Abstract
Terminal restriction fragment analysis is the only method currently available for measuring telomere length in Caenorhabditis elegans. Its limitations include low sensitivity and interference by the presence of interstitial telomeric sequences in the C.elegans genome. Here we report the adaptation of single telomere length analysis (STELA) to measure the length of telomeric repeats on the left arm of chromosome V in C.elegans. This highly sensitive PCR-based method allows telomere length measurement from as few as a single worm. The application of STELA to eight wild-type C.elegans strains revealed considerable strain-specific differences in telomere length. Within individual strains, short outlying telomeres were observed that were clearly distinct from the bulk telomere length distributions, suggesting that processes other than end-replication losses and telomerase-mediated lengthening may generate telomere length heterogeneity in C.elegans. The utility of this method was further demonstrated by the characterization of telomere shortening in mrt-2 mutants. We conclude that STELA appears to be a valuable tool for studying telomere biology in C.elegans.
INTRODUCTION
Telomeres are structures composed of short G-rich tandem repeats and proteins found at the end of eukaryotic chromosomes. A major function of telomeres is to protect chromosome ends from being recognized as double-stranded DNA breaks (1). Telomeres end with a 3′ single-stranded overhang (2–6). In mammals and trypanosomes, this single-stranded overhang loops back and invades the double-stranded region to form a closed structure called the ‘T-loop’ (7,8). This structure with the associated proteins provides a physical cap that protects chromosome ends from degradation and fusion (1,7,9). The length of telomeric repeats may affect the cap structure in two ways. First, a minimal length is required for T-loop formation (7). Second, it has been proposed that telomeres are in a state of equilibrium between capped and uncapped states, and the equilibrium may be shifted towards one of these states depending on telomere length (1). The loss of telomere repeats has been implicated in both tumourigenesis and the ageing process (10–12). Thus, there is considerable interest in accurately measuring telomere length in both humans and model organisms.
The most commonly utilized technology for determining telomere length in various species is terminal restriction fragment (TRF) analysis. This approach involves a standard genomic Southern blot with terminal restriction fragments detected by hybridization to a probe containing telomeric repeats. While TRF analysis is of widespread utility, it has limited resolution and sensitivity (being only capable of determining the mean telomere length of all chromosome ends from a large number of cells). Furthermore, telomere length measurement by TRF analysis can be confounded by the presence of interstitial telomeric sequences (ITS). Another common method for telomere length measurement is fluorescence in situ hybridization (FISH), which involves the hybridization of a fluorescent telomeric PNA probe to either metaphase spreads (13) or interphase chromosomes of intact cells followed by flow cytometry (14), with intensity of fluorescent signals proportional to telomere length. This approach has the resolution and sensitivity to allow the analysis of all chromosome ends. Recently, a new PCR-based method called single telomere length analysis (STELA) has been developed that allows the measurement of single telomeres (15). Designed specifically for measuring the length of the XpYp telomere in human cells, this method allows the accurate measurement of single telomere molecules thereby providing higher resolution than TRF analysis. Provided that telomere-adjacent DNA sequences are available, STELA is in principle applicable to telomeres of other human chromosomes and other species, allowing very accurate estimates of telomere lengths.
Caenorhabditis elegans is a key genetic model organism for the study of many evolutionarily conserved biological processes (such as development, apoptosis, meiosis and ageing) because of its short life cycle, simple anatomy, ease of laboratory manipulation and completed genome sequence. Caenorhabditis elegans should also prove to be an important multicellular model organism for studying telomere function. However, thus far the genome of C.elegans has been largely refractory to detailed telomere length analysis. This is primarily a consequence of the fact that the genome is interspersed with ITS. Hybridization of the telomeric probe to these sequences results in low molecular weight bands that interfere with detection and length measurement of short telomeres during TRF analysis, and greatly complicates the use of FISH-based methods in telomere length measurement in C.elegans. We therefore wished to develop an alternative, more accurate and sensitive method of telomere length determination in C.elegans that was not confounded by the presence of ITS. Here, we describe the development of STELA to determine the telomere length of the VL chromosome from single C.elegans worms. Our results point to considerable heterogeneity in telomere length within individual animals and considerable variation in the average telomere length between strains.
MATERIAL AND METHODS
Strains
Worms were grown at room temperature (19–22°C) and maintained as described (16). The strains used in this study include the following: wild-type strains N2, N2(ancestral), CB3191, CB3192, CB4852, CB4856, CC2 and TR403; CB5348 has the genotype of mrt-2(e2663).
Single telomere length analysis (STELA)
Bulk genomic DNA isolation and ligation
Worms were lysed for 1 h at 55°C in lysis buffer A, which was made by combining 500 μl NTE buffer (100 mM NaCl, 50 mM Tris, 20 mM EDTA), 25 μl of 10% SDS, 10 μl of 10 mg/ml Proteinase K, and 1 μl β-mercaptoethanol. DNA was purified as follows: three extraction steps were performed with equal amounts of phenol/chloroform/isoamyl alcohol (25:24:1) in Phase Lock Gel™ tubes (Eppendorf). DNA was precipitated by addition of 68 μl 7.5 M NH4OAc, 3 μl glycogen and 500 μl 100% ethanol and incubation for 30 min on dry ice in ethanol, washed twice with 70% ethanol, and subsequently resuspended in 10 mM TrisCl (pH 8.5). Ligation was carried out as follows: a mix of 20 ng of DNA (quantitated by PicoGreen® dsDNA Quantitation Kit, Molecular Probes) and 1 μl of 10 μM telorette (Table 1) in a 2 μl volume was incubated at 60°C for 10 min. Ligation was carried out at 35°C for 12–15 h with 0.2 μl of 1 U/μl T4 ligase (Amersham) and 0.4 μl of 10× manufacturer's ligation buffer in a final volume of 4 μl. The ligase was then inactivated by incubation at 70°C for 15 min.
Table 1. Distribution of telomere length in strains used in this study.
| Straina | VL Mean telomere length (kb) | SD |
|---|---|---|
| N2b | 1.25 (±0.05)d | 0.34d |
| N2c | 1.49 (±0.13)e | 0.49e |
| N2 (ancestral) | 1.80 (±0.14)e | 0.48e |
| CB3192 | 1.88 (±0.19)e | 0.56e |
| CB4852 | 2.00 (±0.26)e | 0.73e |
| CB4856 | 2.05 (±0.17)e | 0.48e |
| Mrt-2 F2 | 1.19 (±0.07)f | 0.11f |
| Mrt-2 F5 | 0.86 (±0.05)f | 0.10f |
| Mrt-2 F9 | 0.46 (±0.04)f | 0.08f |
DNA isolation and ligation from five worms or single worms
Five adult worms or a single adult worm were lysed in 5 μl lysis buffer B (1× PCR buffer (500 mM Tris pH 8.3, 350 μM Na2HPO4, 0.5% Tween-20), 1.5 mM MgCl2, 0.06 mg/ml Proteinase K) at 57°C for 60 min. The lysate was diluted with 10 mM Tris-Cl (pH 8.5) and DNA was extracted twice with equal amounts of phenol/chloroform/isoamyl alcohol (25:24:1) using Phase Lock GelTM tubes (Eppendorf). DNA was precipitated by addition of 68 μl 7.5 M NH4OAc, 3 μl glycogen and 500 μl 100% ethanol and incubation overnight at −70°C, washed twice with 70% ethanol, and subsequently resuspended in 2 μl 10 mM Tris (pH 8.5) with 2.5 μM telorette (Table 1). The mixture of genomic DNA and telorette was incubated at 60°C for 10 min, followed by ligation at 35°C for 12–15 h with 0.2 μl of 1 U/μl T4 ligase (Amersham) and 0.3 μl of 10× manufacturer's ligation buffer in a final volume of 3 μl. The ligase was then inactivated by incubation at 70°C for 15 min.
Telomere amplification and detection
Ligated DNA was used as template in original concentration or various dilutions in subsequent PCR reactions. A 15 μl PCR reaction contained the indicated amount of ligated DNA, 1× PCR buffer IV (ABgene), 2 mM MgCl2, 0.5 μM each of the teltail primer and the subtelomeric-region-specific primer (512) (Table 1), 0.3 mM of each dNTP (Amersham) and 1.5 U Extensor Hi-Fidelity PCR Enzyme Mix (ABgene). Thermal cycling conditions were the following: initial denaturation at 94°C for 3 min, 25 cycles of 94°C for 20 s, 64°C for 30 s, and 70°C for 8 min, followed by final elongation at 70°C for 10 min. PCR products were separated on 1% agarose gel, and transferred onto Hybond-N+ membrane (Amersham) according to standard alkali transfer protocol. Membranes were hybridized to either a [α-32P]dCTP random-labelled subtelomeric probe (fragment generated by PCR of N2 genomic DNA with primers 509 and 510; see Figure 1) or a [γ-32P]dATP end-labelled (GCCTAA)5 telomeric oligonucleotide probe. Signals were detected using a Molecular Dynamics Storm 860 PhosphoImager system (Amersham Biosciences).
Figure 1.
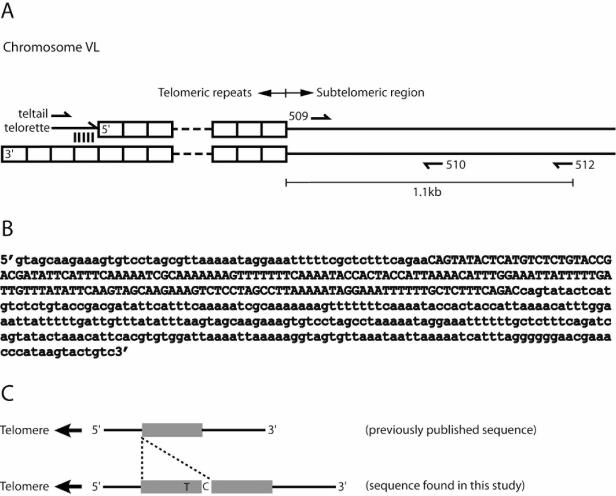
Design of STELA in C.elegans. (A) The diagram shows the telomeric repeats and subtelomeric region of chromosome V Left (VL). The 3′ end of the telorette carries 7 nt that are complementary to the 3′ G-rich overhang at the terminus of the telomere. Primer teltail aligns to the unique sequence of the telorette, and is used in amplification together with primer 512 that is specific to the subtelomeric region of chromosome VL. Primers 509 and 510 are used to generate a fragment for the subtelomeric probe. (B,C) The fragment amplified using the 509 and 510 primers (509/510) contains a duplication that is not present in the previously published sequence of cTel3X. (B) Sequence of 509/510 fragment. The previously published sequence is shown in lowercase, and nucleotides not present in the previously published sequence are in uppercase letters. (C) A schematic representation of the duplication. The grey box represents the duplicated sequence. The two duplicated sequences are separated by a C, and are almost identical except for an insertion of T.
Oligonucleotides
The oligonucleotide sequences were as follows: teltail, 5′-TGCTCCGTGCATCTGGCATC-3′; 509, 5′-GTAGCAAGAAAGTGTCCTAGCG-3′; 510, 5′-GACAGTACTTATGGGTTTCGTTC-3′; 512, 5′-GATGCGCAGCTAACTATAGGAC-3′; telorette 502, 5′-GACAGCTATGACTGCTCCGTGCATCTGGCATCGCCTAAG-3′; telorette 503, 5′-GACAGCTATGACTGCTCCGTGCATCTGGCATCTAAGCCT-3′; telorette 504, 5′-GACAGCTATGACTGCTCCGTGCATCTGGCATCCCTAAGC-3′; telorette 505, 5′-GACAGCTATGACTGCTCCGTGCATCTGGCATCCTAAGCC-3′; telorette 506, 5′-GACAGCTATGACTGCTCCGTGCATCTGGCATCAAGCCTA-3′; telorette 507, 5′-GACAGCTATGACTGCTCCGTGCATCTGGCATCAGCCTAA-3′.
RESULTS AND DISCUSSION
Design of STELA
Figure 1A shows the design of STELA in C.elegans. Chromosome VL was selected for two reasons. First, it has been shown that telomere length on different chromosome ends varies in C.elegans, and VL is among one of the shortest (17). Second, unlike several other chromosome ends, the subtelomeric region of VL contains unique sequences for primer design. Thus, single telomere analysis of this chromosome end should be both specific and well within the range of PCR. The method involves the ligation of an oligonucleotide, termed the telorette, to the 5′ end of the telomere (i.e. the C-rich strand). The 3′ end of the telorette, which is complementary to the G-rich overhang of the telomere, serves to facilitate ligation. The rest of the telorette has a unique sequence that is not found in the C.elegans genome to allow telorette-specific PCR amplifications. The terminus of the C-rich strand of the telomere may end in any of the six nucleotides within a telomeric repeat, which in C.elegans is TTAGGC. Therefore, six telorettes were designed, each carrying one of the six possible frames of a telomeric repeat at its 3′ end. Ligated genomic DNA is then amplified by PCR with the teltail and 512 primers. The primer teltail recognizes specifically the 3′ end of the telorette (15), and the sequence of primer 512 is specific to the subtelomeric region of VL. Using these primers, the telomeric repeats and 1095 bp of subtelomeric sequence of VL are amplified. Such products are expected to hybridize to a subtelomeric probe (generated by primers 509 and 510) as well as a telomeric probe that recognizes telomeric repeats (Figure 1A).
Preliminary analysis with all six telorettes demonstrated that telorette 503, whose 3′ end has the frame of AAGCCT, provided the most efficient amplification, although the other five telorettes also supported amplification but to a lesser extent (data not shown). It is interesting to note that in humans, the telorette that provided the highest efficiency of telomere amplification [telorette 2, (15)], ligated to the C-rich strand in the same frame as 503. It is thus possible that in both humans and C.elegans the C-rich strand preferentially terminates with the sequence 5′-AACCCT-3′ and 5′-AAGCCT-3′, respectively.
The published sequence of cTel3X, which is the cosmid containing the telomere of VL, predicts that PCR would generate a 300 bp product from primers 509 and 510. Our sequencing results revealed a duplication of 170 bp within the 509/510 fragment in the reference wild-type strain N2 and a number of other strains originated from different laboratories (Figure 1B and C). Therefore, we propose that the true sequence of cTel3X in N2 is as shown in Figure 1B and C (GenBank accession AY559143).
STELA from bulk genomic DNA
Figure 2 shows the sensitivity of STELA in measuring telomere length in C.elegans. When ∼300 pg of bulk genomic DNA from the wild-type strain N2 was used in a reaction, a smear was observed. With decreasing amounts of template DNA, the smear was resolved into discrete bands, each band representing a single amplified telomere. As each telomeric molecule contains 1.1 kb of telomere-adjacent DNA, bands below this size should not occur. When DNA was ligated at 5 ng/μL, as little as 10 pg was needed for PCR amplification, thus DNA extracted from a single plate (typically containing 200–300 worms) is more than sufficient for telomere length measurement. There are several features shown in Figure 2 that demonstrate the specificity of STELA in amplifying telomeric sequences. First, almost every band could be detected by hybridization with both the telomeric and subtelomeric probes (Figure 2). Occasional bands were observed that showed differential hybridization with the two probes being only detected by the subtelomeric probe. Such bands were below the threshold length (∼1.2 kb) that facilitated detection by the telomeric probe, presumably as a consequence of insufficient suitable telomere target sequence. Second, the number of bands amplified per reaction was proportional to the amount of DNA template. Third, bands had consistent intensities suggesting that all molecules were amplified from the first round of PCR. These data are consistent with specific amplification from single telomeric molecules.
Figure 2.
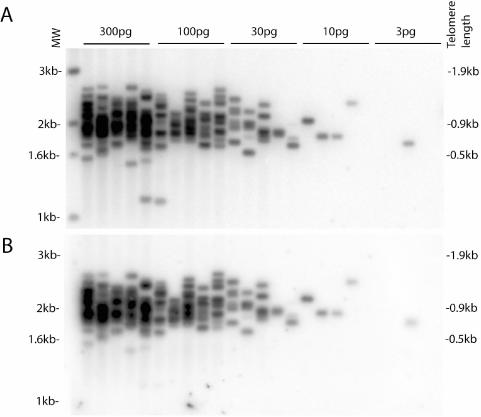
STELA from bulk N2 genomic DNA. 300, 100, 30, 10 or 3 pg of N2 genomic DNA ligated to telorette 503 was amplified with primers teltail and 512. MW is the marker lane showing the molecular weight of the bands. The corresponding telomere length is indicated on the right. The blot was hybridized to (A) the subtelomeric probe, or (B) the telomeric probe.
STELA from five worms or single worms
One goal in developing STELA for C.elegans was to measure telomere length from a few or even single worms. TRF analysis in C.elegans requires on average 5 μg of genomic DNA for each measurement, thus thousands of worms have to be grown for DNA extraction. Mutants with telomere defects are likely to have progressively reducing brood size (18–20), and collecting thousands of such mutants for studying telomere length dynamics may sometimes be impractical. For C.elegans to become a valuable model organism in studying telomere biology, a technique with higher sensitivity than TRF analysis is required. This prompted us to explore the possibility of performing STELA starting from a small number of worms. DNA was extracted from five worms or a single worm and ligated to the telorette (Methods). The rest of the STELA procedures were the same as with bulk genomic DNA. Figure 3 shows that STELA was sensitive enough to allow telomere length measurement from as little as a single adult worm. The majority of VL telomeres were between 1 and 2 kb in our wild-type N2 strain, generating a smear that resembled a TRF analysis (17,20) when DNA from an equivalent of 0.8 worm was used in PCR amplification, and resolved into a single banding pattern upon serial dilution to 0.08 worm per PCR (Figure 3). The variable size of telomere repeats at chromosome VL in single worms is remarkable. The assumption that similar length heterogeneity exists at other chromosome ends within a strain needs to be verified. We did observe a clear bimodal distribution of telomere length in the offspring of crosses between stains with relatively long and short VL telomeres (results not shown) arguing against the observed variation being an artefact of the assay.
Figure 3.
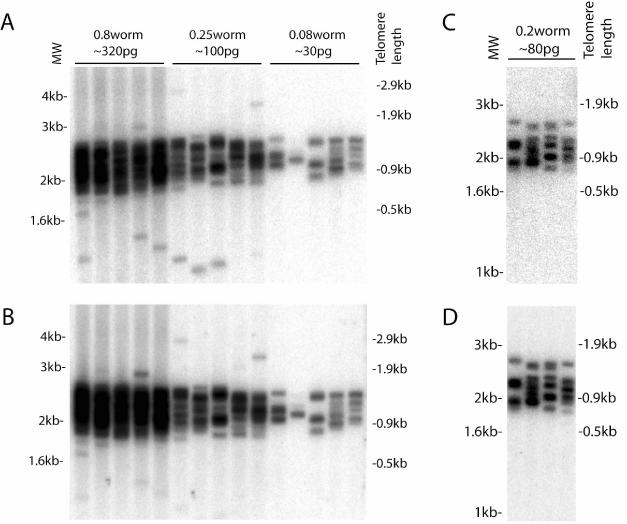
Telomere length measurement from five worms or a single worm by STELA. (A,B) DNA was extracted from five-worm samples independently. After ligation to the telorette, DNA was pooled, and DNA from an equivalent of 0.8 worm, 0.25 worm or 0.08 worm was amplified. Assuming each adult worm contains ∼400 pg of genomic DNA [from 959 somatic cells and ∼1000 germline nuclei per gonad arm; (22,23)], they would represent ∼320, ∼100 and ∼30 pg DNA, respectively, if DNA extraction was 100% efficient. The marker lane is shown on the left and the corresponding telomere length is shown on the right. The blot was hybridized to (A) the subtelomeric probe, or (B) the telomeric probe. (C,D) A single worm is sufficient for telomere length measurement by STELA. DNA was extracted from a single adult worm. After ligation to the telorette, DNA from an equivalent of 0.2 worm was amplified. Assuming each adult worm contains ∼400 pg genomic DNA, it would represent 80 pg of DNA if DNA extraction was 100% efficient. The marker lane is shown on the left and the corresponding telomere length is shown on the right. The blot was hybridized to (C) the subtelomeric probe, or (D) the telomeric probe.
Variations in telomere length in wild-type C.elegans strains
Telomere length of VL in the N2 strain cultured in our laboratory had a mean of 1.25 ± 0.05 kb and a SD of 0.34 (Table 1). To determine how typical the VL telomere length was of wild-type C.elegans, we undertook STELA on a number of different wild-type strains, including the reference wild-type strain N2, obtained from the Caenorhabditis Genetics Center [CGC, Minneapolis, MN; description of the origin of some of the wild-type strains can be found in ref (21)]. DNA was extracted from five worms from each strain and ligated to the telorette and an equivalent of 0.8 worm, 0.25 worm or 0.08 worm was subjected to STELA. Figure 4 shows the variations of VL telomere length in eight wild-type strains. Detailed telomere length analysis revealed that N2 had the shortest mean telomere length (1.49 ± 0.13 kb) among the eight strains (Table 1). CB3192, CB4852, CB4856 and N2 (ancestral) had similar telomere length distribution and slightly longer mean telomere length than N2. In CB3191 and CC2, telomere length was more than twice as long as N2. The subtelomeric region was likely to be invariant among these different wild-type strains because the length of the subtelomeric fragment amplified by primers 509 and 512 was the same as in N2 (data not shown). An exception was TR403, where the 509/512 fragment was about 340 bp shorter than N2 (data not shown). Interestingly, amplification of VL telomere in TR403 resulted in bands that were extremely large (>10 kb). These large bands could be a result of either extremely long telomeres of >10 kb in TR403, or a long insertion between the site of primer 509 and the telomeric repeats. The latter possibility was ruled out because the telomeric probe could hybridize to bands that were >1.6 kb in size, indicating the subtelomeric fragment amplified must be <1.6 kb (Figure 4). Comparison of results from our `in-house' N2 strain with that obtained from the CGC showed a difference in mean VL telomere length of 0.25 kb (Table 1), suggesting telomere length can fluctuate within a specific strain. Our results illustrated that VL telomere length was highly variable (varying from 1.25 kb to >10 kb) among different wild-type C.elegans strains. This variation could be a consequence of genetic or epigenetic differences that affect the balance between telomere shortening and telomere elongation, such as processivity of telomerase and its expression levels, and the affinity of telomere-binding proteins to telomeric DNA (1). Superimposed over the bulk telomere length distributions were additional shorter telomeres; whilst reminiscent of the shortened telomeric outliers observed in human clonal fibroblast cultures (15), the mechanisms underlying the generation of these telomeres are unclear. Although telomere length varies in different wild-type strains, the degree of this additional heterogeneity was similar in all strains except TR403. Therefore, heterogeneity of telomere length in C.elegans may not be generated randomly, but via some tightly controlled mechanism. It is interesting to note that TR403 has a high copy number of the transposable element Tc1 (21). A transposable element inserted close to the telomere might change the chromatin structure near the telomere and thereby affect telomere homeostasis.
Figure 4.
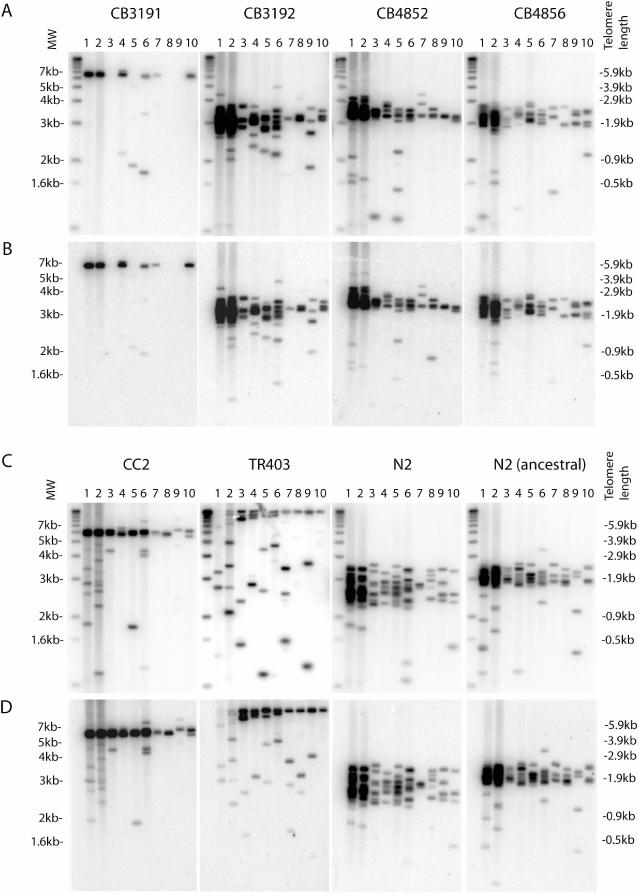
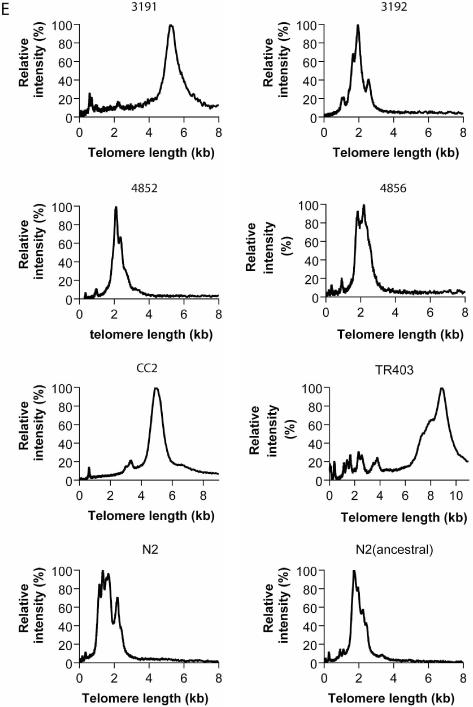
Variations in the length of VL telomeres in different wild-type C.elegans strains. The wild-type strains (A,B) CB3191, CB3192, CB4852, CB4856, and (C,D) CC2, TR403, N2, N2(ancestral) were analysed by STELA. DNA was extracted from five worms for each strain. After ligation to the telorette, DNA from an equivalent of 0.8 worm (lanes 1, 2), 0.25 worm (lanes 3–6) or 0.08 worm (lanes 7–10) was amplified. In TR403, high molecular weight bands could not be amplified in lanes 1 and 2 as in all other lanes. The marker lane is shown on the left and the corresponding telomere length is shown on the right. The blot was hybridized to (A,C) the subtelomeric probe, or (B,D) the telomeric probe. (E) Distribution of telomere length in different wild-type strains. Intensities of signals from lanes 3–10 of each blot from (A) and (C) were measured and added together at each telomere length interval. Relative intensity (compared to the highest intensity) was plotted against telomere length. Relative intensity may represent frequency in this case because the blots were hybridized to the subtelomeric probe and variations in band intensities should be random among all bands.
Use of STELA to measure telomere shortening in mutants
As a further validation of the technique, a mutant with known telomere defects was subjected to STELA. The C.elegans mrt-2 gene is a homolog of the Saccharomyces cerevisiae RAD17 and Schizosaccharomyces pombe rad1+ checkpoint genes. Mrt-2 shows progressively shortening telomeres and frequent end-to-end chromosome fusions (20). STELA revealed that VL telomere shortened by ∼0.7 kb within seven generations in mrt-2 (Table 1, Figure 5). This was comparable to that determined by TRF analysis (20), which measures average telomere length among all telomere ends. This result demonstrates that the dynamics of VL telomere length could represent the average of all telomeres in certain strains. An interesting observation from our analysis was that the distribution of telomere lengths in mrt-2 in any given generation was less heterogeneous than those of the wild-type strains (Table 1, Figures 4 and 5). Furthermore, SD was very similar among the three generations examined in spite of the changes in telomere length (Table 1). Therefore, the mrt-2 gene may contribute to telomere length heterogeneity in C.elegans.
Figure 5.
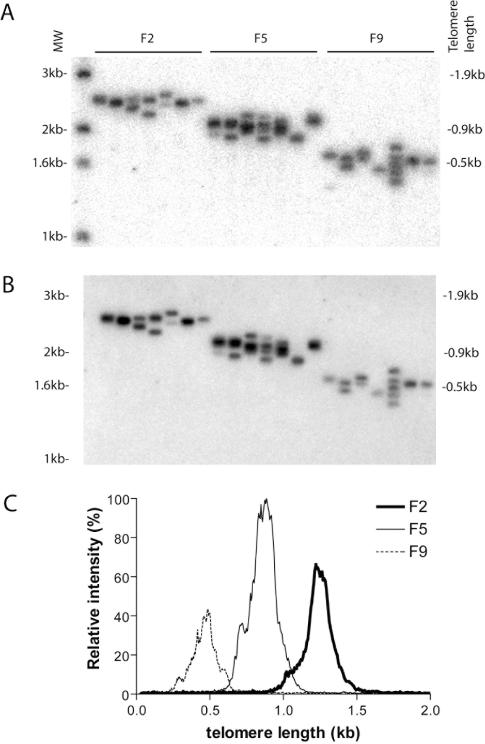
Telomere shortening in mrt-2. (A,B) Telomere length in five worms from F2, F5 and F9 was analysed by STELA. Each lane was amplified from an equivalent of 0.1 worm. The marker lane is shown on the left and the corresponding telomere length is shown on the right. The blot was hybridized to (A) the subtelomeric probe, or (B) the telomeric probe. (C) Distribution of telomere length at each generation in mrt-2. Intensities of signals of each lane in (A) was measured, and those from the same generation were added together at each telomere length interval. Relative intensity was plotted against telomere length. Thick line represents F2, thin line represents F5 and dotted line represents F9. Relative intensity may represent frequency in this case because the blots were hybridized to the subtelomeric probe and variations in band intensities should be random among all bands.
In conclusion, the highly sensitive technique of STELA enables telomere length measurement from as few as a single worm. We have applied STELA to various wild-type strains in addition to the reference strain N2. We observed considerable variation in telomere length among different wild-type strains, which may reflect strain-specific differences in telomere length homeostasis, and suggests that caution is warranted in linking genetic differences to differences in telomere length between stains. We have demonstrated the use of STELA in characterizing shortening of telomeres in mutants. The technique will greatly enhance the potential of C.elegans as a model organism for studying telomere biology.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Shawn Ahmed for providing the mrt-2 mutant strain. The wild-type C.elegans strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Institute of Health National Center for Research Resources. This work is supported by the Terry Fox Foundation and a grant from the Natural Sciences and Engineering Research Council (Canada). I.C. is funded by the Canadian Institutes of Health Research, the Michael Smith Foundation for Health Research and a Roman M. Babicki Fellowship. A.B. held a Canadian Institutes of Health Research postdoctoral research fellowship award. D.M.B. is a Research into Ageing Fellow.
DDBJ/EMBL/GenBank accession no. AY559143
REFERENCES
- 1.Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- 2.Henderson E.R. and Blackburn,E.H. (1989) An overhanging 3′ terminus is a conserved feature of telomeres. Mol. Cell. Biol., 9, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- 4.Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- 5.McElligott R. and Wellinger,R.J. (1997) The terminal DNA structure of mammalian chromosomes. EMBO J., 16, 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright W.E., Tesmer,V.M., Huffman,K.E., Levene,S.D. and Shay,J.W. (1997) Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev., 1, 2801–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 8.Munoz-Jordan J.L., Cross,G.A., de Lange,T. and Griffith,J.D. (2001) t-loops at trypanosome telomeres. EMBO J, 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 10.Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 11.Hastie N.D., Dempster,M., Dunlop,M.G., Thompson,A.M., Green,D.K. and Allshire,R.C. (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature, 346, 866–868. [DOI] [PubMed] [Google Scholar]
- 12.Hackett J.A., Feldser,D.M. and Greider,C.W. (2001) Telomere dysfunction increases mutation rate and genomic instability. Cell, 106, 275–286. [DOI] [PubMed] [Google Scholar]
- 13.Lansdorp P.M., Verwoerd,N.P., van de Rijke,F.M., Dragowska,V., Little,M.-T., Dirks,R.W., Raap,A.K. and Tanke,H.J. (1996) Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet., 5, 685–691. [DOI] [PubMed] [Google Scholar]
- 14.Rufer N., Dragowska,W., Thornbury,G., Roosnek,E. and Lansdorp,P.M. (1998) Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nature Biotechnol., 16, 743–747. [DOI] [PubMed] [Google Scholar]
- 15.Baird D.M., Rowson,J., Wynford-Thomas,D. and Kipling,D. (2003) Extensive allelic variation and ultrashort telomeres in senescent human cells. Nature Genet., 33, 203–207. [DOI] [PubMed] [Google Scholar]
- 16.Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicky C., Villeneuve,A.M., Lauper,N., Codourey,L., Tobler,H. and Muller,F. (1996) Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc. Natl Acad. Sci., USA, 93, 8983–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H.W., Blasco,M.A., Gottlieb,G.J., Horner,J.W., Greider,C.W. and DePinho,R.A. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature, 392, 569–574. [DOI] [PubMed] [Google Scholar]
- 19.Herrera E., Samper,E., Martin-Caballero,J., Flores,J.M., Lee,H.W. and Blasco,M.A. (1999) Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J., 18, 2950–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed S. and Hodgkin,J. (2000) MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature, 403, 159–164. [DOI] [PubMed] [Google Scholar]
- 21.Hodgkin J. and Doniach,T. (1997) Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics, 146, 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulston J. (1988) Cell lineage. In Wood,W.B. (ed.), The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 123–155. [Google Scholar]
- 23.Berry L.W., Westlund,B. and Schedl,T. (1997) Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development, 124, 925–936. [DOI] [PubMed] [Google Scholar]


