Abstract
Bardet-Biedl syndrome (BBS) is a genetically heterogeneous developmental disorder whose molecular basis is largely unknown. Here, we show that mutations in the Caenorhabditis elegans bbs-7 and bbs-8 genes cause structural and functional defects in cilia. C. elegans BBS proteins localize predominantly at the base of cilia, and like proteins involved in intraflagellar transport (IFT), a process necessary for cilia biogenesis and maintenance, move bidirectionally along the ciliary axoneme. Importantly, we demonstrate that BBS-7 and BBS-8 are required for the normal localization/motility of the IFT proteins OSM-5/Polaris and CHE-11, and to a notably lesser extent, CHE-2. We propose that BBS proteins play important, selective roles in the assembly and/or function of IFT particle components. Our findings also suggest that some of the cardinal and secondary symptoms of BBS, such as obesity, diabetes, cardiomyopathy, and learning defects may result from cilia dysfunction.
Keywords: Bardet-Biedl syndrome, BBS proteins, cilia and flagella, Caenorhabditis elegans, basal body, intraflagellar transport
Bardet-Biedl syndrome (BBS) is a rare genetic disorder characterized by a pleiotropic phenotype that includes obesity, rod-cone dystrophy, renal malformations, polydactyly, cognitive impairment, and several other ailments, including congenital heart defects and diabetes (Katsanis et al. 2001). Although six genes associated with BBS have been identified, their sequences have not illuminated the molecular and cellular etiology of the disease. The BBS1, BBS2, and BBS7 genes encode proteins with predicted β-propeller domains (Nishimura et al. 2001; Mykytyn et al. 2002; Badano et al. 2003), which are prevalent supersecondary structures that have multiple functions (Jawad and Paoli 2002). BBS4 (Mykytyn et al. 2001), as well as the newly identified BBS8 protein (Ansley et al. 2003), contain several of the common tetratricopeptide repeats (TPRs) that are involved with protein–protein interactions in proteins displaying various cellular roles (Blatch and Lassle 1999). Lastly, BBS6 encodes a putative molecular chaperone related to the Hsp60/chaperonin family (Katsanis et al. 2000; Slavotinek et al. 2000; Stone et al. 2000). Chaperones assist the biogenesis (e.g., folding, assembly) of many polypeptides (Stirling et al. 2003), but the lack of knowledge regarding the protein substrate(s) of BBS6 precludes any understanding of its cellular function(s).
Although the molecular basis of BBS is unclear, we recently proposed (Ansley et al. 2003) that it may be linked to defects in basal bodies, which are modified centrioles found at the base of cilia and flagella. This hypothesis is supported by several indirect observations. First, mammalian BBS8 is associated with centriolar structures and interacts with PCM-1 (Ansley et al. 2003), a protein that is likely involved in basal body function (Kubo et al. 1999). Second, all four known Caenorhabditis elegans homologs of human BBS genes (bbs-1, bbs-2, bbs-7, and bbs-8) are expressed exclusively in a small subset of cells (sensory neurons) harboring cilia; moreover, these nematode genes all contain DNA-binding sites (X boxes) for DAF-19 (Ansley et al. 2003), a transcription factor required for the expression of several cilia-specific genes in the nematode (Swoboda et al. 2000). Mammalian BBS8 also displays a restricted distribution in tissues consistent with an important role in cilia function (Ansley et al. 2003). Lastly, some of the phenotypes exhibited by BBS patients, including retinal degeneration, skeletal anomalies, situs inversus (left–right axis determination defect), and renal cysts/malformations, bear resemblance to human diseases associated with abnormal cilia function (Pazour and Rosenbaum 2002; Ansley et al. 2003; Zhang et al. 2003).
Interestingly, ciliary diseases can arise from defects in intraflagellar transport (IFT), a microtubule-dependent motility process involving the kinesin-dependent anterograde and dynein-driven retrograde movement of protein complexes (IFT particles and associated cargo), or IFT rafts, between the basal body and distal tips of cilia and flagella (Pazour and Rosenbaum 2002; Rosenbaum and Witman 2002; Scholey 2003). IFT plays a critical role in trafficking cargo and is essential for eukaryotic cilia formation and function (Rosenbaum and Witman 2002; Scholey 2003; Qin et al. 2004). IFT particles are composed of at least 16 polypeptides, which assemble into two complexes, A and B (Piperno and Mead 1997; Cole et al. 1998; Cole 2003). The motor–particle–cargo complex is believed to be loaded onto the ciliary axoneme via transitional fibers emanating from the basal body (Deane et al. 2001). As part of the changeover between anterograde and retrograde transport, the motor–particle–cargo complexes appear to be structurally remodeled at the base and tip of the cilium (Iomini et al. 2001).
One of the best-characterized IFT particle components is the complex B protein, Chlamydomonas IFT-88 or C. elegans OSM-5, whose mutations result in compromised IFT and ciliary defects (Pazour et al. 2000; Haycraft et al. 2001; Qin et al. 2001). Although the mammalian counterpart, Polaris/Tg737, is an essential gene, hypomorphic variants in mice result in many of the phenotypes exhibited by BBS patients (Moyer et al. 1994; Taulman et al. 2001; Pazour et al. 2002; Rosenbaum and Witman 2002; Ansley et al. 2003). It is notable that at least some of the phenotypes associated with IFT protein dysfunction likely result from their involvement in hedgehog signaling in vertebrates (Huangfu et al. 2003). Hedgehog signal transduction is necessary for the proper development and differentiation of many tissue types, and at least two diseases linked to deregulated hedgehog signaling, Palister-Hall syndrome and Smith-Lemli-Opitz syndrome, share polydactyly as well as renal and heart anomalies with Bardet-Biedl syndrome (Bale 2002).
To investigate the potential link between Bardet-Biedl syndrome and the abnormal function of basal bodies and/or cilia, we sought to examine the cellular functions of BBS proteins in C. elegans. We demonstrate that loss-of-function mutations in bbs-7 and bbs-8 result in structural and functional ciliary defects, including shortened/abnormal cilia and chemosensory abnormalities. As with IFT proteins, the BBS proteins localize predominantly at the base of cilia and undergo bidirectional movement along the ciliary axoneme. Our results indicate that BBS-7 and BBS-8 represent members of a novel class of proteins that partake in, and are required for intraflagellar transport.
Results
BBS proteins localize to the base of cilia and undergo IFT-like movement
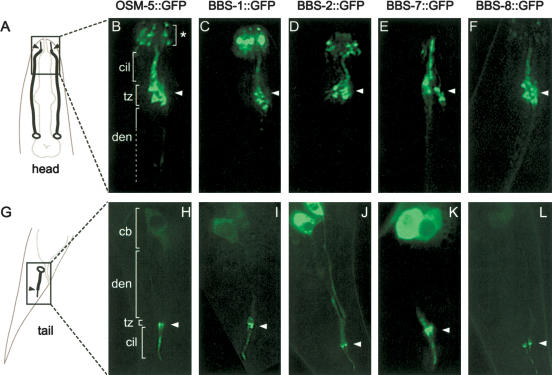
To examine the possible association of C. elegans BBS proteins with ciliary structures, we first determined the cellular localization of GFP-tagged BBS-1, BBS-2, BBS-7, and BBS-8. Expression of the four translational bbs::gfp transgenes in wild-type (N2) worms produced specific GFP signals at the ciliary transition zones (denoted by tz and arrowheads) and along the ciliary axonemes (cil) of both head (Fig. 1C–F) and tail (Fig. 1I–L) ciliated neurons. Transition zones in C. elegans are analogous to basal bodies and consist of microtubule-rich constrictions at the base of cilia (Perkins et al. 1986). Interestingly, the localization of the BBS::GFP proteins overlaps with that of IFT proteins, including OSM-5 (Fig. 1B,H; Haycraft et al. 2001; Qin et al. 2001), OSM-6 (data not shown; Collet et al. 1998), XBX-1 (Schafer et al. 2003), and CHE-13 (Haycraft et al. 2003), all of which show prominent localization at the transition zones and along the ciliary axonemes of ciliated neurons. Discrete GFP fluorescence signals corresponding to transition zones of other neurons, including inner and outer labial neurons, were often observed closer to the anterior end of the animal (Fig. 1B–F; denoted by an asterisk in B).
Figure 1.
C. elegans BBS proteins localize predominantly to the transition zones and axonemes of cilia. (A,G) Schematics of the head (A) and tail (G) indicate the position of cilial structures at the dendritic endings of amphid and phasmid neurons. (B–F,H–L) Fluorescent images of the head (amphids, B–F) and tail (phasmids, H–L) regions of N2 worms expressing GFP-tagged OSM-5 (B,H), BBS-1 (C,I), BBS-2 (D,J), BBS-7 (E,K), and BBS-8 (F,L). The BBS proteins localize specifically to the transition zones (C–F,I–L), and like the IFT protein, OSM-5 (B,H), they are also found along the ciliary axonemes. The ciliary axonemes (cil), transition zones (tz), dendrites (den), and cell bodies (cb) are indicated. An asterisk denotes the ciliated endings of neurons found at the anterior end of the worm (e.g., IL1/2, OLL, CEP, and BAG neurons). Arrowheads in all panels denote the amphid and phasmid transition zones. Note that in all panels, only one set of amphid and phasmid neurons is shown except for the schematic in A, where both sets (one on each side) are shown.
We also noted that, as with OSM-5::GFP, the BBS–GFP fusion proteins exhibited some localization in the cell bodies (cb; Fig. 1H–L), and to a lesser extent, in the neuronal dendrites (den); however, we suspect that these signals may be largely nonspecific, due to overexpression of the transgenes. In the case of OSM-5::GFP, BBS-1::GFP, and BBS-8::GFP, little cell body or dendritic staining was observed. Taken together, these localization studies suggest that the C. elegans BBS proteins function predominantly at the ciliary transition zones and axonemes. This is largely consistent with our previous finding that mammalian BBS8 is found exclusively in the pericentriolar regions of mammalian centrosomes and basal bodies, although immunostaining did not reveal significant ciliary localization in these cultured cells (Ansley et al. 2003).
In accordance with the observation of BBS::GFP staining along the ciliary axonemes, we detected anterograde and retrograde movement of GFP-tagged BBS-1, BBS-2, BBS-7, and BBS-8 in wild-type animals (see Supplementary Movies 1–8). Importantly, the BBS proteins displayed motility comparable to that of GFP-tagged IFT proteins (e.g., OSM-5, OSM-6, XBX-1, and CHE-13; see Supplementary Movies 9, 10 for OSM-5::GFP; see also Qin et al. 2001; Haycraft et al. 2003; Schafer et al. 2003). The rates of anterograde movement of the GFP-tagged BBS-1, BBS-2, BBS-7, and BBS-8 proteins along phasmid cilia were found to be 0.69 ± 0.12 (n = 22), 0.73 ± 0.17 (n = 26), 0.70 ± 0.15 (n = 21), and 0.76 ± 0.14 (n = 25) μm/sec, respectively; retrograde rates were found to be 0.89 ± 0.17 (n = 17), 1.16 ± 0.16 (n = 13), 1.01 ± 0.20 (n = 10), and 1.06 ± 0.18 (n = 12) μm/sec, respectively. These rates are similar to those reported for various C. elegans IFT proteins. For example, Qin et al. (2001) measured the OSM-5 anterograde and retrograde rates to be 0.68 ± 0.09 and 1.10 ± 0.17 μm/sec, respectively.
The common transition zone localization and IFT-like motility of all of the BBS::GFP proteins is strong evidence that these tagged proteins are functional and behave similar to their wild-type counterparts. As demonstrated below, we show that one of the GFP-tagged BBS proteins (BBS-8) rescues the mutant phenotype of its respective gene, validating its observed localization pattern and behavior. On the whole, the specific localization of the BBS proteins to transition zones, as well as their IFT-like motility, suggest that the BBS proteins are associated with IFT rafts and are likely to represent novel, bona fide IFT proteins.
osm-12 and bbs-8 mutants display chemotaxis defects associated with abnormal cilia function
Our finding that the C. elegans BBS proteins display a localization pattern and behavior comparable to that of IFT proteins suggests that they could play an important role in cilia biogenesis and function. To investigate this possibility, we used bioinformatic and mutagenesis approaches to obtain loss-of-function bbs mutants in C. elegans.
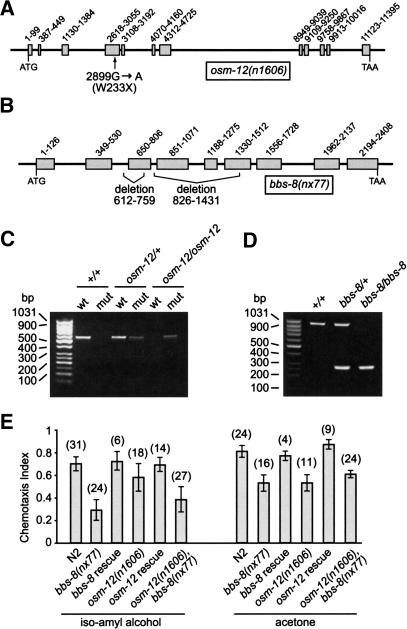
First, we identified the uncloned osm-12(n1606) allele as a candidate bbs-7 mutant on the basis that the bbs-7 gene lies within the osm-12 critical interval, that bbs-7 contains an X box in its promoter (Ansley et al. 2003), and the fact that the osmolarity avoidance (Osm) phenotype is characteristic of ciliary defects in C. elegans (Perkins et al. 1986). Sequencing of the bbs-7 gene from osm-12(n1606) mutant animals revealed a G→A nonsense mutation at nucleotide 2899 (Fig. 2A,C). The n1606 allele is likely to be null, as it contains a stop codon after only 32% of the coding region (henceforth, for simplicity, we often refer to the osm-12(n1606) mutant as the bbs-7 gene mutant). Second, using standard mutagenesis protocols, we generated and identified a strain containing two deletions in bbs-8, of 148 and 606 bp, which span from exons 3 to 6 (Fig. 2B). We isolated a strain homozygous for this allele, which is also likely to be null and is designated bbs-8(nx77) (Fig. 2D). The osm-12(n1606) and bbs-8(nx77) animals were found to be normal with respect to development, lifespan, movement, and brood size (data not shown).
Figure 2.
osm-12(n1606) and bbs-8(nx77) animals bearing mutations in the C. elegans bbs-7 and bbs-8 genes, respectively, exhibit chemotaxis defects. (A,B) Schematics depicting the G→A nonsense mutation at nucleotide 2899 of osm-12(n1606), and the position of two deletions in bbs-8(nx77). Exons are shown as shaded boxes. (C) Single worms containing the osm-12(n1606) nonsense mutation were identified using two PCR reactions that are designed to amplify wild-type (wt), osm-12(+), or mutant (mut) osm-12(n1606) genomic DNA fragments. Shown are the PCR products obtained from +/+, osm-12(n1606)/+, and osm-12(n1606)/osm-12(n1606) animals. (D) Single-worm PCR using primers that flank the 826–1431 deletion in bbs-8(nx77) identifies +/+, bbs-8(nx77)/+, and bbs-8(nx77)/bbs-8(nx77) animals. (E) The osm-12(n1606), bbs-8(nx77), and osm-12(n1606),bbs-8(nx77) mutant worms are defective in chemotaxis (Che) toward isoamyl alcohol and acetone. Rescue of this Che defect was achieved by expressing bbs-7(+) and bbs-8(+) genes in the osm-12(n1606) and bbs-8(nx77) mutant worms, respectively. The number of assays for each chemotaxis data point is indicated in brackets. Error bars, ±S.E.M.
To assess the function of cilia in the osm-12 and bbs-8 mutants, we used a behavioral assay that measures the ability of the worms to chemotax toward two chemical attractants, isoamyl alcohol and acetone. The normal response to these volatile compounds is mediated by the cilia of two chemosensory amphid neurons, AWA and AWC (Bargmann et al. 1993). Using such an assay, we found the two mutants to be chemotaxis defective toward both isoamyl alcohol and acetone (Fig. 2E). This odorant chemotaxis (Odr and Che) defect is directly attributable to disruptions in the bbs-7 and bbs-8 genes, as these phenotypes were fully rescued in osm-12 and bbs-8 mutants expressing wild-type bbs-7(+) and bbs-8(+), respectively (Fig. 2E). Interestingly, the osm-12,bbs-8 double-mutant animals responded similarly to that of the single mutants in our chemotaxis assays (Fig. 2E).
osm-12 and bbs-8 mutants possess an abnormal cilium structure
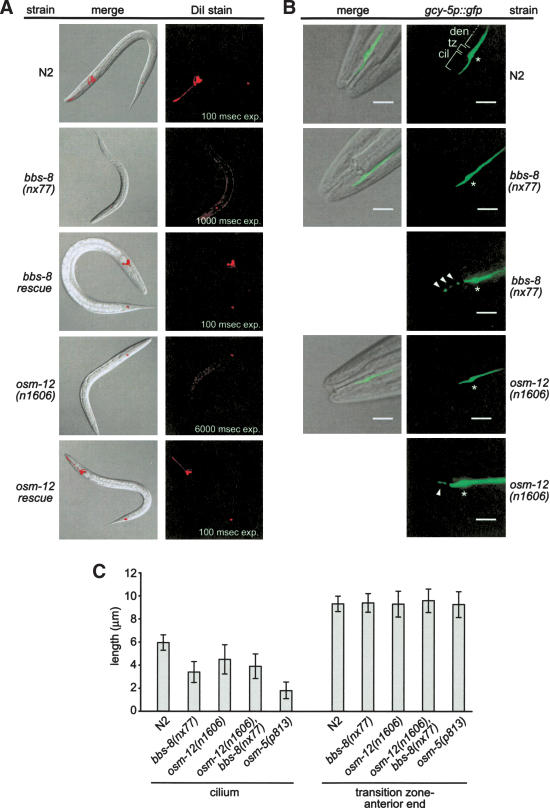
Many C. elegans chemosensory mutants display structural defects in their cilia (Perkins et al. 1986; Starich et al. 1995). We therefore examined the structural integrity of cilia in the osm-12 and bbs-8 mutant strains by using a fluorescent dye (DiI) uptake assay. This dye preferentially fills amphid head neurons and phasmid tail neurons via their exposed ciliated endings (Starich et al. 1995). Compared with N2 controls, osm-12(n1606), bbs-8(nx77), and osm-12(n1606),bbs-8(nx77) animals were found to display little or no detectable DiI uptake, strongly suggesting a defect in cilia structure (Fig. 3A; data not shown for the double mutant). The dye-uptake defective phenotype (Dyf) of the osm-12 and bbs-8 mutants was fully rescued with the respective wild-type bbs-7 and bbs-8 genes (Fig. 3A). Importantly, rescue of Dyf in bbs-8 mutants was also achieved using bbs-8::gfp (data not shown), which indicates that the GFP-tagged protein is functional and displays a localization pattern and IFT-like behavior consistent with that of the wild-type protein.
Figure 3.
osm-12(n1606) and bbs-8(nx77) mutants possess disrupted ciliary structures. (A) osm-12 and bbs-8 animals are severely defective in the ability to incorporate the dye, DiI, into their amphid and phasmid neurons via ciliated endings. The dye-uptake defective (Dyf) phenotype was fully rescued by expressing bbs-7(+) and bbs-8(+) genes in the osm-12 and bbs-8 worms, respectively. Note that barely detectable DiI staining in both bbs gene mutants was observed only after prolonged exposure (exp.) times. (B,C) Using the ASER neuron-specific marker, gcy-5p::gfp, the ASER cilium structure of osm-12(n1606), bbs-8(nx77), and osm-12(n1606),bbs-8(nx77) mutants is observed to be shorter than that of N2 controls. The ciliary axoneme (cil), transition zone (tz), and dendrite (den) structures of ASER are indicated at top, right. An asterisk also denotes the position of the transition zone in each panel. Also shown is the fragmented GFP staining pattern (see arrowheads) that is often observed in the bbs mutants. The cilium morphology data presented in C shows that the ASER cilium structure of osm-12, bbs-8, and osm-12,bbs-8 mutants is significantly shorter (P < 0.05) than N2 controls, but not as short as the ASER cilium in osm-5(p813) mutant animals. Also shown is the identical relative positioning of the ASER transition zone (tz) in the bbs gene mutants compared with N2 worms (measured as tz → anterior end of worm, seen in the DIC–GFP merged images). Each data point represents measurements from 50 different animals. Error bars, ±S.E.M. Bars, 5 μm.
To further investigate cilium morphology in the osm-12 and bbs-8 mutants, we utilized the ASER neuronal cell marker, gcy-5p::gfp, which produces diffuse GFP fluorescence throughout the cell body, dendrite, transition zone, and ciliary axoneme. Analysis of this marker in osm-12 and bbs-8 mutant backgrounds revealed that the transition zones were indistinguishable from that of N2 worms with regard to overall size, shape, and position relative to the nematode anterior end (Fig. 3B,C; the transition zone is denoted with an asterisk). However, as shown in Figure 3C, the GFP-containing ciliary structures of osm-12 and bbs-8 mutants, while emanating normally from the transition zone, appear significantly shorter at 4.51 ± 1.24 μm (n = 50) and 3.38 ± 0.91 μm (n = 50), respectively, compared with 5.94 ± 0.67 μm (n = 50) for N2 animals. The osm-12,bbs-8 double-mutant ASER cilium length of 3.92 ± 1.07 μm (n = 50) is also shorter than wild-type animals (Fig. 3C). Furthermore, in a significant proportion of osm-12, bbs-8, and osm-12,bbs-8 mutant animals (22%–46%), fragmented GFP fluorescence extending from the distal end of the truncated cilium was observed; this suggests that the cilium may in some cases extend to near wild-type lengths, but is structurally perturbed at the distal end (Fig. 3B; see arrowheads). This fragmented staining pattern is never detected in N2 worms, and to our knowledge, has not been reported for other C. elegans mutants with ciliary defects, including osm-5 mutants (data not shown). Interestingly, although shorter than wild type, the ASER cilium length of the bbs mutants is not as short as that of the IFT complex B mutant, osm-5(p813), which possesses a ciliary axoneme that is 1.77 ± 0.71 μm (n = 50) in length (Fig. 3C).
BBS-7 and BBS-8 are required for the proper motility/function of several IFT proteins
Despite their possibly unique ciliary morphology, the overall phenotypes (Che, Odr, Dyf, and structurally perturbed cilia) of the osm-12 and bbs-8 mutants are strikingly similar to those of IFT gene mutants. This suggests that the BBS proteins may influence IFT processes, especially given their localization to transition zones and IFT-like motility. To test this hypothesis, we examined the localization and IFT movement of two IFT complex B proteins (OSM-5/Polaris/IFT-88, CHE-2/IFT-80) and one IFT complex A protein (CHE-11/IFT-140) in the osm-12 and bbs-8 mutants. To ensure comparable expression levels of these GFP markers in the wild-type (N2) and mutant strains, we introduced each gfp transgene from the N2 strain into the osm-12 and bbs-8 mutants by genetic crossing. We reasoned that direct comparisons of the IFT markers in both wild-type and bbs gene mutant backgrounds should reveal any possible IFT defect in the mutant strains.
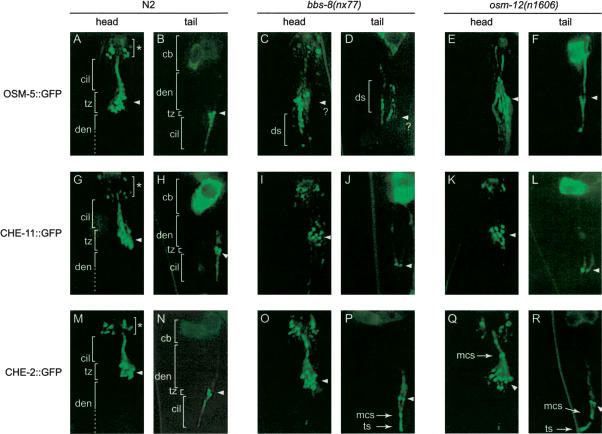
In wild-type animals (N2), GFP-tagged OSM-5, CHE-11, and CHE-2 localize to the transition zones and ciliary axonemes of the ciliated head and tail neurons (Fig. 4A,B,G,H,M,N), and undergo clearly visible, prominent IFT movement in both the anterograde and retrograde directions (see Supplementary Movies 11, 12, 17, 18, 23, 24). However, we observed that when the same osm-5::gfp and che-11::gfp transgenes are expressed in the osm-12 and bbs-8 mutant backgrounds, the GFP-tagged proteins show abnormal localization patterns. In the bbs-8 mutant, the localization of OSM-5::GFP along the ciliary axonomes is highly reduced, and a significant number of worms display disorganized accumulation of this marker within the transition zone region, as well as the dendrites and cell bodies (Fig. 4C,D; ds denotes dendritic staining). Although some OSM-5::GFP appears to enter the ciliary axonemes of a significant number of osm-12 mutant animals, its overall localization pattern appears disorganized, with poorly defined transition zones and a high level of diffuse staining in the dendrites and cell bodies (Fig. 4E,F). The localization of the CHE-11::GFP marker to the ciliary axonemes is also highly reduced in the bbs-8 and osm-12 mutants, but the marker is still present and apparently organized in the transition zone region of both mutants (Fig. 4I–L). Consistent with the defective OSM-5::GFP and CHE-11::GFP localization in the bbs gene mutants, both IFT protein markers were found to undergo a decrease in observable IFT movement in these strains (see Supplementary Movies 13–16, 19–22). Interestingly, the GFP-tagged OSM-5 staining along the ciliary axonemes of the osm-12 mutants (discussed above; Fig. 4E,F) is not indicative of moving IFT particles, but instead, represents abnormal ciliary staining and defective OSM-5 IFT movement in this strain.
Figure 4.
BBS-7 and BBS-8 protein function is required for the proper localization of the GFP-tagged IFT particle components OSM-5 and CHE-11, and to a lesser degree CHE-2. Shown are representative fluorescent images of the anterior (amphids) and posterior (phasmids) ends of N2, bbs-8(nx77) and osm-12(n1606) worms expressing the identical osm-5::gfp, che-11::gfp, and che-2::gfp transgenes. In contrast to the localization of GFP-tagged OSM-5 and CHE-11 to the ciliary axoneme in N2 worms (A,B,G,H), these two IFT proteins generally display abnormal staining along the ciliary axonemes of bbs-8(nx77) (C,D,I,J) and osm-12(n1606) (E,F,K,L) mutant animals. In contrast, there is strong localization of CHE-2::GFP to the ciliary axonemes of the bbs-8(nx77) (O,P) and osm-12(n1606) (Q,R) mutants, similar to that seen in N2 control worms (M,N). The ciliary axonemes (cil), transition zone (tz), and dendrite (den) are indicated in each panel. In some of the bbs gene mutants, spots of GFP fluorescence are often observed in the dendrites (ds), near the midpoint of the ciliary axoneme (mcs) and at the distal end of the ciliary axoneme (tip spot, or ts). Note that only one set of amphid and phasmid cilia are shown in each panel. A question mark denotes the difficulty in precisely identifying the location of the transition zones in panels C and D.
In contrast to the above, the CHE-2::GFP IFT protein marker appears to display a localization pattern in the bbs-8 and osm-12 mutants that is very similar to that of wild type. In both bbs gene mutants, GFP-tagged CHE-2 is found predominantly at the transition zones and along the ciliary axonemes (Fig. 4O–R). Perhaps not surprisingly, anterograde and retrograde IFT movement of the CHE-2::GFP protein in the mutant backgrounds appeared similar to that of wild-type worms (see Supplementary Movies 25–28). However, on the basis of the localization pattern of CHE-2::GFP along the ciliary axonemes, we observed two distinct abnormalities. As is more clearly observed in the phasmid neurons, the bbs mutant cilia often display an accumulation of GFP signal approximately at the midpoint and at the distal ends of the axonemes, neither of which are observed in wild-type cilia (Fig. 4P–R, see indicated mid-ciliary axoneme spot [mcs], and tip spot [ts]). Interestingly, we observed some CHE-2::GFP movement anterior to the mid-ciliary spot in the two bbs gene mutants, suggesting that this section of the axonemes is at least partially intact.
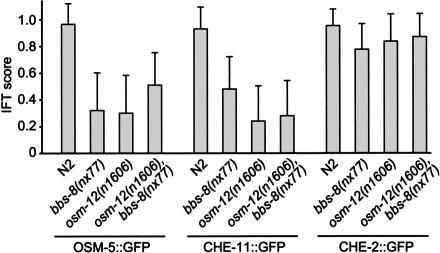
To assess the extent of the IFT defects in the osm-12 and bbs-8 mutants, we quantitated the level of GFP-tagged IFT protein movement in “blind” studies, wherein the observer lacked knowledge of the genotype (wild-type vs. bbs gene mutant) or the nature of the GFP fusion protein. Levels of IFT were expressed on a discontinuous scale of 0 to 1.0, where 0 indicates no observable IFT, 0.25 represents barely detectable IFT (e.g., very weak GFP signals, where particles are sometimes seen moving in either the anterograde or retrograde directions), 0.5 indicates IFT that is weak (e.g., low-level signals of mostly continuous particle movement in both the anterograde and retrograde directions), 0.75 represents a moderate level of IFT movement (e.g., IFT particle movement that is readily observable and continuous in both directions), and 1.0 indicates a strong level of anterograde and retrograde IFT that is highly prominent at both the amphid and phasmid ciliated endings.
The results summarizing our analyses of 40 movies made for each strain are shown in Figure 5. When compared with the wild-type controls (average IFT scores of >0.93 for all three IFT markers), the OSM-5::GFP and CHE-11::GFP proteins possessed considerably reduced levels of IFT in the osm-12 and bbs-8 mutants (average IFT scores of 0.30 and 0.32, respectively, for OSM-5::GFP; and 0.24 and 0.48, respectively, for CHE-11::GFP; n = 40 in each case). As expected from its wild-type-like localization pattern in both bbs mutants (Fig. 4M–R), the CHE-2::GFP protein shows only slightly compromised levels of IFT (see Supplementary Movies 25–28), with average IFT scores of 0.84 (n = 40) and 0.78 (n = 40) in the osm-12 and bbs-8 mutants, respectively (Fig. 5). Although we could not measure rates of IFT for the OSM-5 and CHE-11 markers in the bbs mutants because of the highly compromised IFT movement, the rates of IFT movement along the phasmid ciliary axonemes for CHE-2::GFP in the osm-12 and bbs-8 mutants were found to be normal (anterograde rates of 0.66 ± 0.11 μm/sec [n = 25] and 0.65 ± 0.11 μm/sec [n = 25], respectively, and retrograde rates of 0.97 ± 0.13 μm/sec [n = 10] and 0.96 ± 0.17 μm/sec [n = 10], respectively). These rates are essentially identical to those previously reported for GFP-tagged CHE-2 in the amphid and phasmid neurons of wild-type worms (Qin et al. 2001). Also of note is the fact that the osm-12,bbs-8 double mutants behaved comparably to that of the single mutants, as GFP-tagged OSM-5 and CHE-11 displayed more compromised IFT than the CHE-2 marker (Fig. 5). Taken together, the results from the analyses of GFP-tagged IFT proteins in the bbs mutants show that a loss of BBS protein function results in an overall decrease in the efficiency of the IFT process, with some components being significantly more affected than others. These findings, along with others presented in this study, suggest that the BBS proteins partake directly in the IFT process.
Figure 5.
The C. elegans BBS-7 and BBS-8 proteins are required for efficient IFT. The IFT motility of OSM-5 and CHE-11, but not CHE-2 is significantly compromised in the absence of BBS-7 and BBS-8 protein function. The osm-5::gfp, che-11::gfp, and che-2::gfp transgenes were crossed from wild-type (N2) to each of the bbs single or double mutants (bbs-8(nx77), osm-12(n1606), and osm-12(n1606),bbs-8(nx77) and the amphid (n = 20) and phasmid (n = 20) IFT motilities were recorded in movies (see Materials and Methods). In a blind assay, IFT was scored on a discontinuous scale where 0 equals no detectable IFT, 0.25 equals barely detectable IFT (i.e., extremely weak signals indicating moving particles that are sometimes observed), 0.50 equals weak IFT (i.e., faint signals of continuous particle movement in both the anterograde and retrograde directions), 0.75 equals moderate levels of IFT, and 1 equals IFT that is highly prominent and operating in both the anterograde and retrograde directions. No significant difference between amphid and phasmid IFT was detected; therefore, the scores above are a combination of the amphid and phasmid IFT scores (i.e., n = 40). Error bars, S.E.M.
Discussion
C. elegans BBS proteins are found at the base of cilia and undergo IFT motility
In this study, we have characterized the localization of several C. elegans BBS proteins at the cellular level, and found that they appear to function predominantly at ciliary transition zones, which are akin to basal bodies, and along ciliary axonemes. Such a finding is highly consistent with the remarkably restricted expression pattern of C. elegans bbs genes at the level of the organism, namely within neurosensory cells bearing cilia, and not nonciliated cells (Ansley et al. 2003). Furthermore, our analyses of BBS::GFP fusion proteins indicate that the BBS proteins associate with motor-driven complexes that undergo anterograde and retrograde transport along ciliary axonemes. Although the BBS proteins may, in principle, be transported as particle-associated cargo, destined to become integral parts of the ciliary axoneme and/or membrane, we believe this to be unlikely as most of the BBS proteins localize to transition zones or are undergoing transport—precisely as with the well-characterized core IFT proteins. Instead, we propose that the BBS proteins represent bona fide IFT proteins associated with components of the kinesin or dynein motor complexes, and/or with one or both of the IFT particle complexes A and B. Regarding the latter, although all four C. elegans bbs genes have homologs in Chlamydomonas, the biochemical characterization of the IFT particle in this unicellular organism has not identified any of the BBS proteins as components of IFT complex A or B (Piperno and Mead 1997; Cole et al. 1998). However, as many of the Chlamydomonas IFT particle proteins have yet to be assigned, it is entirely possible that one or more of the BBS proteins may function as part of these protein complexes. Alternatively, the BBS proteins may be peripherally associated with the IFT particles and lost during the isolation of these particles, or are part of different, yet undescribed, motor-associated complexes.
The cilium structure of osm-12(n1606) and bbs-8(nx77) mutants is abnormal
We have isolated mutants of the C. elegans bbs-7 (osm-12) and bbs-8 genes, which afford the possibility of dissecting the functions of the BBS-7 and BBS-8 proteins. Disruption of the bbs genes results in several ciliary structural and functional defects, similar to that observed for mutants in several IFT genes, including abnormal dye filling, cilium length/structure, and chemosensation.
On the basis of the specific ciliary structure phenotypes we observed, it appears that the osm-12 and bbs-8 worms possess cilia that are longer than those of the IFT complex B mutants. Using the gcy-5p::gfp marker, we found that the ASER cilium of osm-5(p813) mutant animals is significantly shorter in length than that of the osm-12 and bbs-8 mutants and often displays ectopic posteriorly directed, cilia-like projections (Fig. 3C; data not shown). These findings are in agreement with electron microscopy and GFP reporter studies, which show that the IFT complex B mutants (e.g., che-2, che-13, osm-1, osm-5, and osm-6) possess cilia that are generally <2 μm in length (Perkins et al. 1986; Collet et al. 1998; Fujiwara et al. 1999; Haycraft et al. 2001). Furthermore, the observation of some CHE-2::GFP IFT movement and staining at the distal ends, combined with an apparent fragmented GFP distal-end staining pattern sometimes observed in the two bbs gene mutants (Fig. 3B), suggest that these bbs gene mutant cilia may be closer to wild-type length and, thus, even more different from complex B mutant cilia.
Although appreciably longer than the cilia of IFT complex B mutants, the length of the bbs mutant cilia appears similar to those of other IFT mutants. Previous studies have shown that the cilia of IFT complex A (e.g., che-11 and daf-10), kinesin (osm-3), dynein (che-3), and dynein light intermediate chain (xbx-1) mutants are often truncated, but not to the same extent as the cilia from IFT complex B mutants (Perkins et al. 1986; Wicks et al. 2000; Schafer et al. 2003). However, in contrast to complex A and dynein motor mutants, which do not show appreciable retrograde movement along the axoneme, such dynein-dependent motility was clearly visualized for CHE-2::GFP in our bbs gene mutants.
Loss of BBS-7 and BBS-8 function results in compromised IFT
Given the IFT protein-like behavior of the BBS proteins, we hypothesized that disrupting the functions of BBS-7 and/or BBS-8 may affect intraflagellar transport, leading to the observed ciliary structure and function defects. Our results indicate that the level of observable IFT for GFP-tagged OSM-5/Polaris and CHE-11 is severely compromised, although not abrogated. Surprisingly, the IFT of CHE-2 was affected to a significantly lower extent.
Taken together, our results provide important insights into the function of BBS-7 and BBS-8. First, that CHE-2::GFP (in particular) enters the cilium and undergoes IFT movement at the expected velocities in the osm-12 and bbs-8 mutants indicates that the kinesin and dynein motors are likely fully functional in the absence of BBS-7 and BBS-8. Second, the relatively normal CHE-2-associated IFT particle movement suggests that the structural platform required for IFT is intact in these mutant strains, although ultrastructural defects are likely to be present in the middle to distal end of the axoneme. Third, as OSM-5 and CHE-11 have highly reduced (but not absent) IFT in the osm-12 and bbs-8 mutants, it is possible that these two IFT proteins inefficiently incororate into IFT particles at the base of the cilium before entering the axoneme.
Model of BBS-7 and BBS-8 protein function
Current models of IFT allow for at least four distinct steps prior to the onset of protein transport along the ciliary axoneme, namely, the targeting or recruitment of all necessary components to the base of the cilium, the assembly of fully intact motor–IFT particle–cargo complexes, docking at the transitional fibers, and loading/translocation onto the axoneme proper (Rosenbaum and Witman 2002; Cole 2003; Scholey 2003). Remarkably, very little is known regarding the molecular mechanisms underlying these processes. For example, the location or order of motor–IFT particle–cargo complex assembly is unclear, although the assembly has been hypothesized to occur predominantly within the basal body and transitional fiber regions (Deane et al. 2001; Scholey 2003). Some preassembly of IFT components may occur upstream of the basal body; in the case of cargo, it has been shown that Chlamydomonas radial spoke proteins partially assemble in the cell body before being transported to the flagellar tip by anterograde IFT (Qin et al. 2004). Furthermore, the full complement of IFT proteins is unlikely to have been uncovered, their precise arrangement in particles is unknown, and the specific functions of the various IFT proteins within the motor–IFT particle complex are unclear.
Targeting of IFT proteins to the base of the cilium is likely a preliminary step in the formation of IFT particles. For example, in the IFT complex B mutant, che-13, the recruitment of GFP-tagged OSM-5 to the transition zone is disrupted, which suggests that targeting (and incorporation into IFT particles) of OSM-5 to the base of cilia is dependent upon CHE-13 function (Haycraft et al. 2003). Because CHE-2 and CHE-11 (and to some extent OSM-5), appear to localize to the transition zone region in the bbs gene mutants, it seems unlikely that BBS-7 and BBS-8 play a direct role in the recruitment of these IFT components to the base of cilia. Loading of fully assembled IFT complexes onto the ciliary axoneme is probably the last major event prior to IFT. Because CHE-2 (and to significantly lesser extents, CHE-11 and OSM-5) associated IFT particles are observed to undergo IFT movement in the osm-12 and bbs-8 mutants, it can be implied that these particles are loaded/translocated correctly onto the axoneme. This suggests that the BBS-7 and BBS-8 proteins are unlikely to play a direct role in the loading process.
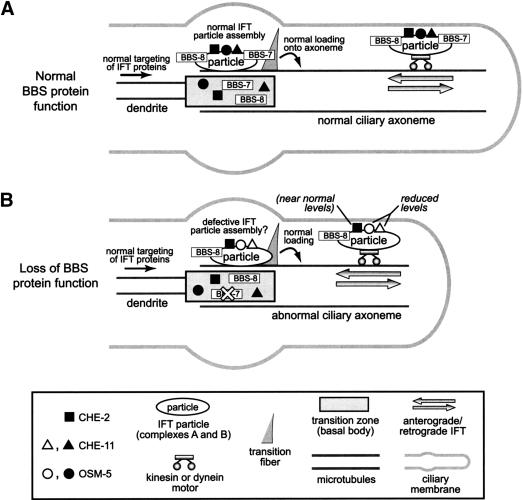
On the basis of our results and the above considerations, we propose a model wherein the BBS-7 and BBS-8 proteins facilitate the assembly of certain IFT protein components into the motor–IFT particle–cargo complex (Fig. 6A). In the absence of BBS protein function, both complex A and B IFT components such as CHE-11 and OSM-5 may largely fail to become assembled, resulting in their compromised ability to undergo IFT (Fig. 6B). Other components, such as CHE-2, may be affected, but to a lesser extent. Because all of the components of the IFT machinery (motors, IFT particle components) very likely assemble prior to loading onto the ciliary axoneme, the BBS proteins may facilitate the incorporation of the IFT proteins into the final motor–IFT particle complex within the transition zone region, including the transitional fibers that appear to play an important role in docking IFT components (Deane et al. 2001). The BBS proteins could conceivably act in the assembly process as molecular chaperones. Interestingly, the first protein linked to Bardet-Biedl syndrome, BBS6, is a putative molecular chaperone related to the HSP60/chaperonin family (Katsanis et al. 2000; Slavotinek et al. 2000; Stone et al. 2000). Alternatively, the BBS proteins may interact directly with different IFT proteins and permit their incorporation into the particles. This is substantiated by the fact that the BBS proteins also undergo IFT-like movement and, therefore, may associate as core or peripheral components of the IFT particles or of the motor complex. Our finding that the IFT-like motility of GFP-tagged BBS-1 is effectively abolished in two IFT mutants (osm-5 and che-11) strongly supports this hypothesis (our unpublshed data).
Figure 6.
Model of BBS-7 and BBS-8 protein function. (A) In wild-type cilia, BBS-7 and BBS-8 proteins, as well as complex A and B IFT particle components (e.g., OSM-5, CHE-11 and CHE-2) are targeted to, and accumulate at the base of the cilium structure. Within the basal body region (including the transitional fibers), the BBS-7 and BBS-8 proteins may play a role in facilitating the selective assembly of IFT protein components into the IFT particle, perhaps by acting as molecular chaperones or directly associating with them. Fully assembled motor–IFT particle–cargo complexes, which likely include the BBS proteins, are subsequently loaded onto the axoneme proper, where they undergo IFT. Note that for simplicity, only one transitional fiber is shown. (B) In cilia where BBS-7 or BBS-8 protein function is lost (shown only for BBS-7), the IFT components are targeted normally to the base of cilia. However, IFT particle assembly within the basal body region may be disrupted, resulting in the failure of certain IFT protein components (e.g., OSM-5 and CHE-11, but not CHE-2) to be efficiently incorporated into the final IFT particle complex and, hence, undergo IFT. This defect results in compromised IFT and ciliary defects (e.g., short and/or structurally abnormal cilia).
In the C. elegans mutants lacking BBS-7 or BBS-8 protein function, the differential incorporation of IFT proteins (OSM-5/CHE-11 vs. CHE-2) into the IFT complexes may reflect an hierarchical or ordered assembly process of IFT components, similar to that suggested by Haycraft et al. (2003). In this latter study with three IFT complex B proteins, CHE-13 was found to be necessary for the incorporation of OSM-5 into IFT particles, whereas OSM-6 incorporated into the particles in the absence of CHE-13. The authors concluded that OSM-6 likely localizes closer to the motor complex than CHE-13, and OSM-5 is a more peripheral component. In light of such a model, our results suggest that BBS proteins may be situated closer to the motor complex in the case of OSM-5 and CHE-11, and farther in the case of CHE-2. Given that CHE-11 and OSM-5 are complex A and B proteins, respectively, the BBS-7 and BBS-8 proteins therefore appear to participate in the assembly/incorporation of components from both IFT particle complexes.
bbs-7 and bbs-8 participate in the same biological process
It is notable that the C. elegans osm-12 and bbs-8 mutants have essentially identical phenotypes, including a differential effect on IFT protein motility (Figs. 2E, 3A–C, 5; see also Supplementary Movies 13–16, 19–22, 25–28). This suggests that the BBS-7 and BBS-8 proteins participate in a common cellular process, a finding consistent with the fact that mutations in different human BBS genes lead to clinically indistinguishable phenotypes (Katsanis et al. 2001). Our above observations that the various phenotypes of the osm-12,bbs-8 double mutants (Figs. 2E, 3C, 5) are comparable to that of the single mutants further suggest that in C. elegans, the two proteins either act sequentially in a biological pathway, where mutations in either or both components would result in the same defect, or that the proteins form a complex where both are equally necessary for its function.
OSM-5/Polaris dysfunction in Bardet-Biedl syndrome
Our discovery that OSM-5::GFP IFT movement is compromised, but not entirely abrogated in C. elegans strains possessing null mutations in two bbs genes is consistent with the comparably more important ciliary roles for C. elegans OSM-5, and its corresponding orthologs in Chlamydomonas (IFT88) and mice (Polaris/Tg737). Specifically, null mutations in the mouse Polaris/Tg737 gene result in embryonic lethality (Murcia et al. 2000), whereas null mutations in the BBS genes do not compromise viability in humans (Katsanis et al. 2001). Furthermore, the cilia of OSM-5/Polaris/IFT88 null mutants are more severely affected than those of the C. elegans bbs gene mutants we have characterized; C. elegans osm-5 mutants possess shorter cilia (Fig. 3C; Perkins et al. 1986), and insertional mutations in Tg737Δ-3β-Gal mice (Murcia et al. 2000) and Chlamydomonas IFT88 (Pazour et al. 2000) result in the complete absence of cilia and flagella, respectively.
Conclusion
The phenotypic overlap between BBS patients and the embryonically viable hypomorphic mouse mutant Tg737orpk (e.g., cystic kidneys, retinal degeneration, laterality defects; Moyer et al. 1994; Katsanis et al. 2001; Taulman et al. 2001; Pazour et al. 2002; Ansley et al. 2003), together with our identification of ciliary defects in two C. elegans bbs gene mutants, lead us to conclude that Bardet-Biedl syndrome is caused, at least in part, by defective Polaris/Tg737 and (more generally) compromised IFT function in humans. On a final note, the C. elegans osm-12 and bbs-8 mutants represent powerful model systems for studying the poorly understood IFT process. These mutants also provide the opportunity to fully investigate the molecular etiology of BBS and the potential link between basal body/ciliary dysfunction and clinical symptoms not previously associated with abnormal cilia function, such as obesity, diabetes, and learning defects.
Materials and methods
Strains
All strains were maintained and cultured using standard techniques. N2 (Bristol); KR3532, dpy-5(e907); MX52, bbs-8(nx77); MX53, osm-12(n1606),bbs-8(nx77); MT3645, osm-12(n1606); MX23, dpy-5(e907);nxEx23[dpy-5(+) + bbs-1::gfp]; MX24, dpy-5(e907); nxEx24[dpy-5(+) + bbs-2::gfp]; MX78, dpy-5(e907); nx25[dpy-5(+) + bbs-7::gfp]; MX27, dpy-5(e907);nxEx27[dpy-5(+) + bbs-8::gfp]. BC11009, dpy-5(e907);sEx11009[dpy-5(+) + gcy-5p::gfp]; MX45, bbs-8(nx77);sEx11009[dpy-5(+) + gcy-5p::gfp]; MX46, osm-12(n1606);sEx11009[dpy-5(+) + gcy-5p::gfp]; MX38, osm-12(n1606), bbs-8(nx77);sEx11009[dpy-5(+) + gcy-5p::gfp]; MX79, osm-5(p813); sEx11009[dpy-5(+) + gcy-5p::gfp]; MX57, bbs-8(nx77);Ex[osm-5::gfp + rol-6(su1006)] (donated by B. Yoder, University of Alabama, Birmingham); MX58, osm-12(n1606);Ex[osm-5::gfp + rol-6(su1006)] (donated by B. Yoder) MX75, osm-12(n1606),bbs-8(nx77);Ex[osm-5::gfp + rol-6(su1006)] (donated by B. Yoder); MX60, N2;myEx10[che-11::gfp + rol-6(su1006)] (donated by M. Barr, University of Wisconsin, Madison). MX61, bbs-8(nx77); myEx10[che-11::gfp + rol-6(su1006)] (donated by M. Barr); MX62, osm-12(n1606);myEx10[che-11::gfp + rol-6(su1006)] (donated by M. Barr); MX76, osm-12(n1606),bbs-8(nx77); myEx10[che-11::gfp + rol-6(su1006)] (donated by M. Barr); MX63, bbs-8(nx77);Ex[che-2::gfp + rol-6(su1006)] (donated by M. Barr); MX64, osm-12(n1606);Ex[che-2::gfp + rol-6(su1006)] (donated by M. Barr); MX77, osm-12(n1606),bbs-8(nx77); Ex[che-2::gfp + rol-6(su1006)] (donated by M. Barr).
Construction of C. elegans bbs::gfp transgenes
Translational gfp fusion constructs containing Y105E8A.5 (bbs-1), F20D12.3 (bbs-2), Y75B8A.12 (bbs-7), and T25F10.5 (bbs-8) were generated as PCR products and transformed into dpy-5(e907) as described previously (Ansley et al. 2003). For bbs-1, bbs-2, and bbs-8, the entire exonic and intronic sequence, along with promoter sequence 5′ to the start codon, was placed upstream of, and in frame with, the coding region of gfp and the unc-54 3′ UTR. Specifically, the bbs-1 gene fragment of 5193 bp included 500 bp of upstream sequence, the bbs-2 gene fragment of 10,438 bp included 6909 bp of upstream sequence, and the bbs-8 gene fragment of 3394 bp included 981 bp of upstream sequence. The bbs-7::gfp transgene was comprised of the full-length cDNA for bbs-7, along with 981 bp of its upstream sequence, fused to gfp as outlined above. The bbs-7 promoter and full-length cDNA was obtained from the pog10 vector construct. The pog10 plasmid contains ∼1 KB of 5′-genomic bbs-7 promoter region fused to the full-length bbs-7 cDNA (gift of the Vidal Lab, Dana Farber Cancer Center) and ∼500 bp of 3′-genomic bbs-7 DNA.
Isolation of a C. elegans bbs-8 deletion mutant
N2 worms were mutagenized using trimethyl psoralen (TMP) and UV irradiation at 365 nm (see www.ko.cigenomics.bc.ca), and worms bearing deletions in T25F10.5 (bbs-8) were identified and isolated from mutagenized worm libraries in liquid culture (see http://info.med.yale.edu/mbb/koelle) using a poison primer approach. Deletions in the bbs-8 gene were identified by two successive rounds of PCR using external (5′-ATGAAAGCAC ACTCTCTTGCCG-3′ and 5′-TGGCGTATGCCATGTGTAA GA-3′) and internal nested primers (5′-TTTTGCATGCTTA TTCCTCCCC-3′ and 5′-CGAATGTGTCACTCGTAACTGG-3′). In addition, a poison primer (5′-CGCCATTGGTTGATC TACTCGA-3′) was included in the first round reaction in order to limit the production of the bbs-8(+) product. Using this primer set-up, the bbs-8(+) product has an expected size of 2615 bp. Once a line bearing a bbs-8 deletion was identified, it was out-crossed to N2 five times.
Chemotaxis assays
Chemotaxis assays were performed on 85-mm round plates as follows. Two points were marked at opposite ends of the plate (5 mm from the edge), to which 1 μL of 1 M NaN3 (anesthetic) was applied. A total of 1 μL of attractant (diluted 1:100 in 95% ethanol) was spotted at one of these points and 1 μL of ethanol was spotted at the other point as a control. Young adult worms (∼100) were washed three times with M9 buffer and once with deionized water, and then applied to the center of the plate, 3.5 cm from the attractant. After removal of the excess liquid, the worms were allowed to partition across the plate for 30 min. The chemotaxis index was calculated as a-b/n, where a equals number of worms within 1.5 cm of the attractant, b equals number of worms within 1.5 cm of the ethanol counter-attractant, and n equals total number of worms in the assay.
Dye uptake assay
Worms were placed in 200 μL of DiI solution (DiIC18(3), Vybrant DiI cell-labeling solution, Molecular Probes; diluted 1:200 with M9 buffer). After a 1-h incubation, the worms were recovered from the dye solution and allowed to destain on seeded NGM plates. Uptake of DiI into the amphid and phasmid neurons was examined in mounted live worms using epifluorescence (Texas Red filter).
Visualization of ASER cilium structure
The ASER cilium structure was visualized using a transcriptional gcy-5p::gfp marker. Specifically, the gcy-5 promoter (gcy-5p) consisted of 2836 bp of upstream sequence and the first 14 bp of the gcy-5 coding sequence. The gcy-5p::gfp construct was expressed as an extrachromosomal array in dpy-5(e907);Ex[dpy-5(+) + gcy-5p::gfp] as outlined previously (Ansley et al. 2003), and subsequently crossed into the bbs-8(nx77), osm-12(n1606), osm-12(n1606),bbs-8(nx77), and osm-5(p813) mutant backgrounds. Cilium length was measured as the distance from the distal end of the transition zone to the cilium tip. Transition zone positioning was determined as the distance from the center of the transition zone to the anterior end of C. elegans.
Microscopy
For live imaging, worms were mounted on agarose pads and immobilized using 15 mM levamisole. Mounted worms were examined on a Zeiss Axioskop compound fluorescent microscope, and images were captured using Northern Eclipse version 6.0 software. For producing movies, individual frames were captured at 500-msec exposure and at 500-msec intervals. The resulting stacked TIFF images were converted into AVI movies, with a frame rate of two frames per second (i.e., real time). For measuring anterograde and retrograde IFT particle rates, the distance traveled by a particle over three consecutive frames of a stacked TIFF movie was determined using Northern Eclipse version 6.0. A minimum of 21 individual particles (14 for BBS-7::GFP) were measured for anterograde rates and a minimum of 10 particles were measured for retrograde rates. No more than two particles were measured for any one worm.
Acknowledgments
We thank Mark Vidal for the gift of a full-length bbs-7 cDNA clone, Mark Edgley for help with construction of the bbs-8(nx77) deletion mutant, and Peter Unrau for helpful comments on the manuscript. We acknowledge the Heart and Stroke Foundation of B.C. and Yukon (M.R.L), the National Institute of Child Health and Development, NIH (N.K.), Genome B.C. and Genome Canada (D.L.B), and Canadian Institute of Health Research (CIHR; W.S.D) for their financial support. M.R.L holds CIHR and Michael Smith Foundation for Health Research (MSFHR) scholar awards, S.R.W. holds an Ellison Medical Foundation Award, M.R.M holds a MSFHR scholarship award, and O.E.B. is the recipient of CIHR and MSFHR fellowship awards.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1194004.
References
- Ansley S.J., Badano, J.L., Blacque, O.E., Hill, J., Hoskins, B.E., Leitch, C.C., Kim, J.C., Ross, A.J., Eichers, E.R., Teslovich, T.M., et al. 2003. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425: 628-633. [DOI] [PubMed] [Google Scholar]
- Badano J.L., Ansley, S.J., Leitch, C.C., Lewis, R.A., Lupski, J.R., and Katsanis, N. 2003. Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am. J. Hum. Genet. 72: 650-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale A.E. 2002. Hedgehog signaling and human disease. Annu. Rev. Genomics Hum. Genet. 3: 47-65. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Hartwieg, E., and Horvitz, H.R. 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515-527. [DOI] [PubMed] [Google Scholar]
- Blatch G.L. and Lassle, M. 1999. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays 21: 932-939. [DOI] [PubMed] [Google Scholar]
- Cole D.G. 2003. Intraflagellar transport in the unicellular green alga, Chlamydomonas reinhardtii. Protist 154: 181-191. [DOI] [PubMed] [Google Scholar]
- Cole D.G., Diener, D.R., Himelblau, A.L., Beech, P.L., Fuster, J.C., and Rosenbaum, J.L. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141: 993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J., Spike, C.A., Lundquist, E.A., Shaw, J.E., and Herman, R.K. 1998. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148: 187-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane J.A., Cole, D.G., Seeley, E.S., Diener, D.R., and Rosenbaum, J.L. 2001. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11: 1586-1590. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Ishihara, T., and Katsura, I. 1999. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126: 4839-4848. [DOI] [PubMed] [Google Scholar]
- Haycraft C.J., Swoboda, P., Taulman, P.D., Thomas, J.H., and Yoder, B.K. 2001. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development 128: 1493-1505. [DOI] [PubMed] [Google Scholar]
- Haycraft C.J., Schafer, J.C., Zhang, Q., Taulman, P.D., and Yoder, B.K. 2003. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 284: 251-263. [DOI] [PubMed] [Google Scholar]
- Huangfu D., Liu, A., Rakeman, A.S., Murcia, N.S., Niswander, L., and Anderson, K.V. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426: 83-87. [DOI] [PubMed] [Google Scholar]
- Iomini C., Babaev-Khaimov, V., Sassaroli, M., and Piperno, G. 2001. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153: 13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad Z. and Paoli, M. 2002. Novel sequences propel familiar folds. Structure 10: 447-454. [DOI] [PubMed] [Google Scholar]
- Katsanis N., Beales, P.L., Woods, M.O., Lewis, R.A., Green, J.S., Parfrey, P.S., Ansley, S.J., Davidson, W.S., and Lupski, J.R. 2000. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat. Genet. 26: 67-70. [DOI] [PubMed] [Google Scholar]
- Katsanis N., Lupski, J.R., and Beales, P.L. 2001. Exploring the molecular basis of Bardet-Biedl syndrome. Hum. Mol. Genet. 10: 2293-2299. [DOI] [PubMed] [Google Scholar]
- Kubo A., Sasaki, H., Yuba-Kubo, A., Tsukita, S., and Shiina, N. 1999. Centriolar satellites: Molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147: 969-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer J.H., Lee-Tischler, M.J., Kwon, H.Y., Schrick, J.J., Avner, E.D., Sweeney, W.E., Godfrey, V.L., Cacheiro, N.L., Wilkinson, J.E., and Woychik, R.P. 1994. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science 264: 1329-1333. [DOI] [PubMed] [Google Scholar]
- Murcia N.S., Richards, W.G., Yoder, B.K., Mucenski, M.L., Dunlap, J.R., and Woychik, R.P. 2000. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left–right axis determination. Development 127: 2347-2355. [DOI] [PubMed] [Google Scholar]
- Mykytyn K., Braun, T., Carmi, R., Haider, N.B., Searby, C.C., Shastri, M., Beck, G., Wright, A.F., Iannaccone, A., Elbedour, K., et al. 2001. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat. Genet. 28: 188-191. [DOI] [PubMed] [Google Scholar]
- Mykytyn K., Nishimura, D.Y., Searby, C.C., Shastri, M., Yen, H.J., Beck, J.S., Braun, T., Streb, L.M., Cornier, A.S., Cox, G.F., et al. 2002. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat. Genet. 31: 435-438. [DOI] [PubMed] [Google Scholar]
- Nishimura D.Y., Searby, C.C., Carmi, R., Elbedour, K., Van Maldergem, L., Fulton, A.B., Lam, B.L., Powell, B.R., Swiderski, R.E., Bugge, K.E., et al. 2001. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum. Mol. Genet. 10: 865-874. [DOI] [PubMed] [Google Scholar]
- Pazour G.J. and Rosenbaum, J.L. 2002. Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 12: 551-555. [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Dickert, B.L., Vucica, Y., Seeley, E.S., Rosenbaum, J.L., Witman, G.B., and Cole, D.G. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151: 709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Baker, S.A., Deane, J.A., Cole, D.G., Dickert, B.L., Rosenbaum, J.L., Witman, G.B., and Besharse, J.C. 2002. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157: 103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.A., Hedgecock, E.M., Thomson, J.N., and Culotti, J.G. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117: 456-487. [DOI] [PubMed] [Google Scholar]
- Piperno G. and Mead, K. 1997. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. 94: 4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Rosenbaum, J.L., and Barr, M.M. 2001. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11: 457-461. [DOI] [PubMed] [Google Scholar]
- Qin H., Diener, D.R., Geimer, S., Cole, D.G., and Rosenbaum, J.L. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164: 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L. and Witman, G.B. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3: 813-825. [DOI] [PubMed] [Google Scholar]
- Schafer J.C., Haycraft, C.J., Thomas, J.H., Yoder, B.K., and Swoboda, P. 2003. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell 14: 2057-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M. 2003. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 19: 423-443. [DOI] [PubMed] [Google Scholar]
- Slavotinek A.M., Stone, E.M., Mykytyn, K., Heckenlively, J.R., Green, J.S., Heon, E., Musarella, M.A., Parfrey, P.S., Sheffield, V.C., and Biesecker, L.G. 2000. Mutations in MKKS cause Bardet-Biedl syndrome. Nat. Genet. 26: 15-16. [DOI] [PubMed] [Google Scholar]
- Starich T.A., Herman, R.K., Kari, C.K., Yeh, W.H., Schackwitz, W.S., Schuyler, M.W., Collet, J., Thomas, J.H., and Riddle, D.L. 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Lundin, V.F., and Leroux, M.R. 2003. Getting a grip on non-native proteins. EMBO Rep. 4: 565-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D.L., Slavotinek, A., Bouffard, G.G., Banerjee-Basu, S., Baxevanis, A.D., Barr, M., and Biesecker, L.G. 2000. Mutation of a gene encoding a putative chaperonin causes McKusick-Kaufman syndrome. Nat. Genet. 25: 79-82. [DOI] [PubMed] [Google Scholar]
- Swoboda P., Adler, H.T., and Thomas, J.H. 2000. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5: 411-421. [DOI] [PubMed] [Google Scholar]
- Taulman P.D., Haycraft, C.J., Balkovetz, D.F., and Yoder, B.K. 2001. Polaris, a protein involved in left–right axis patterning, localizes to basal bodies and cilia. Mol. Biol. Cell 12: 589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S.R., de Vries, C.J., van Luenen, H.G., and Plasterk, R.H. 2000. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 221: 295-307. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Murcia, N.S., Chittenden, L.R., Richards, W.G., Michaud, E.J., Woychik, R.P., and Yoder, B.K. 2003. Loss of the Tg737 protein results in skeletal patterning defects. Dev. Dyn. 227: 78-90. [DOI] [PubMed] [Google Scholar]