Abstract
Methylating agents introduce cytotoxic 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) residues into nucleic acids, and it was recently demonstrated that the Escherichia coli AlkB protein and two human homologues, hABH2 and hABH3, can remove these lesions from DNA by oxidative demethylation. Moreover, AlkB and hABH3 were also found to remove 1-meA and 3-meC from RNA, suggesting that cellular RNA repair can occur. We have here studied the preference of AlkB, hABH2 and hABH3 for single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA), and show that AlkB and hABH3 prefer ssDNA, while hABH2 prefers dsDNA. This was consistently observed with three different oligonucleotide substrates, implying that the specificity for single-stranded versus double-stranded DNA is sequence independent. The dsDNA preference of hABH2 was observed only in the presence of magnesium. The activity of the enzymes on single-stranded RNA (ssRNA), double-stranded RNA (dsRNA) and DNA/RNA hybrids was also investigated, and the results generally confirm the notion that while AlkB and hABH3 tend to prefer single-stranded nucleic acids, hABH2 is more active on double-stranded substrates. These results may contribute to identifying the main substrates of bacterial and human AlkB proteins in vivo.
INTRODUCTION
Alkylating agents introduce a variety of lesions in DNA, leading to cell death and mutagenesis, and many such agents are highly carcinogenic in mammals. Thus, several mechanisms for repairing alkylation damage in DNA have evolved. The current understanding of alkylation repair has mainly emerged from studies performed with Escherichia coli, where the adaptive response to alkylation damage regulates the expression of several alkylation repair proteins through the Ada regulon [reviewed in (1,2)]. When E.coli are exposed to methylating agents, methylphosphotriesters are formed in DNA, and the Ada protein reacts with these moieties, transferring the methyl group to the Cys-38 residue in the N-terminal part of the protein. This self-methylation of Ada converts the protein into an active transcription factor, which turns on the ada-alkB operon as well as the alkA gene. In addition to being a damage sensing transcription factor, Ada is an alkyl transferase which repairs O-alkylated bases, e.g. O6-methylguanine, by direct transfer of the alkyl group to the Cys-321 residue in the C-terminal part of the protein. AlkA is an alkylbase glycosylase which excises the purines alkylated in position 3 and 7, typically 3-methyladenine and 7-methylguanine, from DNA. It was recently discovered that AlkB is an oxidative demethylase which directly reverses 1-methyladenine (1-meA) and 3-methylcytosine lesions [reviewed in (3)]. Interestingly, the three repair functions represented by the Ada, AlkB and AlkA proteins are present in most organisms, including humans, indicating a universal importance of alkylation repair.
AlkB belongs to the superfamily of 2-oxoglutarate and iron(II) dependent oxygenases (4). These enzymes require ferrous iron as a cofactor and 2-oxoglutarate as cosubstrate, and catalyse various oxidation reactions where molecular oxygen is the oxidizing agent (5). In the AlkB reaction, the harmful methyl group in 1-meA or 3-meC is converted to a hydroxymethyl moiety, which is spontaneously released as formaldehyde, regenerating the normal bases, A or C, respectively (6,7). The cosubstrate 2-oxoglutarate is decarboxylated, yielding succinate and CO2. Recent studies have shown that AlkB can dealkylate 1-ethyladenine lesions in DNA (8), as well as 5′ phosphorylated 1-methyldeoxyadenosine mono- and triphosphates (9), albeit at reduced efficiency compared with 1-meA present in DNA. From sequence homology searches it appears that a number of AlkB homologues are present in the human proteome (4,10). However, only two of these, denoted hABH2 and hABH3, have been reported to share the ability of AlkB to demethylate 1-meA and 3-meC (8,11). Interestingly, E.coli AlkB and hABH3 are able to efficiently remove 1-meA and 3-meC lesions also from RNA (11), suggesting that these enzymes may have roles in the repair of both DNA and RNA.
Methylating agents preferentially introduce 1-meA and 3-meC into single-stranded DNA (ssDNA) (12), since the N1 of adenine and N3 of cytosine are involved in Watson–Crick base pairing, and thus are shielded from alkylation in double-stranded DNA (dsDNA). However, when dsDNA substrates were generated by annealing the complementary strand to methylated ssDNA, AlkB, hABH2 and hABH3, were all able to act on such substrates (6–8,11), but the results as to whether ssDNA or dsDNA are the preferred substrates for these enzymes have been somewhat conflicting. Therefore, we have tested in more detail the ssDNA versus dsDNA substrate specificities of AlkB, hABH2 and hABH3, using three oligonucleotide substrates of different sequence composition. In a living cell, double-stranded nucleic acids other than dsDNA are also present. During transcription, RNA/DNA hybrids are formed, and many cellular RNAs contain large segments of double-stranded RNA (dsRNA). Thus, here we have also investigated the activity of AlkB, hABH2 and hABH3 on various RNA-containing substrates, i.e. single-stranded RNA (ssRNA), double-stranded RNA and RNA/DNA hybrids methylated in either of the two strands. Our results indicate that single-stranded nucleic acids are the favored substrates for AlkB and hABH3, whereas hABH2 prefers double-stranded substrates containing a methylated DNA strand.
MATERIALS AND METHODS
Enzymes and reagents
AlkB, hABH2 and hABH3 were expressed and purified as previously described (6,11). [3H]MNU was from Amersham Biosciences. DNA oligonucleotides were from Medprobe (Oslo, Norway) or Invitrogen (Glasgow, Scotland). RNA oligonucleotides were from Dharmacon (Lafayette, CO).
Preparation of [3H]methylated DNA and RNA oligonucleotides
DNA or RNA oligonucleotides (200 μg) were incubated in sodium cacodylate buffer (50 mM, pH 7.0) in the presence of 0.5 mCi N-[3H]methyl-N-nitrosourea ([3H]MNU) for 2 h at 37°C in a total volume of 500 μl. The oligonucleotides were precipitated by the addition of NaCl (final concentration of 1 M) and 1 ml ethanol. After 20 min at 0°C, the substrates were recovered by centrifugation in a benchtop centrifuge (13 000 r.p.m., 10 min). The resulting pellet was washed twice with 70% ethanol, and dissolved in 400 μl TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA). The ethanol precipiation step was repeated once more, followed by dialysis against TE buffer to remove traces of free radioactivity. The resulting [3H]methylated oligonucleotide substrates typically had a specific radioactivity of 10 000 d.p.m./μg. For generation of double-stranded [3H]methylated substrate, the single-stranded [3H]methylated oligonucleotide was incubated in the presence of a 3-fold molar excess of non-methylated complementary strand for 5 min at 37°C in a buffer containing 50 mM Tris–HCl (pH 8.0), 50 mM KCl and 10 mM MgCl2, and then placed on ice. These double-stranded substrates were generated immediately prior to the experiment.
Assay for oxidative demethylation of [3H]methylated DNA and RNA oligonucleotides
When not indicated otherwise, [3H]methylated oligonucleotide (typically 0.1 μg single-stranded substrate, containing 1000 d.p.m. label) was incubated in a 50 μl reaction mixture for 30 min at 37°C in the presence of varying amounts of bacterial or human AlkB proteins in the following buffer: 50 mM Tris pH 8.0, 50 mM KCl, 10 mM MgCl2, 2 mM ascorbic acid, 100 μM 2-oxoglutarate, 40 μM FeSO4, 50 μg/ml BSA. Subsequently, nucleic acids were precipitated by the addition of 7 μl sodium acetate (3 M, pH 5.5), 10 μl glycogen (2 mg/ml) and 170 μl ethanol. After incubation at −70°C for at least 30 min, nucleic acids were pelleted by centrifugation at 13 000 r.p.m. in a benchtop centrifuge for 20 min at 4°C. The supernatant was removed, and subjected to scintillation counting.
RESULTS
Activity of AlkB proteins on different DNA oligonucleotide substrates
We have previously compared the activities of AlkB, hABH2 and hABH3 on ssDNA versus dsDNA, using as substrate an A-rich, methylated oligonucleotide consisting exclusively of A, G and C residues (here denoted AGC-oligo), in the absence or presence of its complementary strand (6,11). Clearly, AlkB and hABH3 were substantially more active on ssDNA than on dsDNA, while the opposite was true for hABH2. However, these results are somewhat in contrast to those reported by Trewick et al. (7), who found that the activity of AlkB on methylated poly(dA) annealed to unmethylated poly(dT) was 3-fold higher than with methylated poly(dA) alone. Furthermore, Duncan et al. (8) found that hABH2 and hABH3 were active on both ssDNA and dsDNA, but did not report any discrimination between these substrates. To resolve these apparent discrepancies, we have here reexamined the ssDNA versus dsDNA specificities of the human and bacterial AlkB proteins using different oligonucleotide substrates. First, we studied an oligonucleotide (denoted AT-oligo) consisting primarily of adenines but containing a few thymine residues, allowing the formation of a double-stranded substrate through hybridization with the complementary strand. Due to the low melting point of this substrate, the repair reactions were incubated at 30°C. Using the AT-oligo, we found that AlkB and hABH2 displayed moderate preferences towards ssDNA and dsDNA, respectively (Figure 1A and B), while hABH3 showed a strong preference for ssDNA (Figure 1C). The assay conditions used in the present study were slightly different from those we have used earlier, so we also reexamined the oligonucleotide (AGC-oligo) described in our previous work (6,11), and the results were similar to those obtained with the AT-oligo (Figure 1D–F). The results were also qualitatively the same when an oligonucleotide with a random sequence containing all four bases (denoted ACGT-oligo) was used (Figure 1G–I). In summary, each of the enzymes AlkB, hABH2 and hABH3 displayed similar ssDNA versus dsDNA preferences on all the substrates tested, indicating that this preference is independent of sequence context, and a general property of the enzyme.
Figure 1.
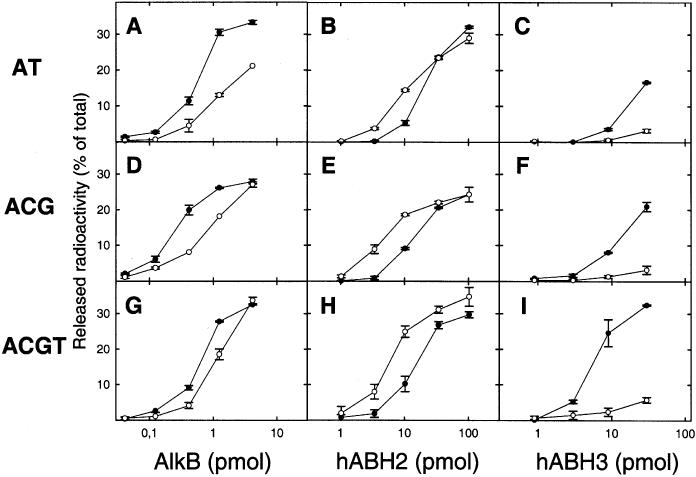
Activity of human and bacterial AlkB proteins on single-stranded and double-stranded DNA substrates. [3H]methylated DNA oligonucleotides were incubated with varying amounts of AlkB (A, D and G), hABH2 (B, E and H) or hABH3 (C, F and I), and the ethanol soluble radioactivity released was measured by scintillation counting. The [3H]methylated substrates used were AT-oligo (A–C) TAAAATAATAAATTAAA; AGC-oligo (D–F) AAAGCAAGAAACGAAAAAGCGAAA; AGCT-oligo (G–I) CATGATAACCGCGACTACACTGAC. Closed symbols indicate single-stranded [3H]methylated DNA oligonucleotides, while open symbols indicate the corresponding double-stranded substrates, generated by association with the unmethylated complementary strand. Error bars represent the range of duplicate measurements.
dsDNA preference of hABH2 is observed only in the presence of magnesium
The structure of DNA is strongly influenced by the presence of Mg2+ ions, and the activity and/or substrate specificity of DNA repair enzymes is often altered by Mg2+, either as a required cofactor, or as a stimulating factor. Thus, when studying the substrate specificity of bacterial and human AlkB proteins we have used reaction buffers containing a concentration of magnesium (10 mM) comparable to that found inside cells, but magnesium-free conditions were used in other studies. We, therefore, considered the possibility that the ssDNA versus dsDNA preference of these proteins may be influenced by the presence of magnesium. In the case of hABH3 and AlkB, we observed a preference for ssDNA also under magnesium-free conditions (data not shown). However, the specificity of hABH2 for dsDNA compared to ssDNA was only observed in the presence of magnesium, but not when magnesium was omitted from the reaction mixture (Figure 2). Apparently, the presence of magnesium stimulated the activity of hABH2 on dsDNA, but inhibited its activity on ssDNA. We tested several different MgCl2 concentrations (1, 3, 10 and 30 mM) and found that a maximal effect was observed in the presence of 10 mM MgCl2 (data not shown).
Figure 2.
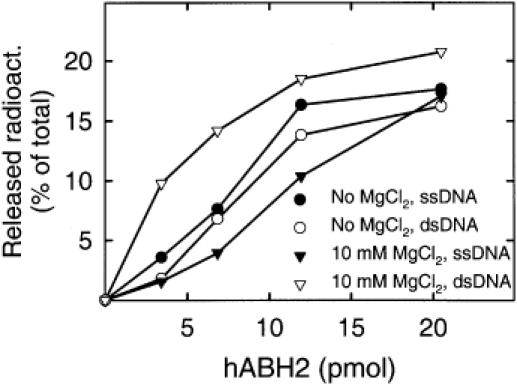
Preference of hABH2 for double-stranded DNA in the presence of magnesium. Single-stranded (closed symbols) or double-stranded (open symbols) [3H]methylated ACGT-oligo was incubated with varying amounts of hABH2 in the absence (circles) or presence (triangles) of 10 mM MgCl2, and the ethanol soluble radioactivity was measured by scintillation counting.
Activity of bacterial and human AlkB proteins on methylated DNA annealed to a complementary RNA strand
Clearly, annealing of the complementary strand to methylated ssDNA increased damage reversal by hABH2, while reversal by AlkB and hABH3 was reduced. We have also investigated the activities of these enzymes on DNA/RNA hybrids where the DNA was methylated prior to annealing to an undamaged complementary RNA strand. The [3H]methylated ACGT-oligo was annealed to complementary RNA, and the activity of AlkB, hABH2 and hABH3 on this substrate (denoted DNA*:RNA; the asterisk indicates the methylated strand) was compared with that on the single-stranded, methylated DNA (ssDNA). In accordance with our previous results, it was observed that hABH2 preferred DNA*:RNA over ssDNA (Figure 3B), while the opposite was true for hABH3 (Figure 3C). In contrast, AlkB displayed similar activity on ssDNA and DNA*:RNA (Figure 3A), which is somewhat surprising, given the observed preference of AlkB for ssDNA over dsDNA.
Figure 3.

Activity of AlkB proteins on a [3H]methylated DNA oligonucleotide associated with a complementary RNA strand. Increasing concentrations of AlkB (A), hABH2 (B) or hABH3 (C) were incubated with [3H]methylated ACGT-oligo (ssDNA, closed symbols), or with the same oligo associated with a complementary RNA strand (DNA*:RNA, open symbols). The liberated ethanol soluble activity was measured by scintillation counting. Error bars represent the range of duplicate measurements.
Activity of AlkB and hABH3 on methylated RNA
While hABH2 showed little or negligible activity on RNA substrates, AlkB and hABH3 were able to efficiently demethylate 1-meA and 3-meC lesions in methylated RNA homopolymers (11). AlkB and hABH3 preferred ssDNA over dsDNA, and we also examined the activity of these enzymes on a methylated RNA oligonucleotide in the single-stranded form (ssRNA), or as a part of RNA*:DNA (the asterisk indicates the methylated strand) or dsRNA duplexes. The results clearly showed that both AlkB and hABH3 had a preference for ssRNA over the RNA*:DNA and dsRNA duplexes, and that the introduction of a complementary DNA or RNA strand had a similar negative effect on the ability of the methylated RNA oligonucleotide to act as a substrate for these enzymes (Figure 4). Since hABH2 displayed a preference for dsDNA over ssDNA, we also tested the activity of this enzyme on the double-stranded substrates containing a methylated RNA strand (RNA*:DNA and dsRNA). However, the activity of hABH2 on these substrates was negligible (data not shown).
Figure 4.
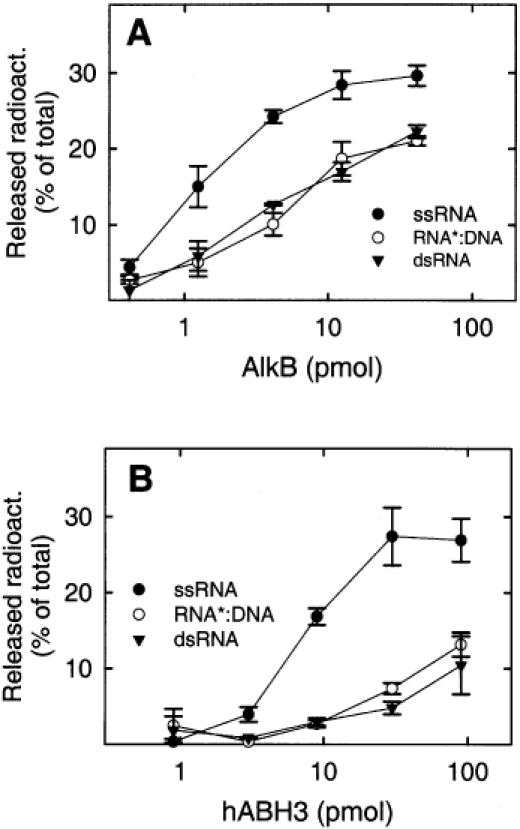
Repair of [3H]methylated RNA oligonucleotides by AlkB and hABH3. Increasing amounts of AlkB (A) or hABH3 (B) were incubated with a [3H]methylated RNA oligonucleotide (ssRNA, closed circles), or with the same oligonucleotide annealed to a complementary DNA strand (RNA*:DNA, open circles) or RNA strand (dsRNA, triangles). The sequence of the [3H]methylated RNA oligonucleotide was CAUGAUAACCGCGACUACACUGAC (corresponding to the DNA sequence of the ACGT-oligo used previously). Error bars represent the range of duplicate measurements.
DISCUSSION
In the present work, we have studied the activities of the E.coli AlkB protein and two human homologues, hABH2 and hABH3, on various single-stranded and double-stranded nucleic acid substrates, and the key findings have been summarized in Table 1. Two main conclusions can be drawn from the data. First, independent of sequence context, AlkB and hABH3 prefer ssDNA over dsDNA, while the opposite is true for hABH2. Second, the same pattern applies in large part also to various RNA-containing substrates.
Table 1. Substrate specificities of AlkB proteins.
| Substrate | EcAlkB | hABH2 | hABH3 |
|---|---|---|---|
| ssDNA | +++ | ++ | +++ |
| dsDNA | ++ | +++ | + |
| DNA*:RNA | +++ | +++ | + |
| ssRNA | ++ | − | +++ |
| dsRNA | + | − | + |
| RNA*:DNA | + | − | + |
+++, preferred substrate; ++, 2–5-fold reduction relative to preferred substrate; +, ∼10-fold reduction; and −, >100-fold reduction.
Detailed insight into the reaction mechanisms of DNA repair enzymes has often been obtained by solving the three-dimensional structure of various enzyme–substrate complexes by X-ray crystallography. The observed differences in substrate specificity between the three enzymes studied here are likely to reflect differences in substrate recognition at the active site, and the information provided in this work may be useful when selecting substrates for structural studies. Until now, the AlkB substrates used for biochemical studies have been produced by treatment of nucleic acids with alkylating agents, generating substrate molecules containing various alkyl lesions, including 1-meA and 3-meC, at random positions. Such heterogeneous substrates are evidently not suitable for structural studies. However, the synthesis of DNA and RNA oligonucleotides containing 1-meA at specific positions has recently been reported (13).
The preference of hABH2 for dsDNA versus ssDNA was observed in the presence of 10 mM magnesium, but not under magnesium-free conditions. Cellular magnesium concentrations have been reported to be in the range of 5–30 mM (14) and it is therefore likely that the preference for dsDNA exists also in vivo. Under physiological conditions of K+ and Mg2+, ∼0.2 mol Mg2+ is bound per mole of phosphate in DNA, stabilizing the double helical structure. It seems plausible that the observed Mg2+-induced increase in hABH2 activity on dsDNA can be attributed to this stabilization. Numerous DNA repair proteins have an absolute requirement for magnesium [reviewed in (14)], and the activity of some repair enzymes are modulated by the presence of magnesium. For example, hNTH is a human DNA glycosylase which removes premutagenic cytosine lesions from DNA, and its activity varies according to the identity of base opposite the lesion. Interestingly, such opposite base dependence, which is thought to be important for the antimutator function, is only observed in the presence of physiological concentrations of Mg2+ (15).
We observed a single exception from the general scheme that AlkB and hABH3 prefer single-stranded nucleic acids while hABH2 favors double-stranded substrates. The activity of AlkB on the DNA*:RNA hybrid was actually similar to that observed with ssDNA, while its activity on the other double-stranded substrates tested (dsDNA, RNA*:DNA and dsRNA) was reduced relative to the corresponding single-stranded methylated molecules. While dsDNA typically forms a helix of the B-form and dsRNA usually is in the A-form, DNA:RNA hybrids tend to adopt an intermediate conformation, though closer to the A-form than to the B-form (16). Conceivably, this conformation allows for a particularly favorable orientation of the methylated base with respect to AlkB-mediated demethylation.
Using an unmethylated poly(dT) annealed to an ∼310 bp methylated poly(dA) substrate, Trewick et al. (7) concluded that AlkB prefers dsDNA over ssDNA, while we, using a different substrate (AGC-oligo), reached the opposite conclusion (6). To resolve this apparent discrepancy, we have used here three different DNA substrates, and the results unequivocally showed that AlkB displayed a moderate preference for ssDNA. The annealing of poly(dA) to poly(dT) may yield a heterogeneous mixture of substrates containing both single- and double-stranded regions, and possibly also substrates containing more than two single-stranded molecules, such as various concatemers and branched molecules. Therefore, this substrate may not be ideal for studying the ssDNA versus dsDNA preference of AlkB, which might explain the slight discrepancies between the results reported by the Sedgwick group and us.
Using a broader range of substrates, we here corroborate our previous findings regarding hABH2 and hABH3, i.e. the only two functional human AlkB homologues confirmed so far (11). These enzymes display very different substrate specificities; while hABH3 strongly favors ssDNA and ssRNA, double-stranded molecules with a damaged DNA strand are preferred substrates for hABH2 (Table 1). In contrast, Duncan et al. (8) did not report any such difference, and concluded that hABH2 and hABH3 are likely to have similar functions in DNA repair. In part, this discrepancy can be explained by our observation that the dsDNA preference of hABH2 was only detected in the presence of magnesium.
In a recent study, Mishina et al. (17) probed the interaction of human and bacterial AlkB proteins with DNA through chemical crosslinking. A conserved, putatively iron-binding histidine residue in AlkB, hABH2 and hABH3 was mutated to cysteine, and the crosslinking of this cysteine to various oligonucleotides containing a modified, thiol-tethered cytosine was studied. In the case of AlkB and hABH2, a similar level of crosslinking was observed with single-stranded and double-stranded substrates, while hABH3 showed preference for ssDNA. Qualitatively, these results agree well with our data, which showed that the discrimination between ssDNA and dsDNA was strongest in the case of hABH3.
The in vitro data presented here are also supported by previous results on the ability of hABH2 and hABH3 to complement an E.coli alkB mutant which is defective in reactivation of methylated DNA and RNA bacteriophages. It was shown that hABH2 reactivated both ssDNA and dsDNA phages, but not an RNA phage, while hABH3 was capable of reactivating ssDNA and RNA phages, but not dsDNA phage (11). Interestingly, hABH2 and hABH3 also display very different subcellular localizations. hABH2, which is confined to the nucleus, is present in replication foci in the S phase, and is concentrated in nucleoli in the other stages of the cell cycle. hABH3 is present both in the cytosol and in the nucleus, but is somewhat excluded from nucleoli, and its localization does not show any cell cycle dependence (11). The observed differences between hABH2 and hABH3 with respect to substrate specificity and subcellular localization, strongly suggest that these enzymes have different roles in the repair of nucleic acids.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Arne Klungland and Hans Krokan for a critical reading of the manuscript. This work was supported by the Norwegian Cancer Society and the Research Council of Norway, and by the Simon Fougner Hartmanns Familiefond (to E.S.).
REFERENCES
- 1.Sedgwick B. and Lindahl,T. (2002) Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene, 21, 8886–8894. [DOI] [PubMed] [Google Scholar]
- 2.Landini P. and Volkert,M.R. (2000) Regulatory responses of the adaptive response to alkylation damage: a simple regulon with complex regulatory features. J. Bacteriol., 182, 6543–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley T.J. and Samson,L.D. (2003) AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem. Sci., 28, 2–5. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L. and Koonin,E.V. (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol., 2, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schofield C.J. and Zhang,Z. (1999) Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol., 9, 722–731. [DOI] [PubMed] [Google Scholar]
- 6.Falnes P.O., Johansen,R.F. and Seeberg,E. (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 419, 178–182. [DOI] [PubMed] [Google Scholar]
- 7.Trewick S.C., Henshaw,T.F., Hausinger,R.P., Lindahl,T. and Sedgwick,B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, 419, 174–178. [DOI] [PubMed] [Google Scholar]
- 8.Duncan T., Trewick,S.C., Koivisto,P., Bates,P.A., Lindahl,T. and Sedgwick,B. (2002) Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA, 99, 16660–16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koivisto P., Duncan,T., Lindahl,T. and Sedgwick,B. (2003) Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J. Biol Chem., 278, 44348–44354. [DOI] [PubMed] [Google Scholar]
- 10.Kurowski M.A., Bhagwat,A.S., Papaj,G. and Bujnicki,J.M. (2003) Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. Bmc Genomics, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aas P.A., Otterlei,M., Falnes,P.O., Vagbo,C.B., Skorpen,F., Akbari,M., Sundheim,O., Bjoras,M., Slupphaug,G., Seeberg,E. et al. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature, 421, 859–863. [DOI] [PubMed] [Google Scholar]
- 12.Singer B. and Grunberger,D. (1983) Molecular Biology of Mutagens and Carcinogens. Plenum Press, New York. [Google Scholar]
- 13.Mikhailov S.N., Rozenski,J., Efimtseva,E.V., Busson,R., Van Aerschot,A. and Herdewijn,P. (2002) Chemical incorporation of 1-methyladenosine into oligonucleotides. Nucleic Acids Res., 30, 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartwig A. (2001) Role of magnesium in genomic stability. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis, 475, 113–121. [DOI] [PubMed] [Google Scholar]
- 15.Eide L., Luna,L., Gustad,E.C., Henderson,P.T., Essigmann,J.M., Demple,B. and Seeberg,E. (2001) Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry, 40, 6653–6659. [DOI] [PubMed] [Google Scholar]
- 16.Gyi J.I., Conn,G.L., Lane,A.N. and Brown,T. (1996) Comparison of the thermodynamic stabilities and solution conformations of DNA center dot RNA hybrids containing purine-rich and pyrimidine-rich strands with DNA and RNA duplexes. Biochemistry, 35, 12538–12548. [DOI] [PubMed] [Google Scholar]
- 17.Mishina Y., Lee,C.H. and He,C. (2004) Interaction of human and bacterial AlkB proteins with DNA as probed through chemical cross-linking studies. Nucleic Acids Res., 32, 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]


