positions +30 to +33 of DPE-dependent core promoters. We therefore introduced the CATA mutation at +30–33 (m30–33) in the DPE-containing core promoters of the E74B gene and Doc retrotransposon. In vitro transcription analysis revealed that the m30–33 mutation essentially abolishes transcription from these DPE-containing core promoters (Fig. 3A).
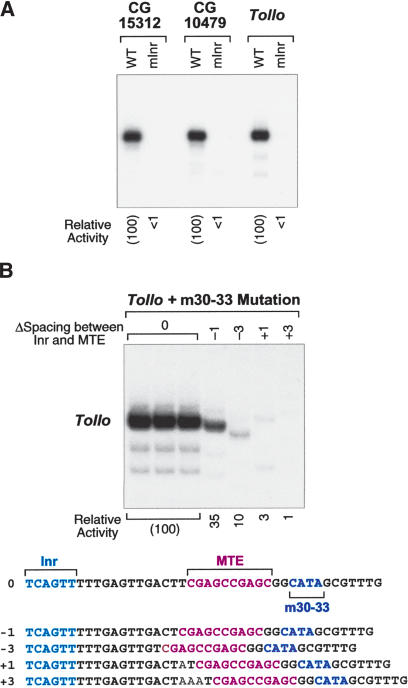
Figure 3.
Nonoverlapping mutations in the motif 10 sequence and the DPE. (A) The +30–33 DPE mutation inactivates DPE-dependent promoters. The +30 to +33 region of the E74B and Doc core promoters was mutated to CATA. The resulting m30–33 promoters were subjected to in vitro transcription analysis in parallel with the corresponding wild-type promoters. (B) Analysis of mutations in the motif 10 sequence (+19 to +29) that do not overlap with the DPE (+28 to +33). A series of progressive substitution mutant versions of the Tollo core promoter was constructed, as depicted at the bottom of the figure. The substitution mutations were chosen to minimize the similarity of the sequences to the motif 10 consensus. The wild-type and mutant promoters were subjected to in vitro transcription analysis with a Drosophila nuclear extract.
Next, we sought to create a mutation that disrupts a motif 10-specific promoter activity but does not overlap with the DPE consensus. Based on the results of the scanning mutational analysis (Fig. 2), we knew that the +17 to +19 region is important for basal transcription from two different motif 10-containing promoters. We therefore generated and analyzed a series of nested substitution mutations in the 5′-region of the motif 10 sequence in the Tollo core promoter. These experiments indicated that mutation of the 5′-region of the Tollo motif 10 sequence results in a substantial reduction in transcription (Fig. 3B).
These findings, combined with those described below, revealed that the 5′-region of the motif 10 sequence contains a promoter activity that does not overlap with the DPE consensus. We will henceforth refer to this core promoter element in the motif 10 sequence as the MTE, for motif ten element. For the analysis of the contribution of the MTE to promoter activity, we chose to use the +18 to +22 substitution mutation (m18–22), because it causes about a sevenfold reduction in transcription and is several nucleotides upstream of the DPE consensus sequence.
To test the contributions of the MTE and the DPE to transcription from MTE-containing promoters, we constructed and analyzed promoters that contain m18–22 (mutant MTE) only, m30–33 (mutant DPE) only, or both m18–22 and m30–33 mutations (Fig. 4A). We performed these analyses with all nine of the MTE-containing promoters shown in Figure 1A, and present the results from three representative promoters in Figure 4B. These studies revealed that both the MTE and the DPE contribute to transcription from the MTE-containing promoters that we have tested. In some promoters, such as Tollo, the MTE appears to be stronger than the DPE, whereas in other promoters, such as CG15695, the DPE appears to be stronger than the MTE. These results suggest that the MTE-containing promoters shown in Figure 4B contain both MTE and DPE elements. In the absence of the DPE, transcription is partially supported by the MTE, and vice versa. Then, in the absence of both the MTE and DPE, transcription is essentially lost.
Figure 4.
The motif ten element, MTE, supports transcription in the absence of the DPE. (A) Diagram of the MTE (m18–22) and DPE (m30–33) mutations. The MTE is depicted from +18 to +27, because this segment of the motif 10 consensus is sufficient to confer MTE activity (see below). (B) The MTE and DPE motifs both contribute to transcription from the Tollo, CG10479, and CG15695 core promoters. The wild-type, m18–22, m30–33, and m18–22/30–33 versions of each core promoter were subjected to in vitro transcription analysis with a Drosophila nuclear extract. (C) The E74B and Doc core promoters lack a functional MTE that can support transcription upon mutation of the DPE. Wild-type and mutant promoters were analyzed, as in B.
For comparison, we performed the same mutational analysis with two DPE-dependent core promoters (Fig. 4C). Consistent with the results shown in Figure 3A, the m30–33 DPE mutation causes the nearly complete loss of transcription from the E74B and Doc promoters. In contrast, the introduction of the m18–22 MTE mutation results in a modest two- to threefold reduction in transcription with these promoters. The strong transcription with the mutant MTE (m18–22) promoters reveals that the E74B and Doc promoters possess strong DPE activity that can function in the absence of an MTE. In contrast, the nearly complete loss of transcription in the mutant DPE (m30–33) promoters indicates that the E74B and Doc promoters lack MTE activity that can support transcription in the absence of a DPE. Thus, the E74B and Doc promoters appear to contain a DPE but not a functionally independent MTE.
Thus, we have identified several promoters in which both the MTE and DPE are important for transcription as well as two promoters, E74B and Doc, in which the DPE is the predominant downstream promoter element. We have not yet identified promoters that appear to be completely dependent on the MTE, although the activity of the m30–33 mutant DPE promoters in Figure 4B suggests that such promoters could theoretically exist.
Lastly, it is useful to indicate that the m18–22 and m30–33 mutations effectively eliminate the respective promoter activities of the DPE and MTE motifs. First, as described above, the m30–33 mutation inactivates the DPE (Figs. 3A, 4C). Then, in the absence of the DPE, transcription from MTE-containing promoters is nearly completely lost upon introduction of the m18–22 mutation (Fig. 4B). We thus conclude that MTE sequences from +18 to +22 are required for MTE-dependent transcription.
Transcription from MTE-containing promoters requires precise Inr–MTE spacing
We had previously shown that the DPE functions with the Inr (Burke and Kadonaga 1996, 1997; Kutach and Kadonaga 2000), and we therefore tested whether the MTE has related properties. With the CG15312, CG10479, and Tollo core promoters, mutation of the Inr results in an essentially complete loss of transcription (Fig. 5A). These results indicate that the Inr is required for transcription from MTE-containing promoters.
Figure 5.
The MTE functions with the Inr in a spacing-dependent manner. (A) The Inr is required for transcription from MTE-containing promoters. In the mutant Inr (mInr) promoters, the Inr sequences (shown in Fig. 1) were mutated to GTGACA. The constructs were subjected to in vitro transcription analysis with a Drosophila nuclear extract. (B) The spacing between the Inr and the MTE is important for core promoter activity. A series of mutant promoters was constructed in which the spacing between the Inr and the MTE was either increased or decreased by one or three nucleotides, as depicted. To ensure that the effects are due to interactions between the Inr and MTE, all of the promoters contain the m30–33 mutation (CATA at +30–33) that inactivates the DPE motif. The constructs were subjected to in vitro transcription analysis with a Drosophila nuclear extract. The transcriptional activity of each mutant promoter is reported relative to that of the m30–33 promoter with wild-type spacing (0) between the Inr and the MTE.
Next, we examined whether the spacing between the Inr and MTE is important for transcriptional activity, as seen in DPE-dependent promoters (Burke and Kadonaga 1997; Kutach and Kadonaga 2000). To this end, we created a series of mutant promoters in which the spacing between the Inr and MTE is increased or decreased by 1 or 3 nt (Fig. 5B). To eliminate the contribution of the DPE to transcriptional activity in these promoters, all of the constructions contain the m30–33 DPE mutation. Transcription of the spacing mutant promoters revealed a strong reduction in transcription upon insertion of one or three nucleotides or deletion of three nucleotides between the Inr and MTE sequences. A more modest threefold reduction in transcription was observed upon deletion of one nucleotide between the Inr and MTE. These results indicate that the proper spacing between the Inr and MTE is important for transcriptional activity.
The MTE is a distinct core promoter element that can compensate for the loss of a DPE
We further sought to investigate the ability of the MTE to act as a distinct core promoter element. In the experiments presented thus far, we have shown that mutation of the MTE results in a decrease in transcription. We therefore examined whether the addition of an MTE can increase transcription. In this regard, we tested the ability of the MTE to substitute for the loss of the DPE. If the functions of the MTE and DPE were interdependent, then the mutation of DPE could not be restored by the addition of an MTE. On the other hand, if the MTE were independent of the DPE, then the addition of an MTE could potentially restore activity that is lost upon mutation of the DPE. The results from the mutational analysis of MTE-containing promoters (Fig. 4B) suggested that the MTE acts independently of the DPE, but it was important to investigate this question to a greater extent.
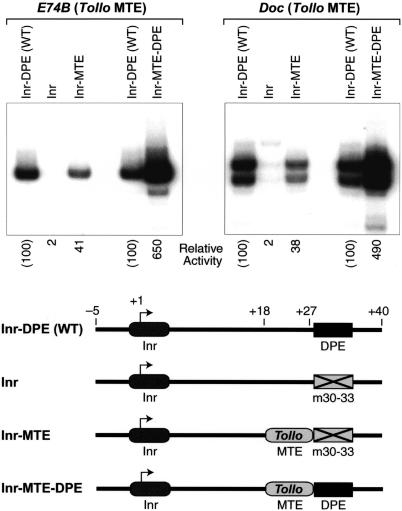
In these experiments, we constructed a series of E74B and Doc core promoters, which are depicted in Figure 6. As seen in Figures 3A and 4C, the introduction of the m30–33 DPE mutation to the E74B promoter or to the Doc promoter results in a near complete loss of transcriptional activity (Fig. 6, cf. Inr–DPE and Inr). If, however, the downstream sequences (+18 to +27) from E74B or Doc are substituted with the analogous sequences (+18 to +27) from the Tollo MTE, then core promoter activity is restored to ∼40% of wild-type activity (Fig. 6, cf. Inr and Inr–MTE). Thus, the +18 to +27 region of the MTE appears to be sufficient to compensate for the loss of a DPE in the E74B and Doc core promoters. (Note that the +28 and +29 sequences of E74B and Doc [TG and AG, respectively] are not identical to those of Tollo [GG] or to the motif 10 consensus [GS].)
Figure 6.
The MTE can compensate for the loss of a DPE. The diagram depicts the four variants of the E74B and Doc core promoters that were tested. In the constructs that contain an MTE (Inr–MTE and Inr–MTE–DPE), the +18 to +27 segment of each wild-type promoter is replaced by the MTE sequence (from +18 to +27) of the Tollo core promoter. In the Inr and Inr–MTE constructs, the DPE sequence is mutated to CATA at positions +30 to +33. These E74B and Doc promoter sets were subjected to in vitro transcription analysis with a Drosophila nuclear extract.
We also examined whether there might be synergy between the MTE and DPE motifs. To this end, we constructed variants of the E74B and Doc promoters in which the wild-type sequences (from +18 to +27) are replaced with the analogous sequences from the Tollo MTE. These experiments revealed potent synergy between the DPE and MTE motifs (Fig. 6, cf. Inr–DPE and Inr–MTE with Inr–MTE–DPE). For example, with the E74B promoter, the activity of the MTE- and DPE-containing promoter (Inr–MTE–DPE) is greater than four times the sum of the activities of the Inr–DPE (WT) and the Inr–MTE promoters. Thus, when combined in a single promoter, the MTE and DPE motifs can function synergistically to facilitate transcription. It might also be noted that the synergism seen in the artificial constructs shown in Figure 6 resembles the effects seen in the mutational analysis of the natural MTE-containing promoters (Fig. 4B). In those experiments, the transcriptional activity of each wild-type promoter was found to be greater than the sum of the activities of the Inr–DPE (m18–22) and the Inr–MTE (m30–33) versions of the promoter.
Hence, the MTE can substantially restore the transcriptional activity that is lost upon mutation of the DPE. The results suggest that the +18 to +27 sequences of the motif 10 consensus can confer MTE activity to heterologous core promoters. In addition, the MTE can function synergistically with the DPE. These findings indicate that the MTE is a distinct core promoter element that can function both independently of the DPE as well as in collaboration with the DPE.
The MTE can compensate for the loss of a TATA-box
We then sought to determine whether the MTE possesses core promoter activity that can compensate for the loss of a TATA-box. To this end, we constructed a series of hybrid promoters in which the TATA-box and Inr of the hbP2 core promoter is fused to the MTE-containing downstream promoter region of Tollo or CG10479, as depicted in Figure 7. The hbP2 promoter contains a tandemly repeated Inr that directs transcription initiation at two sites that are designated “+1” and “+5.” The downstream regions of Tollo and CG10479 were fused to hbP2 such that the MTE is aligned with the Inr that yields the +1 start site. In addition, all of the promoter constructions contained the m30–33 mutation to eliminate the contribution of the DPE to transcriptional activity.
Figure 7.
The MTE can compensate for the loss of a TATA-box. Two sets of hybrid promoters were constructed by fusing the -36 to +10 region of the hbP2 promoter to the +16 to +40 region of either the Tollo or the CG10479 promoter. The hbP2 core promoter contains repeated Inr motifs that direct initiation at +1 and +5. In the hybrid promoters, the spacing of the MTE motifs is aligned with the Inr at +1. To eliminate the contribution of the DPE, all of the promoters contain the m30–33 mutation (CATA at +30 to +33) that inactivates the DPE. The Inr and Inr–MTE constructs contain the mTATA mutation, in which the hbP2 TATA-box, TATATAAA, is replaced by ACGTCCGT. The TATA–Inr and Inr constructs contain the m18–22 mutation (ATCCA from +18 to +22), which inactivates the MTE. The hybrid promoter sets were subjected to in vitro transcription analysis with a Drosophila nuclear extract, and the transcriptional activity of each hybrid promoter is reported relative to that of the TATA–Inr (“wild-type” hbP2) promoter.
The TATA–Inr promoters are essentially equivalent to the wild-type hbP2 promoter, because the TATA–Inr promoters consist of the hbP2 TATA and Inr fused to downstream sequences of Tollo or CG10479 with mutant versions of the MTE and DPE. The TATA–Inr promoters yield transcripts that initiate from the +1 and the +5 start sites at approximately equal levels. The Inr promoters are identical to the TATA–Inr promoters except that the TATA-box sequences are mutated. As a consequence, the Inr promoters exhibit significantly less transcriptional activity than the TATA–Inr promoters. This loss of activity due to mutation of the TATA-box can be rescued by the addition of an MTE, as seen with the Inr–MTE promoters. The Inr–MTE promoters contain either the Tollo MTE or the CG10479 MTE in a downstream position in alignment with the Inr motif that directs the +1 start site of the hbP2 promoter. As seen in Figure 7 (cf. TATA–Inr, Inr, and Inr–MTE), the Tollo MTE can almost completely restore the transcriptional activity that is lost upon mutation of the TATA-box. The CG10479 MTE is able to compensate partially (∼43%) for the loss of the TATA-box. In addition, there is a strong preference for transcription from the +1 site relative to the +5 site in the Inr–MTE promoters. This bias for the +1 site is most likely due to the alignment of the MTE sequences, which exhibit a strict Inr–MTE spacing requirement (see Fig. 5B), with the +1 Inr in the hbP2 core promoter. Thus, these results indicate that the MTE, in the absence of a DPE, can compensate for the loss of a TATA-box.
We were also interested in testing whether there is transcriptional synergism between the TATA and MTE motifs. We observed strong synergism between the hbP2 TATA box and the Tollo MTE, as the hybrid promoter that contains both the TATA and the MTE (TATA–Inr–MTE) possesses greater than four times the sum of the activities of the TATA only (“TATA–Inr”) and the MTE only (Inr–MTE) promoters (Fig. 7; cf. TATA–Inr, Inr–MTE, and TATA–Inr–MTE for the hbP2–Tollo hybrid promoter). We did not, however, observe synergism between the hbP2 TATA and the CG10479 MTE. Based on its ability to rescue the loss of the TATA-box and to function synergistically with the TATA-box, it seems likely that the Tollo MTE has stronger activity than the CG10479 MTE.
Thus, the MTE can compensate for the loss of the TATA-box as well as function synergistically with the TATA-box.
The MTE appears to affect the interaction of TFIID with the core promoter
TFIID is a key factor in the recognition of the TATA, Inr, and DPE core promoter motifs. We therefore tested whether the MTE affects the binding of purified Drosophila TFIID to an MTE-containing core promoter. To this end, we carried out DNase I footprinting experiments with the Tollo core promoter. Three versions of the Tollo promoter were tested: wild-type, m30–33 (mutant DPE), and m18–22/30–33 (mutant MTE and DPE). We were particularly interested in the effect of the MTE upon TFIID binding in the absence of the DPE (i.e., comparison of the binding of TFIID to the m30–33 promoter relative to the m18–22/30–33 promoter). In these experiments, we observed a weak interaction of native Drosophila TFIID (of ∼50% purity) to the MTE-containing Tollo promoter (Fig. 8, lanes 1–4). Mutation of the MTE resulted in a reduction in TFIID-induced hypersensitive bands (arrow) as well as the loss of some sites of weak DNase I protection (dots) in the vicinity of the Inr (Fig. 8, cf. lanes 3,4 and 5,6). Notably, as seen in the binding of TFIID to DPE-dependent promoters (Burke and Kadonaga 1996), mutation of the downstream MTE sequences affects the binding of TFIID to the Inr region. These results suggest that the MTE contributes to the binding of TFIID to the core promoter. The weak interaction of TFIID with the Tollo promoter could be due to an intrinsically low affinity of TFIID to the promoter or to low DNA-binding activity of our preparations of native TFIID. It is also possible that other factors function to augment the binding of TFIID to TATA-less, MTE-containing promoters.
Figure 8.
The MTE appears to affect the interaction of TFIID with the core promoter. The wild-type, m18–22 (mutant MTE), and m18–22/30–33 (mutant MTE and DPE) versions of the Tollo core promoter were subjected to DNase I footprinting analysis with purified Drosophila TFIID. The mutation of the MTE in the absence of the DPE results in the decrease of a TFIID-induced hypersensitive site (arrow) as well as the loss of sites of weak protection by TFIID (dots) in the vicinity of the Inr (cf. lanes 3,4 and 5,6).
The MTE is recognized by human transcription factors
To investigate whether MTE function is conserved from Drosophila to humans, we tested the ability of human transcription factors in HeLa cells to recognize the Drosophila Tollo MTE (Fig. 9). First, we carried out in vitro transcription reactions with wild-type and mutant Tollo core promoters. As seen in Figure 9A, mutation of the MTE results in an eightfold decrease in transcription with a HeLa nuclear extract. In addition, we attempted to test the activity of the Tollo core promoter by transient transfection analysis in HeLa cells (Fig. 9B). In these experiments, the wild-type core promoter exhibited a low level of activity that was only about three- to fourfold higher than that of the promoterless vector. These results reflect the difficulty of studying basal transcription in vivo. Mutation of the core promoter elements resulted in a lower level of activity, but it was not possible to obtain an accurate assessment of the magnitude of the effects.
Figure 9.
Human transcription factors recognize the MTE. (A) The MTE in the Drosophila Tollo core promoter is recognized by human basal transcription factors. The Tollo promoter constructs described in Figure 4 were transcribed with a HeLa nuclear extract. (B) Transient transfection analysis of wild-type and mutant Tollo core promoters in HeLa cells. The wild-type, m18–22, m30–33, and m18–22/30–33 versions of the Tollo–luc reporter constructs as well as the promoterless vector (pGL3-Basic; “vector only”) were transiently transfected in HeLa cells, and the relative activities were determined. (C) The MTE exhibits synergy with the TATA-box as well as weak activity in the absence of the TATA and DPE motifs with human transcription factors. The hbP2–Tollo hybrid promoter constructs shown in Figure 7 were transcribed with a HeLa nuclear extract. (D) The MTE can compensate for the loss of the DPE and exhibits synergy with the DPE with human transcription factors. The E74B-based constructs shown in Figure 6 were transcribed with a HeLa nuclear extract. (E) The human sterol C5 desaturase-like (SC5DL) gene has an MTE-dependent promoter. The wild-type, m18–22, m30–33, and m18–22/30–33 versions of the SC5DL promoter were subjected to in vitro transcription analysis with a HeLa nuclear extract.
Next, we examined whether the MTE can compensate for the loss of the TATA-box with human transcription factors. To this end, we used the hbP2–Tollo hybrid promoter series, as in Figure 7. Transcription of these promoters with human factors revealed that the addition of the Tollo MTE is able to restore partially the loss of transcription upon mutation of the hbP2 TATA box (Fig. 9C; cf. TATA–Inr, Inr, and Inr–MTE). In addition, we observed a modest synergism between the TATA and MTE in transcription reactions with human factors (Fig. 9C; cf. TATA–Inr, Inr–MTE, and TATA–Inr–MTE).
We also tested whether the MTE can restore the loss of promoter activity that occurs upon mutation of the DPE in transcription reactions with human factors. In these experiments, we used the E74B (Tollo MTE) promoter series that is shown in Figure 6. With the human factors, the MTE is able to compensate fully for the loss of the DPE (Fig. 9D; cf. Inr–DPE, Inr, and Inr–MTE). We additionally observed potent synergy between the MTE and DPE (Fig. 9D; cf. Inr–DPE, Inr–MTE, and Inr–MTE–DPE), as seen with the Drosophila factors (Fig. 6).
Lastly, we identified a human core promoter that appears to contain a functional MTE. As depicted in Figure 9E, the human sterol C5 desaturase-like (SC5DL; also known as SC5D) promoter (Nishi et al. 2000; Sugawara et al. 2001) has sequences with similarity to the Drosophila Inr and MTE (see Fig. 1) but not to the DPE. By primer extension analysis of poly(A)+ RNA from HeLa cells, we confirmed that the +1 site shown in Figure 9E is used as a transcription start site in vivo (data not shown). We then constructed and analyzed wild-type and mutant versions of the human SC5DL core promoter. These experiments revealed that the m18–22 MTE mutation results in about a 6.5-fold decrease in transcription, whereas the m30–33 DPE mutation causes less than a twofold decrease in transcription (Fig. 9E). These data collectively indicate that the MTE is conserved from Drosophila to humans.
Discussion
The MTE (motif ten element) is a new core promoter element for RNA polymerase II. Motif 10 was initially identified as a downstream, overrepresented sequence in a computational analysis of Drosophila core promoters (Ohler et al. 2002). In this study, we determined that the motif 10 sequence contains the MTE, a distinct downstream core promoter motif that functions with the Inr element to promote transcription. The MTE acts independently of the DPE and TATA motifs, but can also work synergistically with the DPE or the TATA-box. In addition, the MTE appears to be conserved from Drosophila to humans.
It is important to note that the function of the MTE as a core promoter element is more clearly demonstrated by its ability to increase transcription when added to a heterologous core promoter (i.e., gain-of-function experiments, as in Figs. 6, 7) than by the loss of transcription upon mutation of the MTE sequences (i.e., loss of function experiments, as in Fig. 4). The mutational analysis of promoter sequences is complicated by the nonexistence of transcriptionally neutral nucleotides—hence, there is a good probability that any given nucleotide change will have some effect (most likely negative, if it is in a core promoter region) upon transcriptional activity. (However, the near complete loss of core promoter activity upon mutation of both the MTE and DPE, as in Fig. 4B, does strongly suggest a function of the MTE in the core promoter.) It is less likely that an alteration of the core promoter (i.e., the addition of an MTE, as in Figs. 6, 7) will significantly increase transcription. Also, the strict spacing requirement between the Inr and MTE (Fig. 5B) and the synergy between the MTE and DPE (Fig. 6) as well as between the TATA and MTE (Fig. 7) demonstrate a functional linkage between the TATA, Inr, and DPE core promoter elements and the MTE.
The motif 10 consensus sequence encompasses +18 to +29 relative to A+1 in the Inr element, whereas the DPE is from +28 to +33 (Burke and Kadonaga 1996, 1997; Kutach and Kadonaga 2000). We have found, however, that the motif 10 sequences from +17 to +22 are most important for MTE transcriptional activity (Figs. 2, 3), and that the +18 to +27 motif 10 sequences are sufficient to confer MTE activity to heterologous core promoters (Fig. 6). These findings suggest that the 3′-end of the motif 10 sequence is not essential for the basal transcription function of the MTE. In fact, the overlap of the 3′-end of the motif 10 consensus sequence with the 5′-end of the DPE consensus (i.e., positions +28 and +29) may be an artifact of the motif identification algorithm in the computational analysis (Ohler et al. 2002). For instance, an unintended bias at the 3′-end of motif 10 could have arisen from the close and constant spacing between the MTE and DPE and the frequent occurrence of both motifs in the same core promoters.
One intriguing feature of motif 10 is the presence of the tandemly repeated AACGGAACGG motif in the consensus sequence. Mutation of this repetitive sequence has little effect on basal transcriptional activity (Fig. 2; data not shown). It is possible, for instance, that this motif is a recognition site for a sequence-specific DNA-binding factor that does not participate directly in the basal transcription process. Thus, the motif 10 sequence may comprise two different elements—the MTE and the AACGGAACGG motif.
It is useful to compare the properties of the MTE and DPE. For instance, there is a strict spacing requirement between the Inr and MTE (Fig. 5B) as well as between the Inr and DPE (Burke and Kadonaga 1997; Kutach and Kadonaga 2000). In addition, the loss of transcription upon mutation of the TATA-box can be restored by the addition of an MTE (Fig. 7) or a DPE (Burke and Kadonaga 1996). The subregion of the MTE that is most important for basal transcription is the 5′-end of the element in the vicinity of +17 to +22 (Figs. 2, 3). Thus, the regions of the MTE and DPE that are most important for basal transcription are separated by ∼10 or 11 bp, which is roughly one turn of the DNA helix. This arrangement of the MTE and DPE suggests that there may be key contacts of TFIID with the same face of the DNA helix at the MTE and DPE. It will be important to analyze further the interaction of TFIID and possibly other factors to the MTE.
It is also useful to note differences between the MTE and DPE. The mutational studies (Fig. 4B) and the replacement experiments (Fig. 6) indicate that the MTE can function independently of the DPE as well as substitute for the loss of the DPE. In addition, we observed strong synergism between the MTE and the TATA-box (Fig. 7) but not between the DPE and the TATA-box (Burke and Kadonaga 1996). It is possible, however, that there are combinations of TATA and DPE motifs that exhibit strong synergism.
The MTE-containing promoters that were identified in this study lack a TATA-box, but it seems likely that some core promoters will contain both MTE and TATA elements. In general, it will be important to investigate further the frequency of occurrence of the MTE as well as the contexts in which it functions in different core promoters.
In conclusion, the MTE is a new addition to the relatively small set of core promoter elements for RNA polymerase II. There is, of course, much more to be learned about the MTE and the other known core promoter motifs. For instance, it will be interesting to determine whether some transcriptional enhancers act specifically with the MTE, as seen with the TATA and DPE (Butler and Kadonaga 2001). In addition, it is likely that there are many other core promoter elements that remain to be identified. The core promoter is a critical gateway in the path leading to gene expression, and it is likely that we have yet to realize fully the diversity of core promoter function. In this respect, the core promoter is reemerging as an exciting frontier for new discoveries in gene regulation.
Materials and methods
Core promoter sequences and plasmids
The initial search for MTE-containing promoters was carried out with a sample of potential MTE-containing promoter sequences that were obtained in the course of the computational analysis of Drosophila core promoters (Ohler et al. 2002). After initial studies with these promoters, we performed a search of the entire Drosophila genome database (Berkeley Drosophila Genome Project; http://www.fruitfly.org) by using the JDSA program (available at http://www-biology.ucsd.edu/labs/kadonaga/JDSA.html). To identify potential MTE-containing core promoters, we searched for the MTE consensus sequence, CSARCSSAACGS (Ohler et al. 2002), at precisely +18 to +29 relative to the A+1 position of a canonical Inr consensus sequence.
The plasmids that were used to map the in vitro start sites (Fig. 1B) were constructed by PCR amplification of Drosophila genomic DNA with primers that yielded a 300-bp fragment of each promoter from -150 to +150 of the expected RNA start site. These promoter fragments were then cloned into the pGEMT-easy vector (Promega). The minimal core promoter templates, which were used in Figures 2, 3, 4, 5, 6, 7, 8, 9, were constructed by insertion of double-stranded oligonucleotides into the XbaI and PstI sites in the polylinker of pUC119. Typically, the minimal core promoters include sequences from -10 to +40 relative to the A+1 site. In the mInr mutants, the native Inr sequences were replaced with GTGACA. The Tollo minimal promoter spacing mutants were generated by insertion of A between T+16 and T+17 (+1), insertion of AAA between T+16 and T+17 (+3), deletion of T+16 (-1), and deletion of A+14 through T+16 (-3). The hybrid hbP2–Tollo and hbP2–CG10479 promoters were constructed as follows: First, double-stranded oligonucleotides comprising the -36 to +10 sequences of the hbP2 promoter were inserted into the XbaI and PstI sites of pUC119 to give the pUChbP2 plasmid; then, double-stranded oligonucleotides comprising the +16 to +40 sequences of Tollo or CG10479 were inserted into the PstI site of the pUChbP2 plasmid. The plasmids used in transient transfection assays were constructed by subcloning the Tollo minimal core promoters into the pGL3-Basic expression vector (Promega).
In vitro transcription analysis
Transcription reactions were carried out as described previously (Wampler et al. 1990; Kraus and Kadonaga 1999) by using supercoiled DNA templates (250 ng or 500 ng) with either Drosophila nuclear extracts (Soeller et al. 1988) or HeLa (human) cell nuclear extracts (Dignam et al. 1983). The resulting transcripts were subjected to primer extension analysis with the M13 reverse sequencing primer (AGCGGATAACAATTTCA CACAGGA). Quantitation of reverse transcription products was carried out with a PhosphorImager (Molecular Dynamics). All experiments were performed a minimum of three times to ensure reproducibility of the data.
DNase footprinting analysis
DNase I footprint probes were prepared by PCR amplification of each promoter with unlabeled M13 universal primer (upstream) and 5′-32P-labeled M13 reverse sequencing primer (downstream) flanking the promoter region. The PCR amplification products were purified with Microspin S300HR columns (Amersham Pharmacia Biotech). TFIID was purified to ∼50% homogeneity from Drosophila embryos by using a combination of conventional and immunoaffinity chromatography. Footprinting reactions were carried out as described previously (Burke and Kadonaga 1996).
Transient transfection and reporter gene assays
HeLa cells were cultured in DMEM supplemented with 10% FBS and were transfected at ∼50% confluence in 60-mm plates by a calcium phosphate transfection procedure. For each experiment, the cells were cotransfected with 3 μg of the Tollo promoter–luciferase constructs as well as the promoterless vector (pGL3-Basic) and 1 μg of a β-galactosidase expression vector. Cells were harvested and assayed for luciferase and β-galactosidase activities 48 h posttransfection by using the Luciferase Assay and β-galactosidase Enzyme Assay systems as specified by the manufacturer (Promega). To correct for transfection efficiency, the luciferase activity of each sample was normalized over the corresponding β-galactosidase activity. The reported activities are the averages of three independent experiments performed in triplicate, and are expressed relative to the activity of the wild-type promoter, which was defined to be 100%.
Acknowledgments
We are especially grateful to Gerry Rubin for stimulating and enjoyable interactions that led to this study. We thank Alexandra Lusser, Tammy Juven-Gershon, Dmitry Fyodorov, Jer-Yuan Hsu, Barbara Rattner, and Debra Urwin for critical reading of the manuscript. This work was supported by a grant from the NIH to J.T.K. (GM41249). C.Y.L. is a recipient of the National Science Scholarship from the Agency for Science, Technology, and Research (ASTAR), Singapore.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1193404.
References
- Brivanlou A.H. and Darnell Jr., J.E. 2002. Signal transduction and the control of gene expression. Science 295: 813-818. [DOI] [PubMed] [Google Scholar]
- Burke T.W. and Kadonaga, J.T. 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 10: 711-724. [DOI] [PubMed] [Google Scholar]
- ____. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 11: 3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.E.F. and Kadonaga, J.T. 2001. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 15: 2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 16: 2583-2592. [DOI] [PubMed] [Google Scholar]
- Chalkley G.E. and Verrijzer, C.P. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAFII250–TAFII150 complex recognizes the Initiator. EMBO J. 18: 4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. and Manley, J.L. 2003. Core promoter elements and TAFs contribute to the diversity of transcriptional activation in vertebrates. Mol. Cell. Biol. 23: 7350-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M.L. 1979. “Sequence analysis of Drosophila histone genes.” Ph.D. thesis, Stanford University, Stanford, CA.
- Hirose Y. and Manley, J.L. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415-1429. [PubMed] [Google Scholar]
- Hochheimer A. and Tjian, R. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes & Dev. 17: 1309-1320. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. 2002. The DPE, a core promoter element for transcription by RNA polymerase II. Exp. Mol. Med. 34: 259-264. [DOI] [PubMed] [Google Scholar]
- ____. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA-binding factors. Cell 116: 247-257. [DOI] [PubMed] [Google Scholar]
- Kaufmann J. and Smale, S.T. 1994. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes & Dev. 8: 821-829. [DOI] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga, J.T. 1999. Ligand- and cofactor-regulated transcription with chromatin templates. In Nuclear receptors: A practical approach (ed. D. Picard), pp. 167-189. Oxford University Press, Oxford/New York.
- Kutach A.K. and Kadonaga, J.T. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20: 4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T., Kapanidis, A.N., Tang, H., Reinberg, D., and Ebright, R.H. 1998. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes & Dev. 12: 34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. and Young, R.A. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34: 77-137. [DOI] [PubMed] [Google Scholar]
- Levine M. and Tjian, R. 2003. Transcription regulation and animal diversity. Nature 424: 147-151. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499-506. [DOI] [PubMed] [Google Scholar]
- Myers L.C. and Kornberg, R.D. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69: 729-749. [DOI] [PubMed] [Google Scholar]
- Näär A.M., Lemon, B.D., and Tjian, R. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70: 475-501. [DOI] [PubMed] [Google Scholar]
- Nishi S., Nishino, H., and Ishibashi, T. 2000. cDNA cloning of the mammalian sterol C5-desaturase and the expression in a yeast mutant. Biochim. Biophys. Acta 1490: 106-108. [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao, G., Niemann, H., and Rubin, G.M. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3: research0087.1–research0087.12. [DOI] [PMC free article] [PubMed]
- Orphanides G. and Reinberg, D. 2002. A unified theory of gene expression. Cell 108: 439-451. [DOI] [PubMed] [Google Scholar]
- Purnell B.A., Emanuel, P.A., and Gilmour, D.S. 1994. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes & Dev. 8: 830-842. [DOI] [PubMed] [Google Scholar]
- Smale S.T. 1997. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta 1351: 73-88. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Core promoters: Active contributors to combinatorial gene regulation. Genes & Dev. 15: 2503-2508. [DOI] [PubMed] [Google Scholar]
- Smale S.T. and Baltimore, D. 1989. The `initiator' as a transcription control element. Cell 57: 103-113. [DOI] [PubMed] [Google Scholar]
- Smale S.T. and Kadonaga, J.T. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449-479. [DOI] [PubMed] [Google Scholar]
- Soeller W.C., Poole, S.J., and Kornberg, T. 1988. In vitro transcription of the Drosophila engrailed gene. Genes & Dev. 2: 68-81. [DOI] [PubMed] [Google Scholar]
- Struhl K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1-4. [DOI] [PubMed] [Google Scholar]
- Sugawara T., Fujimoto, Y., and Ishibashi, T. 2001. Molecular cloning and structural analysis of human sterol C5 desaturase. Biochim. Biophys. Acta 1533: 277-284. [DOI] [PubMed] [Google Scholar]
- Verrijzer C.P., Yokomori, K., Chen, J.-L., and Tjian, R. 1994. Drosophila TAFII150: Similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science 264: 933-941. [DOI] [PubMed] [Google Scholar]
- Wampler S.L., Tyree, C.M., and Kadonaga, J.T. 1990. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J. Biol. Chem. 265: 21223-21231. [PubMed] [Google Scholar]