Abstract
We show that intracellular transcription of G-rich regions produces novel DNA structures, visible by electron microscopy as large (150–500 bp) loops. These G-loops are formed cotranscriptionally, and they contain G4 DNA on one strand and a stable RNA/DNA hybrid on the other. G-loop formation requires a G-rich nontemplate strand and reflects the unusual stability of the rG/dC base pair. G-loops and G4 DNA form efficiently within plasmid genomes transcribed in vitro or in Escherichia coli. These results establish that G4 DNA can form in vivo, a finding with implications for stability and maintenance of all G-rich genomic regions.
Keywords: G-quartet, G-tetraplex, immunoglobulin switch region, telomere/transcription bubble
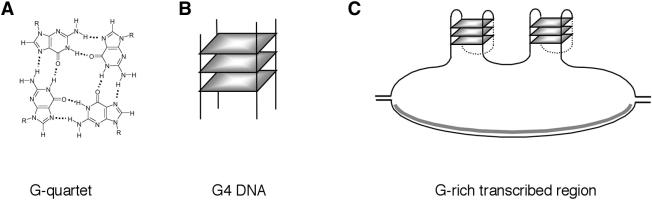
Genomic DNA is normally maintained as a Watson-Crick duplex, but DNA has considerable potential to form other structures. One sequence motif especially prone to form structures in vitro is a guanine run (Sen and Gilbert 1988, 1990). Oligonucleotides containing runs of guanines readily and spontaneously assemble into four-stranded structures stabilized by G-quartets, planar arrays of four hydrogen-bonded guanines (Gellert et al. 1962). These structures, referred to as G-tetraplex, G-quadruplex, or G4 DNA, may involve intramolecular or intermolecular interactions, and the phosphodiester backbones of the four participating strands may be in parallel or antiparallel orientation (Fig. 1A,B; for review, see Gilbert and Feigon 1999). Intramolecular G4 DNA readily forms in short oligonucleotides carrying at least four runs of guanines, in which each run is at least 3 nt in length, such as (TTAGGG)4, a 4-mer telomeric repeat; and longer G-runs support formation of even more stable structures, which are tolerant of single-base interruptions within the G-runs. G4 DNA is very stable once formed. This reflects hydrogen bonding within G-quartets, stacking of the large, planar G-quartets on one another, and charge coordination by monovalent cations (especially K+) within the central cavity.
Figure 1.
G4 DNA and G-quartets. (A) Four guanines assemble in a planar ring to form a G-quartet. (B) G4 DNA is a stable structure composed of stacked G-quartets (shaded squares). (C) Diagram of predicted structure formed upon transcription of G-rich DNA. The G-rich strand contains G4 DNA, stabilized by G-quartets; and the C-rich template strand is hybridized to the RNA transcript (gray). For simplicity, only two regions of G4 DNA structure are diagrammed, although loops formed upon transcription of a long G-rich region could, in principle, contain many structured regions.
Certain chromosomal regions are G-rich and contain sequences that readily form G4 DNA in vitro. This has led to considerable interest in the possibility that G4 DNA might form in vivo and have specific functions in regulation of gene expression or genetic stability. G-rich chromosomal domains include four classes of repetitive regions—telomeres, rDNA, immunoglobulin heavy chain switch regions, and G-rich minisatellites. Telomeric repeats in almost all eukaryotes contain runs of Gs (Saccharomyces cerevisiae, TG1–3; Tetrahymena, TTT TGGGG; mammals, TTAGGG), and telomeric sequences form G4 DNA in vitro (Parkinson et al. 2002; Neidle and Parkinson 2003). The rDNA repeats are G-rich on the nontemplate DNA strand, both within the region that is transcribed into mature rRNA and within the spacers (e.g., Hanakahi et al. 1999). The immunoglobulin heavy chain switch (S) regions, which are required for class-switch recombination, consist of degenerate G-rich repeats that are from 2 to 10 kb in length and conform to a loose consensus. Examples of S-region consensus sequences in four of the seven murine S regions are as follows: Sμ, GCTGAGCTGGGGTGAGC TGA; Sγ3, CTGGGCAGCTCTGGGGGAGCTGGGG TAGGTTGGGAGTGTGGGGACCAGG; Sγ2b, CCTA GCAGCTGTGGGGGAGCTGGGGAAGGTGGGAGT GTGAGGGACCAGA; and Sα, TGAGCTGGGATAG GCTGAGCTGGGCTGGA (Arakawa et al. 1993; Dunnick et al. 1993). The human genome is replete with repetitive minisatellites, and among those that exhibit the most pronounced instability are G-rich sequences capable of forming G4 DNA, including the MS1 minisatellite (D1S7), with repeat unit AGGGTGGAG; D4S43, repeat unit GGGGAGGGGGAAGA; the insulin-linked hypervariable repeat, ACAGGGGTGTGGGG; and longer repeats such as MS32 (29 bp), CEB1 (37–43 bp), D1Z2 (40 bp), and MS205 (45–54 bp; Buroker et al. 1987; Wong et al. 1987; Jeffreys et al. 1988; Weitzmann et al. 1997).
G4 DNA resists attack by enzymes that target single-stranded or duplex DNA, but cells do contain factors that actively cleave and unwind G4 DNA. GQN1 is a nuclease identified in mammalian cells that specifically targets G4 DNA with striking specificity, cleaving in the single-stranded region 5′ of the stacked G-quartets (Sun et al. 2001). A distinct activity, the KEM1/SEP1 nuclease of S. cerevisiae, cleaves both G4 DNA and G4 RNA, and appears to function in RNA processing (Hsu and Stevens 1993; Liu and Gilbert 1994). Helicases in the highly conserved RecQ family are essential to genomic stability in organisms from Escherichia coli to humans (Shen and Loeb 2000; Nakayama 2002; Harrigan et al. 2003; Hickson 2003; Khakhar et al. 2003). Members of this family including human BLM (Sun et al. 1998) and WRN (Fry and Loeb 1994; Mohaghegh et al. 2001), S. cerevisiae Sgs1p (Sun et al. 1999), and E. coli RecQ (Wu and Maizels 2001) can unwind G4 DNA and other structured substrates (for review, see Bachrati and Hickson 2003).
The potential for DNA to form structures in vivo is dramatically illustrated by the human neurodegenerative diseases caused by triplet repeat expansion (McMurray 1999; Sinden 1999; Cummings and Zoghbi 2000; Bowater and Wells 2001). Among the diseases associated with repeat expansion are Huntington's disease (CAG repeat), fragile X syndrome (CGG repeat), myotonic dystrophy (CTG repeat), and Friedreich's ataxia (GAA repeat). Repeat expansion is due to formation of structures on the lagging strand during DNA replication, and the ability of a triplet repeat-sequence motif to form hairpin (CAG, CTG), triplex (GAA), or G4 DNA (CGG) in vitro correlates with expansion of that sequence motif in vivo (McMurray 1999; Sinden 1999; Sakamoto et al. 1999). Structures formed by repeats are targets for the mismatch repair apparatus, and deficiency in mismatch repair factors leads to repeat instability in yeast (Strand et al. 1993; Sia et al. 2001) and in human tumors (Kolodner 1995; Modrich and Lahue 1996).
Despite the readiness with which G-rich sequences form G4 DNA in vitro, the biological significance of G4 DNA has remained in question pending direct demonstration that G4 DNA forms in living cells. We have developed a novel assay that uses electron microscopy (EM) to visualize structures formed within individual DNA molecules. This assay is based upon results from other laboratories demonstrating that a stable RNA/DNA hybrid forms upon transcription of a C-rich template strand (Reaban and Griffin 1990; Reaban et al. 1994; Daniels and Lieber 1995; Tian and Alt 2000; Mizuta et al. 2003; Yu et al. 2003). If the complementary G-rich strand forms G4 DNA, then transcription of a G-rich region within a plasmid would be predicted to produce a loop in which one strand contains the RNA/DNA hybrid, and the other strand contains G4 DNA, as diagrammed in Figure 1C. To ask whether such structures form, we have visualized individual DNA molecules using EM, and probed their structures with proteins that specifically cleave or bind G4 DNA. We have now shown that characteristic loops do form efficiently upon transcription of G-rich regions, both in vitro and in vivo; and that these loops contain G4 DNA. Formation of G4 DNA is dependent upon transient denaturation of the DNA duplex, which is induced by transcription of the G-rich region within the model substrates. These results establish that G4 DNA formation can occur upon transcription in living cells, and suggest that G4 DNA may also form during transient denaturation, which normally accompanies recombination and replication.
Results
Transcription of G-rich DNAs causes stable loops to form
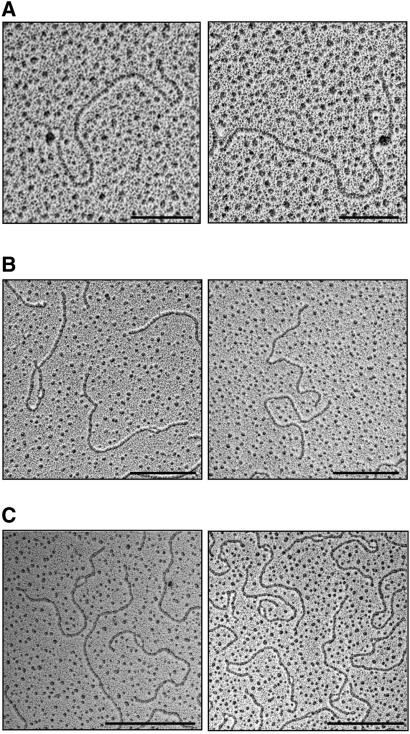
We assayed G4 DNA formation in plasmid templates that carry G-rich regions downstream of the T7 promoter in pBluescript (Fig. 2A). The G-rich regions were 350–1050 bp in length and derived from the mammalian immunoglobulin Sμ and Sγ3 switch regions, and the TTAGGG telomeric repeat (Fig. 2B). Following in vitro transcription, free RNA was digested with RNase A, and the reaction deproteinized and visualized by Kleinschmidt spreading (Kleinschmidt and Zahn 1959; Kleinschmidt 1968). EM revealed distinct loops in the transcribed plasmids, as shown in the examples in Figure 2C. The loops mapped to the G-rich transcribed region within the plasmids (Fig. 2D).
Figure 2.
Loops form in transcribed G-rich DNAs. (A) Map of plasmid templates, which carry G-rich sequences downstream of a T7 promoter. (B) G-rich regions within the plasmid templates analyzed by EM. (C) Examples of loops formed upon transcription of pRX15F (Sμ repeat; left), pPH600 (Sγ3; middle), and pHumtel (telomeric repeat, right). DNA was transcribed in vitro, linearized with AflIII, and visualized by EM. Bar, 500 nm. (D) Maps of loops visualized in 15 randomly selected pRX15F (left), pPH600 (middle), and pHumtel (right) templates. Maps are drawn to scale.
Loop formation was very efficient. Following transcription of Sμ (pRX15F, pRX15F3), Sγ3 (pPH600), or telomeric (pHumtel) sequences, loops were evident in 42%–63% of the template DNA molecules (Table 1). This number may be an underestimate, because loops smaller than 150 bp are not readily visualized by electron microscopy. Loop formation required a G-rich template, and transcription of an immunoglobulin Vλ1 region (pTT1, Table 1), or other sequences of random base composition produced no loops visible by EM. Loops ranged in size from 0.05 to 0.17 μm (150–500 bp). Loops of comparable size were produced by in vitro transcription with E. coli RNA polymerase from the lactose promoter in the plasmid backbone (data not shown). No loops were visualized in nontranscribed plasmids, demonstrating that transcription is essential for the formation of loops (Table 1). In addition, loop formation was dependent upon strand orientation and required that the G-rich strand be the top or nontemplate DNA strand for transcription (cf. plasmids pRX15F and pRX15R, Table 1).
Table 1.
Loop formation upon transcription in vitro
| Plasmida | Insert | Insert Size (bp) | Transcribed | nb | Loops (%)c |
|---|---|---|---|---|---|
| pRX15F | Sμ repeat | 350 | + | 108 | 42 |
| pRX15R | Sμ repeat | 350 | + | 31 | 0 |
| pRX15F3 | Sμ repeat | 1050 | + | 100 | 60 |
| pPH600 | Sγ3 | 604 | + | 99 | 63 |
| pPH600 | Sγ3 | 604 | — | 51 | 0 |
| pHumtel | (TTAGGG)n | 810 | + | 108 | 60 |
| pTT1 | Vλ1 | 550 | + | 50 | 0 |
For descriptions of plasmids and inserts, see Materials and methods
Number of plasmids analyzed, n
Fraction of plasmids containing loops was determined by EM analysis
Loops contain a stable RNA/DNA hybrid formed cotranscriptionally
One arm of each loop contained a stable RNA/DNA duplex, as assayed by two criteria. First, inclusion of digoxygenin-UTP in transcription reactions allowed the digoxygenin label to be visualized in one arm of the loop by decoration with gold beads coupled to antidigoxygenin antibodies (Fig. 3A). Second, transcribed molecules were treated with RNase H, which specifically digests the RNA strand of an RNA/DNA hybrid. RNase H treatment destroyed the loops (among 134 molecules visualized, none contained loops; e.g., Fig. 3B, right).
Figure 3.
Loops contain an RNA/DNA hybrid formed cotranscriptionally. (A) Examples of loops formed in pHumtel transcribed with Dig-UTP and visualized with anti-Dig/gold beads. Bar, 200 nm. (B) Loops are destroyed by RNase H. Transcribed pRX15F before (left) and after (right) RNase H treatment. No loops were identified among 134 molecules visualized following treatment with RNase H. Bar, 500 nm. (C) Loops form cotranscriptionally. Treatment of pPH600 templates with 20 μg/mL RNase A during (example on left) or after (example on right) transcription did not alter loop formation; loops were evident in 58% (53/90) and 60% (60/99) of molecules, respectively. Bar, 500 nm.
To ask whether loops form cotranscriptionally or as a result of an invasion of a DNA duplex by an RNA molecule synthesized on a different template, RNase A was included during the transcription reaction to digest transcripts that had been freed from the template, at a concentration (20 μg/mL) determined to be sufficient to digest all single-stranded RNA produced in the transcription reaction. The presence of RNase A did not significantly affect the frequency of loop formation— loops were apparent in 58% of the molecules (52/90) when transcription was carried out in the presence of RNase A, and 60% of the molecules (60/99) when RNase A was added after transcription (e.g., Fig. 3C). Loops therefore form cotranscriptionally.
G4 DNA forms upon transcription of G-rich regions
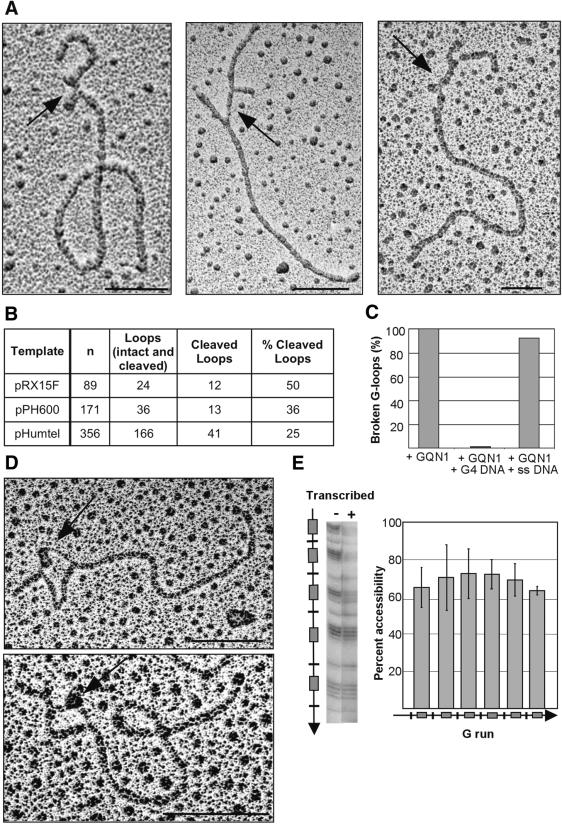
To ask whether G4 DNA forms within the transcribed plasmids, we assayed sensitivity of loops to GQN1, an endonuclease that specifically cleaves molecules containing G4 DNA in the single-stranded region 5′ of the stacked G-quartets (Sun et al. 2001). GQN1 cleavage is diagnostic for the presence of G4 DNA, because GQN1 cleaves a variety of G-quadruplex structures independent of DNA sequence, but it does not cleave duplex DNA, single-stranded DNA, Holliday junctions, or G4 RNA. Cleavage was assayed in a series of independent reactions with each of these three templates by scoring cleaved loops following GQN1 digestion (Fig. 4B). In the absence of GQN1 treatment, <0.4% of molecules contained a broken loop (1 of 282 molecules). GQN1 cleaved within the loops of transcribed plasmid templates bearing G-rich Sμ, Sγ3, or telomeric repeats, to produce characteristic opened loops (e.g., Fig. 4A). The cleavage efficiency ranged from 25% to 50% (Fig. 4B). Because only templates containing opened loops with visible ends were scored, this may be an underestimate of the efficiency of loop cleavage. Specificity of GQN1 for G4 DNA was confirmed by competition experiments, which showed that cleavage was inhibited by 10-fold molar excess of competitor G4 DNA formed from a synthetic oligonucleotide, but not by 1000-fold molar excess of single-stranded competitor DNA (Fig. 4C). Thus, G4 DNA formed upon transcription of Sμ, Sγ3, or telomeric sequences.
Figure 4.
Loops contain G4 DNA. (A) Examples of GQN1 cleavage of loops in transcribed pRX15F (Sμ; left), pPH600 (Sγ3; middle), and pHumtel (telomeric repeat, right). Arrows mark cleaved G-loops. Bar, 200 nm. (B) Effect of GQN1 treatment on transcribed pRX15F, pPH600, and pHumtel templates. DNAs were visualized by EM, and loops were scored as cleaved if they were opened and contained a clearly visible arm or ends, as in Figure 4A. (n) Number of molecules scored. (C) Effect of competitor G4 DNA (10-fold excess) or single-stranded DNA (1000-fold excess) on GQN1 cleavage of G-loops in transcribed plasmids, normalized to 100% cleavage in the absence of competitor. (D) G4 DNA recognized by recombinant biotinylated Nucleolin-428/streptavidin gold beads. Arrows indicate beads bound at loops. Bar, 200 nm. (E) DMS footprinting analysis verifies the presence of G-quartets. (Left) Example of a DMS footprint of pRG4F (left, mock transcribed; right, transcribed). (Right) compiled results of six independent footprinting experiments showing ratio of DMS accessibility of each G-run in the transcribed sample relative to the untranscribed control. Diagram indicates positions of G-runs (boxes) and single Gs (lines), and direction of transcription (arrows). Bars, S.D.
We used an additional, independent method to verify G4 DNA formation in individual molecules. Although antibodies have been described that interact with G4 DNA (Brown et al. 1995, 1998), some recombinant proteins bind to G4 DNA with much higher affinity and specificity than any of these antibodies. One especially useful probe for G4 DNA is Nucleolin-428, a recombinant, truncated derivative of nucleolin that binds tightly (kD = 0.5 nM) and specifically to G4 DNA (Hanakahi et al. 1999). G-rich plasmids were transcribed, incubated with biotinylated Nucleolin-428, then with streptavidin-coated gold beads, and visualized by EM. This procedure clearly revealed Nucleolin-428/bead complexes decorating one arm of the loop (e.g., Fig. 4D). In control reactions, in which transcribed DNAs were incubated with streptavidin gold beads in the absence of Nucleolin-428, no loops were bound by beads, among >100 molecules visualized. Note that the arm of the loop containing the extended RNA/DNA hybrid appears thicker than the arm containing G4 DNA in the images shown in Figure 4D and in most other micrographs. Differential thickness of nucleic acid filaments has also been observed in experiments that used RNA/DNA hybrid formation to map transcribed regions (Chow and Broker 1989).
We also demonstrated formation of G4 DNA in transcribed G-rich DNA molecules by dimethylsulfate (DMS) footprinting. DMS footprinting has been used routinely to identify G-quartets and G4 DNA formed in synthetic oligonucleotides, because DMS attacks the N7 of guanine, which is accessible in single-stranded and duplex DNA but not if paired with the exocylic amino group of a neighboring guanine in a G-quartet (Sen and Gilbert 1988; Fig. 1A). Plasmid pRGF4 carries a short (104-bp) G-rich insert, in which runs of guanines that can participate in G4 DNA formation are interspersed with single guanines, which cannot. This plasmid was subjected to DMS footprint analysis after mock or bona fide transcription in vitro. Transcription resulted in a decrease of DMS sensitivity of guanines within G-runs; in six independent experiments, protection averaged 30% (Fig. 4E). DMS footprinting assays G-quartet formation throughout the population of molecules. Thus, foot-printing not only verifies the presence of G4 DNA within transcribed G-rich regions, but also further confirms that G4 DNA formation is efficient.
Identification of G4 DNA in plasmids propagated in E. coli
We next asked whether G4 DNA forms in vivo by propagating plasmids in an E. coli strain (NM232) lysogenic for the λDE3 prophage, which expresses T7 RNA polymerase under the control of the lactose repressor (Studier and Moffatt 1986). Transcription was induced by addition of IPTG, and low molecular weight circular DNA was recovered, deproteinized, linearized, and visualized by EM. No intact loops were evident in molecules recovered from that strain. E. coli RNase H digests the RNA strand of an RNA/DNA hybrid and could destabilize the loops; and E. coli RecQ helicase actively unwinds G4 DNA (Wu and Maizels 2001) and could eliminate G4 DNA that formed in vivo. We therefore tested recovery of loops from plasmids propagated in strain NM256 [AB1157 (λDE3) rnh::cat recQ::kan], which lacks both RNase H and RecQ helicase activities. Following induction of transcription in NM256, loops were readily recovered. In three independent experiments, intact loops were identified in 9%–18% of DNA molecules (e.g., Fig. 5A). A small fraction of molecules recovered from E. coli (3%) contained opened loops, which may have been broken within the cell or during DNA isolation. Loops formed by intracellular transcription were sensitive to GQN1 nuclease, which produced typical cleaved structures (e.g., Fig. 5B), and GQN1 cleaved essentially all loops in molecules recovered following intracellular transcription (Fig. 5C). G4 DNA therefore formed upon transcription of G-rich regions in vivo.
Figure 5.
Loops containing G4 DNA form in plasmids transcribed in E. coli. (A) Examples of loops formed in pPH600 (Sγ3) transcribed in E. coli strain NM 256 [AB1157 (λDE3) rnh::cat recQ::kan]. Bar, 200 nm. (B) Examples of GQN1 cleavage of loops formed in pPH600 (Sγ3) transcribed in E. coli NM 256. Arrows indicate broken loops. Bar, 200 nm. (C) Sensitivity of loops formed in E. coli to GQN1 cleavage. Loops were analyzed by EM and were scored as cleaved if they were opened and contained clearly visible DNA ends. (n) Number of molecules scored.
Results presented in Figures 4 and 5 show that G4 DNA can be detected within the characteristic loops formed upon transcription of a G-rich region. We call these novel structures G-loops, to emphasize that their structure is determined by the G-richness of the DNA, which allows formation of a stable RNA/DNA hybrid on the template strand and G4 DNA on the nontemplate strand.
The stability of the rG/dC base pair permits G-loop formation
What causes G-loops and G4 DNA to form? Loop formation requires a C-rich template strand (Table 1). A hybrid of G-rich RNA and C-rich DNA would be predicted to be quite stable, reflecting the high stability of the rG/dC base pair and stacking of adjacent guanines in the RNA (Sugimoto et al. 1995). Like guanine, inosine (I) base pairs with C, but an I/C pair contains only two hydrogen bonds and is therefore significantly less stable than a G/C pair. Therefore, we asked whether G-loops formed upon transcription in a reaction in which ITP was substituted for GTP. Substitution of ITP for GTP has been shown to reduce both transcriptional initiation and elongation, in part by enhancing polymerase pausing (Morgan and Shatkin 1980; Chanda et al. 1983; Shaevitz et al. 2003). Comparable to results reported by others, we found that transcription was reduced sixfold in ITP-containing reactions, as measured by net incorporation of 32P-labeled UTP; and although average transcript length was somewhat reduced, transcripts were sufficiently long to produce loops visible by EM if stable hybrids did form (data not shown). In parallel control reactions containing GTP, 60% of molecules contained loops after transcription. We examined 262 molecules recovered from ITP-containing reactions for the presence of loops, and observed no loops. Making the limiting assumption that the sixfold reduction in transcript levels in the ITP-containing reactions would cause a sixfold decrease in molecules containing loops, one would predict that loops would have been evident in 10% (one-sixth of 60%) of molecules in the ITP-containing reactions, or 26 molecules. As no loops were evident, we conclude that the stability of the rG/dC base pair is critical for transcription-dependent formation of G-loops.
G-loops form upon transcription of supercoiled, relaxed, or linear templates
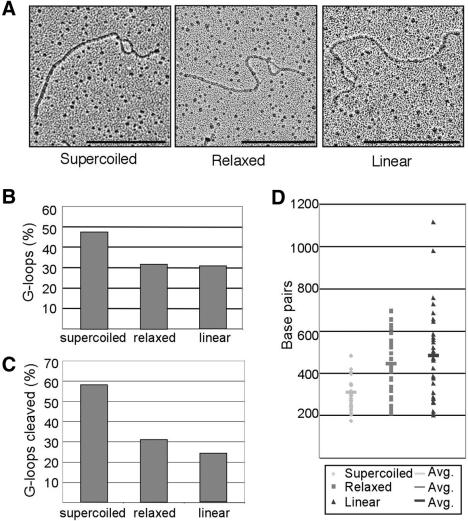
DNA supercoiling affects the potential for DNA denaturation and structure formation, and local supercoiling of DNA can vary in vivo (Wang 2002). We asked whether G-loop formation depends upon topology by comparing formation of loops and G4 DNA on supercoiled, relaxed, and linearized pPH600 plasmid templates. Loops were readily visualized on all these topoisomers (e.g., Fig. 6A). The efficiency of loop formation was determined by scoring loops in 400 molecules of each topoisomer, which showed that loops were present in 48% of supercoiled templates, 32% of relaxed circles, and 31% of linear molecules (Fig. 6B). Thus, loops form more efficiently on supercoiled templates. GQN1 sensitivity of loops was assayed by scoring broken loops in 250 molecules of each topoisome in parallel reactions in which GQN1 was included or omitted prior to visualization. GQN1 cleaved 58% of loops in supercoiled templates, 31% of loops in relaxed templates, and 24% of loops in linear templates (Fig. 6C). The greater sensitivity of G-loops in super-coiled templates to GQN1 cleavage may reflect more efficient formation of G4 DNA in supercoiled templates, or more efficient cleavage of supercoiled molecules by GQN1. The variation of loop size with topology was determined by measuring loop sizes in 35 molecules of each topoisomer. This showed that larger loops formed in the linear and relaxed circular templates (Fig. 6D), suggesting that loop size is constrained by super-coiling.
Figure 6.
Effect of topology on G-loop formation. (A) Examples of loops formed on supercoiled, relaxed, and linearized pPH600 templates. Bar, 500 nm. (B) Effect of plasmid topology on efficiency of loop formation. Loops were visualized by EM, and at least 400 molecules of each topoisomer were scored. (C) Effect of plasmid topology on G4 DNA formation. Templates of each topoisomer were treated with GQN1 and visualized by EM to assay cleavage; 200 GQN1-treated and 200-untreated molecules of each topoisomer were visualized. (D) Effect of plasmid topology on loop size. Loop size was measured following transcription of supercoiled, relaxed, or linear pPH600 templates, by EM visualization of 35 molecules of each topoisomer.
Discussion
We have shown that G4 DNA forms within G-rich regions in transcribed plasmid genomes in vitro and in vivo. In our experiments, we used EM to identify and characterize G-loops, novel structures produced upon transcription of G-rich regions in individual molecules. G-loops contain a stable RNA/DNA hybrid on one strand and G4 DNA on the other, as diagrammed in Figure 1C. G-loops form very efficiently in vivo. Following a brief induction of transcription in rapidly proliferating E. coli cells, from 9% to 18% of recovered plasmids contained G-loops, and essentially all G-loops contained G4 DNA, as assayed by GQN1 sensitivity (Fig. 5C). This is the first direct evidence for the formation of G4 DNA in living cells, and it resolves the long-standing question of whether G4 DNA can form in vivo. Pathways of genomic expression and maintenance are remarkably conserved throughout evolution, and identification of G4 DNA in E. coli provides a compelling precedent for the likely biological importance of G4 DNA in higher cells.
Analysis of structures produced upon in vitro transcription provided insights into the parameters that govern G-loop and G4 DNA formation. Following in vitro transcription of a supercoiled template, loops were evident in 42%–63% of molecules (Table 1; Fig. 6B), and 25%–58% of loops contained G4 DNA, as assayed by GQN1 cleavage (Figs. 4A,B, 6B). G-loops and G4 DNA also formed within relaxed and linear templates (Fig. 6B,C). Formation of G-loops required a C-rich template strand and a G-rich nontemplate strand. G-loops did not form in a sequence of random base composition, or in G-rich sequences transcribed in opposite orientation (Table 1). Other laboratories have shown that RNA/DNA hybrids form in transcribed G-rich DNAs from the immunoglobulin S regions (Reaban and Griffin 1990; Reaban et al. 1994; Daniels and Lieber 1995; Tian and Alt 2000; Mizuta et al. 2003; Yu et al. 2003), S. cerevisiae mitochondrial DNA (Xu and Clayton 1995), and the E. coli rrnB operon (Masse et al. 1997). G-loops did not form in reactions in which ITP is substituted for GTP, so loop formation appears to depend upon the stability of the rG/dC base pair. The RNA/DNA hybrid within a G-loop forms cotranscriptionally, not posttranscriptionally, because the hybrids were unaffected by the presence of RNase A in the transcription reaction (Fig. 3C).
We refer to the loops formed upon transcription of a G-rich region as “G-loops” to distinguish them from R-loops, a term used to describe any stable RNA/DNA hybrid formed within duplex DNA (Thomas et al. 1976; Chow and Broker 1989). This distinction is important. G-loops form cotranscriptionally, whereas the RNA/DNA hybrid in an R-loop may be formed either cotranscriptionally or as a result of invasion of DNA by an RNA molecule that was synthesized on the same or a different template (Thomas et al. 1976). Furthermore, G-loop formation depends upon the base composition of the template, which allows an RNA/DNA hybrid to form on one strand and G4 DNA on the other, whereas R-loop formation is independent of base composition.
A G-loop resembles the transcription bubble that advances with RNA polymerase along the DNA template, because it contains an RNA/DNA hybrid and the displaced nontemplate strand. However, the transcription bubble spans 17 ± 1 bp of DNA, as estimated by topological approaches (Gamper and Hearst 1982, 1983), whereas G-loops are much larger, ranging from 150 bp (the lower limit for EM visualization) to 500 bp in length. More importantly, unlike the RNA/DNA hybrid within the advancing transcription bubble, the hybrid within a G-loop is very unlikely to reflect persistent interactions between the nascent RNA and the DNA template strand. In both single- and multisubunit RNA polymerases, the nascent RNA is forced to exit through a topological tunnel, making it unlikely that an RNA polymerase could generate a long RNA/DNA hybrid simply by failing to pry apart the RNA/DNA hybrid within the advancing transcription bubble. Structural and biochemical analyses of T7 polymerase elongation complexes show that a specific α-helix within the T7 polymerase thumb domain pries the transcript from the template, forcing it through a tunnel that shields the newly made RNA from the template DNA strand (Huang and Sousa 2000; Tahirov et al. 2002; Yin and Steitz 2002, 2004; for review, see Steitz 2004). The nascent RNA/template hybrid in the T7 elongation complex is 8 bp in length, and the tunnel shields an additional 4 nt of RNA from the DNA template (Huang and Sousa 2000; Tahirov et al. 2002; Yin and Steitz 2002). The multisubunit RNA polymerases also use a specific structure, the rudder, to pry off the nascent transcript, which is then forced through an exit pore to prevent reassociation with the template (Gnatt et al. 2001; Westover et al. 2004). In light of this understanding of RNA polymerases, a plausible model for cotranscriptional formation of G-loops would be that the elongating polymerase pries apart the RNA/DNA hybrid as usual and forces the RNA out through the tunnel, but that the extruded RNA invades the duplex to initiate G-loop formation. The G-loop would then increase in size behind the advancing transcription bubble, as RNA exiting from the tunnel hybridizes with the template. In this model, only a very short stretch of RNA would not be stably hybridized, consistent with the observed insensitivity of G-loop formation to the presence of RNase A in the transcription reaction (Fig. 3C); proximity to the polymerase might help protect this short stretch of RNA from nuclease attack. Formation of a stable RNA/DNA hybrid within the G-loop will leave unpaired guanines in the nontemplate strand free to associate into G-quartets and intramolecular G4 DNA structures. The presence of stable G4 DNA in the nontemplate strand might then further impede renaturation of the DNA duplex.
The fact that both single- and multisubunit RNA polymerases have evolved to carry specific structures that separate nascent transcripts from the template DNA suggests that a newly made transcript has considerable tendency to invade the template DNA. In support of this, deficiencies in subunits of THO/TREX complex in S. cerevisiae are associated with reduced efficiency of transcriptional elongation, enhanced recombination between flanking direct repeats, and an increased accumulation of RNA/DNA hybrids (Huertas and Aguilera 2003). THO/TREX is essential for production of specific classes of transcripts, particularly long or GC-rich RNAs (Chavez et al. 2001). The THO/TREX complex is conserved (Strasser et al. 2002), suggesting that transcript release may therefore be an important and potentially regulatable step in RNA processing in many organisms.
Potential biological significance of G4 DNA
Our experiments have documented the formation of G4 DNA upon transcription of G-rich sequences derived from two chromosomal domains, the immunoglobulin S regions and the TTAGGG telomeric repeat. There have been numerous suggestions that the regulated formation of G4 DNA may contribute to telomere biology, for example, by preventing degradation of the telomere tails or by inhibiting telomere replication by telomerase (Rhodes and Giraldo 1995; Rezler et al. 2002; Neidle and Parkinson 2003). For immunoglobulin class switch recombination to occur in an activated B cell, both S regions targeted for recombination must be transcribed in a unique orientation, using the C-rich strand as the transcriptional template (Shinkura et al. 2003). This is the orientation that supports formation of G-loops and G4 DNA, and regulated formation of G4 DNA could create a structural target for specific factors involved in DNA recombination. Nucleases and helicases that attack G4 DNA could participate in such a structure-specific recombination pathway.
Other G-rich sequence motifs would also be predicted to form G4 DNA upon transient denaturation of the DNA duplex. These include the rDNA, which is transcribed using the C-rich strand as template, G-rich minisatellites, and G-rich regions within single copy genes. Unregulated formation of G4 DNA within these regions could lead to genetic instability. One example of instability specific to G-rich genomic regions has been documented in the nematode, where large deletions occur at long polyguanine tracts in lines mutant in dog-1 (Cheung et al. 2002). This gene encodes a putative helicase, which is thought to function to unwind structures that form during lagging strand replication. Other examples are evident in the human genome, where some of the most unstable of the variable number tandem repeats (VNTRs) are G-rich sequences capable of forming G4 DNA include the minisatellites D1S7 (MS1); D4S43; MS32, CEB1, D1Z2; MS205; and the insulin-linked hypervariable repeat (Buroker et al. 1987; Wong et al. 1987; Jeffreys et al. 1988; Weitzmann et al. 1997). New variants at these highly unstable loci arise due to complex recombination events such as intra-allelic rearrangement and interallelic gene conversions (Berg et al. 2003). Instability of these sequences cannot be ascribed to replication slippage due to hairpin formation, because these repeats do not form stable hairpins. However, they are strongly predicted to form G4 DNA.
The methods we have developed allow G4 DNA to be inducibly generated in vitro and in living cells. We anticipate that further study of G-rich sequences will enhance our understanding of the dynamics of G4 DNA formation and its functions in vivo.
Materials and methods
Plasmid templates and transcription
pHumtel contains a 810-bp fragment of the TTAGGG mammalian telomeric repeat (Zhong et al. 1992) in the pSP73 (Promega) backbone. Other plasmid templates were pBluescript KS+ (Stratagene) derivatives. pRX15F contains a 350-bp insert comprised of 14 iterations of the 25-mer murine immunoglobulin heavy chain Sμ consensus repeat, GCTGAGCTGGGGTG AGCTGA; pRX15R is identical to pRX15F, except that the insert orientation is reversed; pRX15F3 contains a 1.1-kb insert comprised of 42 iterations of this same Sμ consensus repeat; pPH600 contains a 604-bp PvuII–HindIII fragment of the murine Sγ3 switch region; and pTT1 contains a 550-bp insert spanning the murine Vλ1 region (Elenich et al. 1996). pRG4F carries a small (104-bp) insert comprised of four repeats of a 26-bp sequence containing two runs of four Gs with a lone G within the region separating each G-run, GGGGTCAGCTAGGGGATTC TAGACTA. This facilitates footprinting, because the Gs within runs can participate in G4 DNA formation, whereas the lone Gs cannot, and thus provide an internal control for normalizing the signal within a lane. All buffers contained at least 20 mM KCl to maintain stability of G4 DNA.
Plasmids were prepared for in vitro transcription using the Qiagen maxiprep procedure (Qiagen). Transcription was carried out for 15 min at 37°C in reactions containing 40 μg/mL supercoiled plasmid DNA, 1 mM each NTP, and 50 U/mL T7 RNA polymerase (NEB) in the manufacturer's buffer supplemented with 40 mM KCl. Free RNA was digested by incubation with 20 μg/mL of RNase A for 15 min at 37°C. RNase H digestion was carried out at 37°C for 30 min with 17 U/mL of RNase H (Roche Molecular Biochemicals). For transcript labeling, digoxygenin (Dig)-UTP (Roche Molecular Biochemicals) was included at the ratio 1:2 Dig-UTP: UTP, and DNA incubated with 3.6 μg/mL of anti-Dig gold beads (5 nm; Ted Pella, Inc.) in 5 mM EDTA (pH 8.0), 100 mM NaCl at 37°C for 1 h, and cross-linked in 1% glutaraldehyde for 30 min at 37°C before spreading. Relaxed templates were produced by incubation of 1.5 μg of plasmid DNA and 6 U Topoisomerase I (Invitrogen) in a 50-μL reaction containing 50 mM Tris-Cl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA, 30 μg/mL BSA for 2 h at 37°C; topology was verified by chloroquine gel electrophoresis. DNA was diluted 1:10 into restriction buffer and linearized with AflIII (NEB), which cuts at a unique restriction site either upstream (pSP73 backbone) or downstream (pBluescript backbone) of the T7 promoter. DNA was purified by phenol/chloroform extraction followed by ethanol precipitation and two 70% ethanol washes.
G4 DNA formation in vivo was studied in plasmids propagated in NM232 [E. coli AB1157 (λDE3)] and NM256 [E. coli AB1157 (λDE3) rnh::cat recQ::kan]. These strains were constructed by lysogenization of E. coli AB1157 and AB1157 recQ::kan (Mendonca et al. 1993) with the λDE3 prophage (Novagen), which expresses T7 RNA polymerase under control of the lactose repressor, followed by P1 transduction of the rnh::cat marker from E. coli strain MIC1020 (Itaya and Crouch 1991). Expression of T7 polymerase was induced by addition of 2 mM IPTG to log-phase cells, and 5 min later, cultures were chilled for 20 min on ice, 5 mL of cells were pelleted, and plasmids isolated by a neutral lysis procedure to avoid altering DNA structures. Cells were resuspended in 0.5 mL of 10 mM Tris-Cl, 1 mM EDTA at pH 8.0 (TE) containing 2 mg/mL of lysozyme and 20 μg/mL of RNaseA, incubated 5 min at 4°C, then 1 mL of lysis buffer (0.5 M potassium acetate, 75 mM EDTA, 44 mM sodium hydroxide, 1% SDS at pH 5.3) was added, the mixture gently swirled and cleared by centrifugation at 4°C for 20 min at 13 K. Plasmid DNA was purified over a nucleobond AX20 column (BD Biosciences-Clontech miniprep kit).
GQN1 cleavage and Nucleolin-428 binding
GQN1 cleavage was carried out in 25 mM Hepes-NaOH (pH 7.5), 5 mM MgCl2, 50 mM KCl, 10 mM DTT, 7.5% glycerol, 10 μg/mL transcribed plasmid DNA, for 30 min at 37°C (Sun et al. 2001). Following GQN1 treatment, reactions were deproteinized with 1 mg/mL of proteinase K at 50°C for 20 min, extracted with phenol/chloroform, precipitated with ethanol, and resuspended in sterile water. Loops were scored as cleaved if they were opened and an arm or end was clearly visible. Competitor DNAs added were G4 DNA formed from the TP oligonucleotide (Sun et al. 1998); or single-stranded HMD3 oligonucleotide (AG GACTTCTGGTCCCTACATTCCCACCTTCCCCAGCTCCC CCATAGCTGC). Recombinant Nucleolin-428 (Hanakahi et al. 1999) was biotinylated with the Sulfo-NHS biotinylation kit according to the manufacturer's protocol (Pierce), and transcribed plasmid DNA was incubated with 15 nM biotinylated Nucleolin-428 in 20 mM Hepes-NaOH (pH 7.4), 100 mM KCl, 0.2 mg/mL of BSA, 1 mM DTT at 37°C for 30 min, cross-linked in 1% glutaraldehyde for 30 min at 37°C, DNA linearized, purified, and incubated with 1 μg/mL of strepatividin gold beads (10 nm; Ted Pella, Inc.) in TE containing 50 mM NaCl for 30 min at 37°C, then spread.
Transmission electron microscopy
DNA was spread in 40% formamide (Electron Microscopy Sciences), 0.1 mg/mL cytochrome C (Sigma-Aldrich), 100 mM Tris-Cl, 10 mM EDTA at pH 8.5 (Kleinschmidt 1968). DNA was lifted onto copper grids coated with 2% parlodion in amylacetate (Electron Microscopy Sciences), stained with uranyl acetate, shadowed with evaporated platinum/palladium, and further stabilized with evaporated carbon. DNA was visualized using a JEOL 1010 transmission electron microscope at 40 kV. Images were captured using a Gatan ultrascan camera and acquired using Gatan Digital Micrograph software. DNA lengths were measured using NIH Image 1.62 software.
Dimethylsulfate footprinting
Footprinting with DMS (Sigma-Aldrich) was carried out as described (Eversole and Maizels 2000), except that 50 mM KCl was included in all buffers. Transcribed DNAs were treated with RNase A prior to addition of DMS. Following the removal of DMS, DNA was heated at 65°C for 20 min in water, and treated with RNase H (17 U/mL) and PstI (NEB) for 30 min at 37°C in NEB3 buffer, and then with BstXI (NEB) at 55°C for 1 h to excise the insert. DNA was precipitated with ammonium acetate and then ethanol, washed three times, dried, and then cleaved with piperidine. DNA was electrotransferred to Hybond N+ nylon membrane (Amersham Pharmacia Biotech), cross-linked using the autocross-link option on a Stratalinker 2400 (Stratagene), and hybridized with 32P-labeled oligonucleotide probes complementary to unique sequences immediately 5′ (HMD35, TG CAGCCCGACCCCTAGT) or 3′ (HMD33, GCCGCCACCGC GAGATCTCC) of the G-rich insert at 60°C for 4 h in 0.34 M Na2HPO4, 0.16 M NaH2PO4, 1 mM EDTA (pH 8.0), 7% SDS, and 5 μg/mL BSA. Membranes were washed and scanned by PhosphorImager, and signals quantified using Imagequant Software (Amersham Pharmacia Biotech). DMS accessibility was determined by first normalizing signals from each G-run to the adjacent lone G, and then calculating the ratio of the normalized signals at each G-run in the transcribed and untranscribed samples.
Acknowledgments
We thank Bob Crouch and Steve Matson for providing E. coli strains and Gerry Smith and other colleagues for valuable discussions. This research was supported by NIH R01 grants GM39799 and GM65988 to N.M.; A.F.T. was supported by NIH GM31693 (P.I., Gerald R. Smith); and M.D. was supported by NIH Genetics Training Program T32 HD07149.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1200804.
References
- Arakawa H., Iwasato, T., Hayashida, H., Shimizu, A., Honjo, T., and Yamagishi, H. 1993. The complete murine immunoglobulin class switch region of the α heavy chain gene-hierarchic repetitive structure and recombination breakpoints. J. Biol. Chem. 268: 4651-4655. [PubMed] [Google Scholar]
- Bachrati C.Z. and Hickson, I.D. 2003. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem. J. 374: 577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg I., Neumann, R., Cederberg, H., Rannug, U., and Jeffreys, A.J. 2003. Two modes of germline instability at human minisatellite MS1 (locus D1S7): Complex rearrangements and paradoxical hyperdeletion. Am. J. Hum. Genet. 72: 1436-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R.P. and Wells, R.D. 2001. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 66: 159-202. [DOI] [PubMed] [Google Scholar]
- Brown B.A., Lin, Y., Roberts, J.F., and Hardin, C.C. 1995. Antibodies specific for the DNA quadruplex [d(CGC G4 GCG)4] isolated from autoimmune mice. Nucleic Acids Symp Ser 33: 134-136. [PubMed] [Google Scholar]
- Brown B.A., Li, Y., Brown, J.C., Hardin, C.C., Roberts, J.F., Pelsue, S.C., and Shultz, L.D. 1998. Isolation and characterization of a monoclonal anti-quadruplex DNA antibody from autoimmune `viable motheaten' mice. Biochemistry 37: 16325-16337. [DOI] [PubMed] [Google Scholar]
- Buroker N., Bestwick, R., Haight, G., Magenis, R.E., and Litt, M. 1987. A hypervariable repeated sequence on human chromosome 1p36. Hum. Genet. 77: 175-181. [DOI] [PubMed] [Google Scholar]
- Chanda P.K., Roy, J., and Banerjee, A.K. 1983. In vitro synthesis of genome length complementary RNA of vesicular stomatitis virus in the presence of inosine 5′-triphosphate. Virology 129: 225-229. [DOI] [PubMed] [Google Scholar]
- Chavez S., Garcia-Rubio, M., Prado, F., and Aguilera, A. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I., Schertzer, M., Rose, A., and Lansdorp, P.M. 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 31: 405-409. [DOI] [PubMed] [Google Scholar]
- Chow L.T. and Broker, T.R. 1989. Mapping the genetic organization of RNA by electron microscopy. Meth. Enzymol. 180: 239-261. [DOI] [PubMed] [Google Scholar]
- Cummings C.J. and Zoghbi, H.Y. 2000. Trinucleotide repeats: Mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 1: 281-328. [DOI] [PubMed] [Google Scholar]
- Daniels G.A. and Lieber, M.R. 1995. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: Implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 23: 5006-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W., Hertz, G.Z., Scappino, L., and Gritzmacher, C. 1993. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 21: 365-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenich L.A., Ford, C.S., and Dunnick, W.A. 1996. The γ1 heavy chain gene includes all of the cis-acting elements necessary for expression of properly regulated germ-line transcripts. J. Immunol. 157: 176-182. [PubMed] [Google Scholar]
- Eversole A. and Maizels, N. 2000. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol. 20: 5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M. and Loeb, L.A. 1994. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. 91: 4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H.B. and Hearst, J.E. 1982. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell 29: 81-90. [DOI] [PubMed] [Google Scholar]
- ____. 1983. Size of the unwound region of DNA in Escherichia coli RNA polymerase and calf thymus RNA polymerase II ternary complexes. Cold Spring Harb. Symp. Quant. Biol. 47: 455-461. [DOI] [PubMed] [Google Scholar]
- Gellert M., Lipsett, M.N., and Davies, D.R. 1962. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. 48: 2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.E. and Feigon, J. 1999. Multistranded DNA structures. Curr. Opin. Struct. Biol. 9: 305-314. [DOI] [PubMed] [Google Scholar]
- Gnatt A.L., Cramer, P., Fu, J., Bushnell, D.A., and Kornberg, R.D. 2001. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science 292: 1876-1882. [DOI] [PubMed] [Google Scholar]
- Hanakahi L.A., Sun, H., and Maizels, N. 1999. High affinity interactions of nucleolin with G-G paired rDNA. J. Biol. Chem. 274: 15908-15912. [DOI] [PubMed] [Google Scholar]
- Harrigan J.A., Opresko, P.L., von Kobbe, C., Kedar, P.S., Prasad, R., Wilson, S.H., and Bohr, V.A. 2003. The Werner syndrome protein stimulates DNA polymerase β strand displacement synthesis via its helicase activity. J. Biol. Chem. 278: 22686-22695. [DOI] [PubMed] [Google Scholar]
- Hickson I.D. 2003. RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer 3: 169-178. [DOI] [PubMed] [Google Scholar]
- Hsu C.L. and Stevens, A. 1993. Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. and Sousa, R. 2000. T7 RNA polymerase elongation complex structure and movement. J. Mol. Biol. 303: 347-358. [DOI] [PubMed] [Google Scholar]
- Huertas P. and Aguilera, A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711-721. [DOI] [PubMed] [Google Scholar]
- Itaya M. and Crouch, R.J. 1991. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol. Gen. Genet. 227: 424-432. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Royle, N.J., Wilson, V., and Wong, Z. 1988. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332: 278-281. [DOI] [PubMed] [Google Scholar]
- Khakhar R.R., Cobb, J.A., Bjergbaek, L., Hickson, I.D., and Gasser, S.M. 2003. RecQ helicases: Multiple roles in genome maintenance. Trends Cell Biol. 13: 493-501. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt A. 1968. Monolayer techniques in electron microscopy of nucleic acid molecules. Meth. Enzymol. 12b: 361-377. [Google Scholar]
- Kleinschmidt A. and Zahn, R. 1959. Naturforsch 14b: 770. [Google Scholar]
- Kolodner R.D. 1995. Mismatch repair: Mechanisms and relationship to cancer susceptibility. Trends Biochem. Sci. 20: 397-401. [DOI] [PubMed] [Google Scholar]
- Liu Z. and Gilbert, W. 1994. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: Implication of in vivo functions for this novel DNA structure. Cell 77: 1083-1092. [DOI] [PubMed] [Google Scholar]
- Masse E., Phoenix, P., and Drolet, M. 1997. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J. Biol. Chem. 272: 12816-12823. [DOI] [PubMed] [Google Scholar]
- McMurray C.T. 1999. DNA secondary structure: A common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. 96: 1823-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca V.M., Kaiser-Rogers, K., and Matson, S.W. 1993. Double helicase II (uvrD)-helicase IV (helD) deletion mutants are defective in the recombination pathways of Escherichia coli. J. Bacteriol. 175: 4641-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta R., Iwai, K., Shigeno, M., Mizuta, M., Ushiki, T., and Kitamura, D. 2003. Molecular visualization of immunoglobulin switch region RNA/DNA complex by atomic force microscope. J. Biol. Chem. 278: 4431-4434. [DOI] [PubMed] [Google Scholar]
- Modrich P. and Lahue, R. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101-133. [DOI] [PubMed] [Google Scholar]
- Mohaghegh P., Karow, J.K., Brosh Jr., R.M., Bohr, V.A., and Hickson, I.D. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29: 2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M.A. and Shatkin, A.J. 1980. Initiation of reovirus transcription by inosine 5′-triphosphate and properties of 7-methylinosine-capped, inosine-substituted messenger ribonucleic acids. Biochemistry 19: 5960-5966. [DOI] [PubMed] [Google Scholar]
- Nakayama H. 2002. RecQ family helicases: Roles as tumor suppressor proteins. Oncogene 21: 9008-9021. [DOI] [PubMed] [Google Scholar]
- Neidle S. and Parkinson, G.N. 2003. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 13: 275-283. [DOI] [PubMed] [Google Scholar]
- Parkinson G.N., Lee, M.P., and Neidle, S. 2002. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417: 876-880. [DOI] [PubMed] [Google Scholar]
- Reaban M.E. and Griffin, J.A. 1990. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348: 342-344. [DOI] [PubMed] [Google Scholar]
- Reaban M.E., Lebowitz, J., and Griffin, J.A. 1994. Transcription induces the formation of a stable RNA-DNA hybrid in the immunoglobulin a switch region. J. Biol. Chem. 269: 21850-21857. [PubMed] [Google Scholar]
- Rezler E.M., Bearss, D.J., and Hurley, L.H. 2002. Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol. 2: 415-423. [DOI] [PubMed] [Google Scholar]
- Rhodes D. and Giraldo, R. 1995. Telomere structure and function. Curr. Opin. Struct. Biol. 5: 311-322. [DOI] [PubMed] [Google Scholar]
- Sakamoto N., Chastain, P.D., Parniewski, P., Ohshima, K., Pandolfo, M., Griffith, J.D., and Wells, R.D. 1999. Sticky DNA: Self-association properties of long GAA · TTC repeats in R · R · Y triplex structures from Friedreich's ataxia. Mol. Cell 3: 465-475. [DOI] [PubMed] [Google Scholar]
- Sen D. and Gilbert, W. 1988. Formation of parallel four-stranded complexes by guanine rich motifs in DNA and its implications for meiosis. Nature 334: 364-366. [DOI] [PubMed] [Google Scholar]
- ____. 1990. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 344: 410-414. [DOI] [PubMed] [Google Scholar]
- Shaevitz J.W., Abbondanzieri, E.A., Landick, R., and Block, S.M. 2003. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426: 684-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J.C. and Loeb, L.A. 2000. The Werner syndrome gene: The molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 16: 213-220. [DOI] [PubMed] [Google Scholar]
- Shinkura R., Tian, M., Smith, M., Chua, K., Fujiwara, Y., and Alt, F.W. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 4: 435-441. [DOI] [PubMed] [Google Scholar]
- Sia E.A., Dominska, M., and Petes, T.D. 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21: 8157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R.R. 1999. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 64: 346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T.A. 2004. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 14: 4-9. [DOI] [PubMed] [Google Scholar]
- Strand M., Prolla, T.A., Liskay, R.M., and Petes, T.D. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. 365: 274-276. [DOI] [PubMed] [Google Scholar]
- Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304-308. [DOI] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt, B.A. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189: 113-130. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Nakano, S., Katoh, M., Matsumura, A., Nakamuta, H., Ohmichi, T., Yoneyama, M., and Sasaki, M. 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211-11216. [DOI] [PubMed] [Google Scholar]
- Sun H., Karow, J.K., Hickson, I.D., and Maizels, N. 1998. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273: 27587-27592. [DOI] [PubMed] [Google Scholar]
- Sun H., Bennett, R.J., and Maizels, N. 1999. The S. cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 27: 1978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Yabuki, A., and Maizels, N. 2001. A human nuclease specific for G4 DNA. Proc. Natl. Acad. Sci. 89: 12444-12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov T.H., Temiakov, D., Anikin, M., Patlan, V., McAllister, W.T., Vassylyev, D.G., and Yokoyama, S. 2002. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature 420: 43-50. [DOI] [PubMed] [Google Scholar]
- Thomas M., White, R.L., and Davis, R.W. 1976. Hybridization of RNA to double-stranded DNA: Formation of R-loops. Proc. Natl. Acad. Sci. 73: 2294-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M. and Alt, F.W. 2000. Transcription-induced cleavage of immunogloublin switch regons by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 275: 24163-24172. [DOI] [PubMed] [Google Scholar]
- Wang J.C. 2002. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell. Biol. 3: 430-440. [DOI] [PubMed] [Google Scholar]
- Weitzmann M.N., Woodford, K.J., and Usdin, K. 1997. DNA secondary structures and the evolution of hypervariable tandem arrays. J. Biol. Chem. 272: 9517-9523. [DOI] [PubMed] [Google Scholar]
- Westover K.D., Bushnell, D.A., and Kornberg, R.D. 2004. Structural basis of transcription: Separation of RNA from DNA by RNA polymerase II. Science 303: 1014-1016. [DOI] [PubMed] [Google Scholar]
- Wong Z., Wilson, V., Patel, I., Povey, S., and Jeffreys, A.J. 1987. Characterization of a panel of highly variable minisatellites cloned from human DNA. Ann. Hum. Gene. 51: 269-288. [DOI] [PubMed] [Google Scholar]
- Wu X. and Maizels, N. 2001. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 29: 1765-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B. and Clayton, D.A. 1995. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell. Biol. 15: 580-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y.W. and Steitz, T.A. 2002. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298: 1387-1395. [DOI] [PubMed] [Google Scholar]
- ____. 2004. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116: 393-404. [DOI] [PubMed] [Google Scholar]
- Yu K., Chedin, F., Hsieh, C.L., Wilson, T.E., and Lieber, M.R. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4: 442-251. [DOI] [PubMed] [Google Scholar]
- Zhong Z., Shiue, L., Kaplan, S., and de Lange, T. 1992. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 12: 4834-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]