Abstract
The Aquifex aeolicus αβ-LeuRS is the only known heterodimeric class Ia aminoacyl-tRNA synthetase. In this study, we investigated the function of the β subunit which is believed to bind tRNALeu. A yeast three-hybrid system was constructed on the basis of the interaction of the β subunit with its cognate tRNALeu. Then, seven mutated β subunits exhibiting impaired tRNA binding capacities were selected out from a randomly mutated library. Two mutations were identified in the class Ia-helix-bundle-domain, which might interact with the D-hairpin of the tRNA analogous to other class Ia tRNA:synthetases complexes. The five other mutations were found in the LeuRS-specific C-terminal domain of which the folding is still unknown. tRNA affinity measurements and kinetic analyses performed on the isolated β subunits and on the co-expressed αβ-heterodimers showed for all the mutants an effect in tRNA affinity in the ground state. In addition, an effect on the transition state of the aminoacylation reaction was observed for a 21-residues deletion mutant of the C-terminal end. These results show that the genetic approach of the three hybrid system is widely applicable and is a powerful tool for the investigation of tRNA:synthetase interactions.
INTRODUCTION
The aminoacyl-tRNA synthetases (aaRSs) catalyze the activation of their cognate amino acids and transfer them to the relevant tRNA molecules. The precise recognition of cognate tRNAs and amino acids by aaRSs guarantees the high fidelity of protein synthesis (1). The specific recognition of tRNAs by their cognate aaRSs depends on a set of identity determinants that are mostly located at the two distal extremities: the anticodon loop and the amino acid accepting stem. Most determinants are in directed contact with cognate synthetases (2), nevertheless, the aminoacylation fidelity is controlled by kinetic differences more than by binding differences (3).
The twenty aaRSs can be classified into two families of ten members each on the basis of conserved sequences and characteristic structural motifs (4). Leucyl-tRNA synthetase (LeuRS), together with arginyl-tRNA synthetase (ArgRS), cysteinyl-tRNA synthetase (CysRS), isoleucyl-tRNA (IleRS), methionyl-tRNA (MetRS) and valyl-tRNA synthetase (ValRS), comprise the class Ia aaRSs. The resolution of at least one crystal structure for each class Ia synthetase shows that the overall structure consists of the typical Rossmann fold and of a specific class Ia α-helical domain called ‘helix-bundle-domain’ (5–15). The helix-bundle-domain binds the tRNA anticodon and can be considered as the signature of the class Ia enzymes. Critical residues involved in anticodon recognition have been identified in ArgRS (16,17), IleRS (18), MetRS (19–25) and ValRS (26). It has also been shown that communication signals between the active site and the anticodon-binding domain are essential for the catalysis (17,27–29). Several idiosyncratic domains are found appended to this class Ia-scaffold of which several are remarkable. For example, a specific N-terminal domain is found in all ArgRSs that binds the D-loop of tRNAArg (5,8). Large enzymes (IleRS, LeuRS and ValRS) share CP1 domains involved in tRNA editing (9,13,30). Some bacterial MetRSs exhibit an OB-fold homologous to Trbp111, EMAPII, p43 and Arc1p. Moreover, IleRS, LeuRS and ValRS contain C-terminal domains which recognize the anticodon loop and arm [IleRS, (13)], or the D-loop [ValRS, (26)]. The function and the three-dimensional (3D) structure of the C-terminal domain of LeuRS has still to be determined (7).
The crystallographic structure of Thermus thermophilus LeuRS has been solved in the presence of leucine, leucyl-adenylate analog (7) and several analogs of the distinct pre- and post-tranfer editing substrates (30). The overall architecture is similar to that of IleRS and ValRS except that the editing domain is inserted at a different position of the primary structure and differs by the presence of an additional domain called ‘leucyl-specific’ domain which is located in the catalytic core just before the KMSKS sequence. It has been shown that this domain is essential for the aminoacylation reaction (31). Until now, no structure of the tRNALeu: LeuRS complex is available, and data on tRNALeu recognition by LeuRS mainly result from mutational studies on the tRNA identity elements. Unlike in other class Ia aaRSs systems, the anticodon triplet is mostly not recognized in LeuRS system. The main identity elements of the tRNALeu, which is a class II tRNA with an extra-long variable region, consist of the discriminator base A73 and tertiary interactions between bases from the D- and T-loops (32–39).
In the hyperthermophilic bacterium Aquifex aeolicus, leucyl-tRNA synthetase (αβ-LeuRS) is found as a heterodimer αβ resulting from the spilt of the canonical monomeric LeuRS into two polypeptide chains. In analogy with the T.thermophilus LeuRS of which the 3D structure is known, the split occurs in the leucyl-specific domain (40,41). We previously showed that the fusion of the α and β subunits resulted in a product with an activity comparable to the native αβ-LeuRS (31). Whereas the α subunit cannot be correctly folded when expressed alone, the isolated β subunit composed of 289 amino acid residues (Mr = 33.5 kDa) is stable in vivo and in vitro. Although the β subunit is catalytically inactive, it can bind and recognize tRNALeu (40,41). In analogy to the T.thermophilus LeuRS, the β subunit includes half of the leucyl-specific domain, the second half of the Rossmann fold, the helix-bundle-domain and the C-terminal domain (41). In theory, these domains, or parts of them, should be involved in tRNALeu recognition. In this study, we examine the contribution of these domains of the β subunit to the tRNALeu binding in the ground state, and to the stabilization of the transition state of the aminoacylation reaction. For this we used the yeast three-hybrid system (3HS) developed for the in vivo study of RNA-protein interactions (42,43). A functional 3HS system that uses A.aeolicus β subunit of LeuRS and ![]() was set up. A selection of loss-of-binding mutants was subsequently used to identify the β subunit residues involved in tRNALeu binding. Mutants were further analyzed after reconstitution of the whole αβ-LeuRS enzymes. This work is the first report of a functional 3HS implicating a tRNA and an aminoacyl-tRNA synthetase. It also shows that the 3HS can be used as a new tool for studying tRNA recognition in vivo.
was set up. A selection of loss-of-binding mutants was subsequently used to identify the β subunit residues involved in tRNALeu binding. Mutants were further analyzed after reconstitution of the whole αβ-LeuRS enzymes. This work is the first report of a functional 3HS implicating a tRNA and an aminoacyl-tRNA synthetase. It also shows that the 3HS can be used as a new tool for studying tRNA recognition in vivo.
MATERIALS AND METHODS
Materials
l-Leucine, dithiothreitol (DTT), ATP, CTP, UTP, GTP, 5′-GMP, inorganic pyrophosphatase and 3-amino-1, 2, 4-triazole (3-AT) were purchased from Sigma (USA). [α-32P]GTP, [14C]-leucine (300 Ci/mol) and 32P-labeled tetrasodium pyrophosphate were obtained from Amersham Biosciences (England). GF/C filters were from Whatman Company (Germany). T4 DNA ligase, and restriction endonucleases were obtained from Sangon Company (Canada, Shanghai Branch). T7 RNA polymerase was purified from an overproducing strain in our laboratory (44).
Strains and plasmids
Plasmids were isolated in Escherichia coli strain DH10B. Plasmid pSBETb (45) and pET-15b (Novagen) both with the T7 promoter were used for protein expression in BL21 (DE3). The 3HS procedures were carried out in the Saccharomyces cerevisiae strain L40coat (46). The shuttle plasmids pACTII with a LEU2 marker, and pIIIA-MS2-1, pIIIA-MS2-2 with a URA3 marker were used to express the hybrid protein and hybrid RNA molecules, respectively, in yeast (46). pIIIA-MS2-1 and pIIIA-MS2-2 differ only in the reverse orientations of the SmaI/XmaI site and in the location of the MS2-binding sites. Hybrid RNAs were transcribed from the yeast RNase P RNA (RPR1) promoter.
Construction of the yeast 3HS
By PCR, using the two primers 5′-CAGGAAACAGACCCCGGGAATT-3′ and 5′-CAAAACAGCCCGGGTTGCATGCCT-3′ (SmaI sites indicated in italic), the genes for E.coli ![]() ,
, ![]() , and A.aeolicus
, and A.aeolicus ![]() were amplified from the plasmids carrying the corresponding tRNA genes (42,43). Then, the SmaI digested DNA fragments were inserted into the same site of the plasmid pIIIA-MS2-1 and pIIIA-MS2-2. The recombinant plasmids derived from pIIIA-MS2-1 are named pIIIA-MS2-
were amplified from the plasmids carrying the corresponding tRNA genes (42,43). Then, the SmaI digested DNA fragments were inserted into the same site of the plasmid pIIIA-MS2-1 and pIIIA-MS2-2. The recombinant plasmids derived from pIIIA-MS2-1 are named pIIIA-MS2-![]() , pIIIA-MS2-
, pIIIA-MS2-![]() and pIIIA-MS2-
and pIIIA-MS2-![]() . Plasmids derived from pIIIA-MS2-2 are named as pIIIA-
. Plasmids derived from pIIIA-MS2-2 are named as pIIIA-![]() -MS2, pIIIA-
-MS2, pIIIA-![]() -MS2 and pIIIA-
-MS2 and pIIIA-![]() -MS2. The two series of vectors differ in the position of the tRNA gene relative to the MS2 RNA, for instance in pIIIA-MS2-
-MS2. The two series of vectors differ in the position of the tRNA gene relative to the MS2 RNA, for instance in pIIIA-MS2-![]() the MS2 RNA is located at the 5′ end of the tRNALeu. The series of pIIIA-MS2-derived plasmids were used to express the hybrid RNAs in yeast 3HS. On the other hand, the gene encoding the LeuRS β-subunit was inserted between the NcoI and BamHI sites of plasmid pACTII; the resulting plasmid was named pACT-LRSB. Then, the yeast strain L40coat was co-transformed with the pIIIA-MS2-derived plasmids and plasmid pACT-LRSB. Transformants were grown at 30°C on synthetic medium (YNB) lacking uracil, histidine and leucine for selection of the URA3, HIS3 and LEU2 marker genes, respectively. Testing of reporter gene activation was carried out as described (47). Colonies that appeared 5–6 days later were further analyzed for lacZ expression by plating on medium lacking uracil and leucine and supplemented with 80 μg/mL 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). To test for HIS3 activation, drops (5 μL) of yeast double transformants grown to an OD600 = 0.1 were applied to YNB plates deprived of uracil, leucine, histidine and supplemented with different concentrations (0–100 mM) of the competitive His3p inhibitor 3-amino-1, 2, 4-triazole (3-AT). To determine levels of lacZ expression, β-galactosidase activity was measured in extracts from L40coat cells expressing both the β subunit and the hybrid RNA as described (47).
the MS2 RNA is located at the 5′ end of the tRNALeu. The series of pIIIA-MS2-derived plasmids were used to express the hybrid RNAs in yeast 3HS. On the other hand, the gene encoding the LeuRS β-subunit was inserted between the NcoI and BamHI sites of plasmid pACTII; the resulting plasmid was named pACT-LRSB. Then, the yeast strain L40coat was co-transformed with the pIIIA-MS2-derived plasmids and plasmid pACT-LRSB. Transformants were grown at 30°C on synthetic medium (YNB) lacking uracil, histidine and leucine for selection of the URA3, HIS3 and LEU2 marker genes, respectively. Testing of reporter gene activation was carried out as described (47). Colonies that appeared 5–6 days later were further analyzed for lacZ expression by plating on medium lacking uracil and leucine and supplemented with 80 μg/mL 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). To test for HIS3 activation, drops (5 μL) of yeast double transformants grown to an OD600 = 0.1 were applied to YNB plates deprived of uracil, leucine, histidine and supplemented with different concentrations (0–100 mM) of the competitive His3p inhibitor 3-amino-1, 2, 4-triazole (3-AT). To determine levels of lacZ expression, β-galactosidase activity was measured in extracts from L40coat cells expressing both the β subunit and the hybrid RNA as described (47).
Expression and purification of A.aeolicus LeuRS and its β subunit
The genes of the α and the β subunit of A.aeolicus LeuRS were inserted into the plasmid pSBET and pET-15b, respectively. The resulting plasmids pSBET-LRSA and pET-LRSB were used to overexpress the α and β subunit. To purify the β subunit, E.coli BL21(DE3) cells were transformed with plasmid pET-LRSB. To purify the heterodimeric form αβ-LeuRS, the two plasmids pSBET-LRSA and pET-LRSB were co-introduced into the E.coli BL21(DE3) cells. Transformants harboring the two recombinant plasmids were grown in 1 l of LB with 100 μg/ml of ampicillin and 50 μg/ml of kanamycin at 37°C. When the cell density reached A600 = 0.6, the expression was induced by adding 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) and cells were grown for an additional 4 h. Since all the mutated proteins were thermostable, a heating step at 75°C for an hour was employed to precipitate most of the bacterial proteins. Then, the mutated β subunits and the αβ-LeuRSs were purified according to the procedure used for the wild-type protein (41).
Binding assay of the A.aeolicus LeuRS and its β subunit with tRNA
Labeled A.aeolicus ![]() transcripts were prepared as follows: T7 transcripts were generated in a 50 μl reaction mixture, containing 40 mM Tris–HCl (pH 8.0), 5 mM DTT, 10 mM MgCl2, 1 mM of each ATP, UTP and CTP, 10 μM [α-32P] GTP (3000 Ci/mmol), 10 mM 5′-GMP, 1 μg of BstNI-digested template DNA, 1000 U/ml RNasin (Takara, Japan), 1 U/ml of inorganic pyrophosphatase and 2 mg/ml of pure T7 RNA polymerase, at 37°C for 4 h. The transcripts were purified by 15% denaturating polyacrylamide gel electrophoresis and folded by heating at 75°C for 5 min in the presence of 5 mM MgCl2 and slowly cooled down to 30°C. Protein-tRNA interactions were examined by a gel-shift assay. Wild-type LeuRS or mutated β subunits were mixed with the labeled tRNALeu in a 5 μl volume containing 100 mM Tris–HCl buffer (pH 6.8), 30 mM KCl, 12 mM MgCl2 and 0.1 mM EDTA at 0°C for 15 min. A 20% sucrose solution (3 μl) containing tracer dyes was added immediately before loading. The final mixture was then loaded on a 1 mm thick 6% native polyacrylamide gel in Tris–glycine buffer (50 mM Tris–base, 50 mM glycine) and was run for 1 h at 100 V at 8°C in the same buffer. After electrophoresis the gel was dried and radioactive bands were analyzed by phosphorimaging.
transcripts were prepared as follows: T7 transcripts were generated in a 50 μl reaction mixture, containing 40 mM Tris–HCl (pH 8.0), 5 mM DTT, 10 mM MgCl2, 1 mM of each ATP, UTP and CTP, 10 μM [α-32P] GTP (3000 Ci/mmol), 10 mM 5′-GMP, 1 μg of BstNI-digested template DNA, 1000 U/ml RNasin (Takara, Japan), 1 U/ml of inorganic pyrophosphatase and 2 mg/ml of pure T7 RNA polymerase, at 37°C for 4 h. The transcripts were purified by 15% denaturating polyacrylamide gel electrophoresis and folded by heating at 75°C for 5 min in the presence of 5 mM MgCl2 and slowly cooled down to 30°C. Protein-tRNA interactions were examined by a gel-shift assay. Wild-type LeuRS or mutated β subunits were mixed with the labeled tRNALeu in a 5 μl volume containing 100 mM Tris–HCl buffer (pH 6.8), 30 mM KCl, 12 mM MgCl2 and 0.1 mM EDTA at 0°C for 15 min. A 20% sucrose solution (3 μl) containing tracer dyes was added immediately before loading. The final mixture was then loaded on a 1 mm thick 6% native polyacrylamide gel in Tris–glycine buffer (50 mM Tris–base, 50 mM glycine) and was run for 1 h at 100 V at 8°C in the same buffer. After electrophoresis the gel was dried and radioactive bands were analyzed by phosphorimaging.
Kinetic assays
ATP-PPi exchange and aminoacylation activities of LeuRS were measured at 37°C as described (41). The aminoacylation activity was determined at 37°C in the reaction mixture containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 4 mM ATP, 0.5 mM DTT, 10 μM tRNALeu, appropriate amounts of [14C]leucine and enzymes. The kinetic constants of enzymes were determined using various concentrations of the relevant substrates.
RESULTS
Construction of the yeast 3HS based on the interaction of the β subunit of A.aeolicus LeuRS and its cognate tRNA
The first step to develop a functional yeast 3HS is to find a couple of protein and RNA partners able to interact as hybrids with the activation domain of HIS3-GAL and with the MS2-RNA respectively (Figure 1A). The β subunit of A.aeolicus LeuRS was chosen for this assay since it binds its cognate tRNA in vitro (41,48). It was also shown that this complex could be displaced by the E.coli ![]() (41). In consequence, we tested both A.aeolicus
(41). In consequence, we tested both A.aeolicus ![]() and E.coli
and E.coli ![]() , and E.coli
, and E.coli ![]() which is chargeable by the A.aeolicus αβ-LeuRS up to one-fourth of the kcat for the charging of the native tRNA (41). Six recombinant pIIIA-MS2-derived plasmids producing the different hybrid RNAs (see Materials and Methods) were introduced into the L40coat cells containing plasmid pACT-LRSB. Transformants were then replicated on plates supplemented with X-gal. After 3 days, we found that only the L40coat cells transformed with pACT-LRSB and pIIIA-tRNAAquiLeu-MS2 exhibited a blue coloration, which reveals an interaction between the RNA and protein partners. The five other pIIIA-derivatives failed to give a productive interaction in the 3HS, suggesting that the complexes between the β subunit and the E.coli tRNAArg and tRNALeu cannot occur in vivo (Figure 1B). The authenticity of the observed interaction was verified in order to exclude unspecific interactions between the activation domain and the A.aeolicus
which is chargeable by the A.aeolicus αβ-LeuRS up to one-fourth of the kcat for the charging of the native tRNA (41). Six recombinant pIIIA-MS2-derived plasmids producing the different hybrid RNAs (see Materials and Methods) were introduced into the L40coat cells containing plasmid pACT-LRSB. Transformants were then replicated on plates supplemented with X-gal. After 3 days, we found that only the L40coat cells transformed with pACT-LRSB and pIIIA-tRNAAquiLeu-MS2 exhibited a blue coloration, which reveals an interaction between the RNA and protein partners. The five other pIIIA-derivatives failed to give a productive interaction in the 3HS, suggesting that the complexes between the β subunit and the E.coli tRNAArg and tRNALeu cannot occur in vivo (Figure 1B). The authenticity of the observed interaction was verified in order to exclude unspecific interactions between the activation domain and the A.aeolicus ![]() or between the MS2 RNA and the β subunit (for this we used the empty pIIIA-MS2 and pACTII). Then, the expression level of the reporter gene HIS3 was assayed by growth on plates lacking histidine and supplemented with the inhibitor 3-AT. The highest permissive concentration of 3-AT was obtained for the β subunit and its cognate tRNALeu (50 mM) that is higher than the 25 mM measured for the positive control iron responsive element (IRE) and iron regulatory protein (IRP) (Figure 1B, lane 6) (46). Thus, we concluded that the β subunit interacts with its cognate
or between the MS2 RNA and the β subunit (for this we used the empty pIIIA-MS2 and pACTII). Then, the expression level of the reporter gene HIS3 was assayed by growth on plates lacking histidine and supplemented with the inhibitor 3-AT. The highest permissive concentration of 3-AT was obtained for the β subunit and its cognate tRNALeu (50 mM) that is higher than the 25 mM measured for the positive control iron responsive element (IRE) and iron regulatory protein (IRP) (Figure 1B, lane 6) (46). Thus, we concluded that the β subunit interacts with its cognate ![]() in the yeast 3HS. The interaction is very specific since active substrates like E.coli
in the yeast 3HS. The interaction is very specific since active substrates like E.coli ![]() and
and ![]() cannot form the hybrid, and the interaction can only occur when the tRNA is located at the 5′ end of the hybrid RNA molecule (pIIIA-
cannot form the hybrid, and the interaction can only occur when the tRNA is located at the 5′ end of the hybrid RNA molecule (pIIIA-![]() -MS2). Previous studies have reported functional three-hybrid interactions between an aminoacyl-tRNA synthetase and an intron (49) and between a tRNA and intranucleolar component of the nuclear tRNA export machinery (50), but to our knowledge, this is the first report of a functional 3HS based on the interaction of a tRNA and an aminoacyl-tRNA synthetase.
-MS2). Previous studies have reported functional three-hybrid interactions between an aminoacyl-tRNA synthetase and an intron (49) and between a tRNA and intranucleolar component of the nuclear tRNA export machinery (50), but to our knowledge, this is the first report of a functional 3HS based on the interaction of a tRNA and an aminoacyl-tRNA synthetase.
Figure 1.
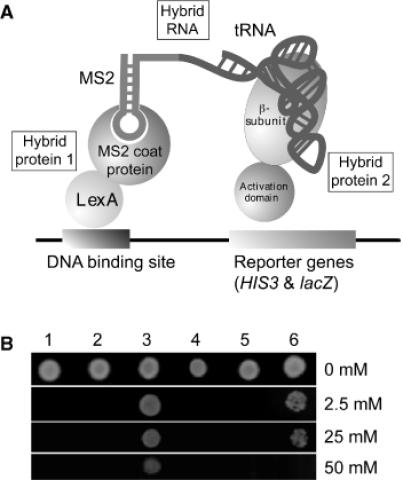
Three-hybrid formation with the A.aeolicus β subunit and tRNALeu. (A) Schematic presentation of the yeast 3HS adapted to a tRNA bait. The two hybrid proteins are the LexA-MS2 coat protein and the LeuRS β subunit in fusion with the GAL4 activation domain. The hybrid RNA consists of the MS2 RNA and the A.aeolicus tRNALeu. Three-hybrid formation leads to expression of the two reporter genes HIS3 and lacZ. (B) Assay of the expression level of HIS3 by drop test. The L40coat yeast cells containing pACT-LRSB and pIIIA- -MS2 were grown on 3-AT-containing medium. The highest permissive concentration of 3-AT (50 mM) was measured for the β subunit with its cognate tRNA (lane 3). The positive control is IRP with IRE (25 mM, lane 6). HIS3 activation was not detectable when the GAL4 activation domain alone was expressed with the A.aeolicus tRNALeu bait (lane 1), or when the β subunit was expressed with the MS2 RNA alone (lane 2), or when the β subunit was expressed with the E.coli tRNAArg (lane 4), or when the β subunit was proposed to the E.coli tRNALeu (lane 5).
-MS2 were grown on 3-AT-containing medium. The highest permissive concentration of 3-AT (50 mM) was measured for the β subunit with its cognate tRNA (lane 3). The positive control is IRP with IRE (25 mM, lane 6). HIS3 activation was not detectable when the GAL4 activation domain alone was expressed with the A.aeolicus tRNALeu bait (lane 1), or when the β subunit was expressed with the MS2 RNA alone (lane 2), or when the β subunit was expressed with the E.coli tRNAArg (lane 4), or when the β subunit was proposed to the E.coli tRNALeu (lane 5).
Selection of β subunits with impaired tRNA binding capacity by the 3HS
We used the three-hybrid system to identify mutants of the β subunit presenting impaired tRNA binding capacity. The loss-of-binding mutants were identified by reduced expression levels of the reporter genes HIS3 and lacZ. Mutants were created by random mutagenesis and site-directed mutagenesis.
First, a library of randomly mutated β subunit genes was screened in order to identify residues that lost tRNA binding when mutated. The randomly mutated pACT-LRSB library was produced by hydroxylamine treatment (47). L40coat cells containing plasmid pIIIA-![]() -MS2 were transformed with the pACT-LRSB library and plated on selective medium lacking uracil and leucine. As before, the plates were replicated onto plates containing X-gal and checked for the coloration. The rare white colonies that appeared among a majority of blue ones were further analyzed. We screened ∼100 000 transformants and isolated 90 white colonies in the first round. A second screening was carried out in order to exclude the false positive clones that resulted from mutations located in the yeast replicon, in the promoter, or in the activation domain of the fusion gene. For this, the mutagenized pACT-LRSB plasmids from the 90 white colonies were isolated and the genes encoding the β subunits were re-cloned in plasmid pACTII. These new pACT-LRSB plasmids were introduced into L40coat cells containing the plasmid pIIIA-
-MS2 were transformed with the pACT-LRSB library and plated on selective medium lacking uracil and leucine. As before, the plates were replicated onto plates containing X-gal and checked for the coloration. The rare white colonies that appeared among a majority of blue ones were further analyzed. We screened ∼100 000 transformants and isolated 90 white colonies in the first round. A second screening was carried out in order to exclude the false positive clones that resulted from mutations located in the yeast replicon, in the promoter, or in the activation domain of the fusion gene. For this, the mutagenized pACT-LRSB plasmids from the 90 white colonies were isolated and the genes encoding the β subunits were re-cloned in plasmid pACTII. These new pACT-LRSB plasmids were introduced into L40coat cells containing the plasmid pIIIA-![]() -MS2 and were checked again for the blue/white coloration. After this second round of selection only 35 colonies were white from the 90 original colonies. The 35 genes encoding the β subunits from these white clones were inserted into the plasmid pET-15b and expressed in E.coli BL21(DE3). Only 14 genes were successfully expressed and their DNAs were sequenced. Most of genes that did not give any expression product contained premature stop codons at various positions. This agrees well with previous findings, which show that the easiest way to inactivate a protein is to cause truncated forms by means of premature stop codons (16). Among the expressed proteins, two contained nonsense mutations at residues Q269 (Q269stop) and V286 (V286stop) leading to deletions of 21 and 4 residues, respectively, at their C-terminal ends (the whole β subunit contains 289 residues). The other 11 expressed mutants contained the following single mutations: A156V, K160N, Q234H, G237D, and L283F that was found seven times. These mutants are located in the characteristic class Ia α-helix-bundle-domain or in the C-terminal domain which was disordered in the crystal of T.thermophilus LeuRS (7).
-MS2 and were checked again for the blue/white coloration. After this second round of selection only 35 colonies were white from the 90 original colonies. The 35 genes encoding the β subunits from these white clones were inserted into the plasmid pET-15b and expressed in E.coli BL21(DE3). Only 14 genes were successfully expressed and their DNAs were sequenced. Most of genes that did not give any expression product contained premature stop codons at various positions. This agrees well with previous findings, which show that the easiest way to inactivate a protein is to cause truncated forms by means of premature stop codons (16). Among the expressed proteins, two contained nonsense mutations at residues Q269 (Q269stop) and V286 (V286stop) leading to deletions of 21 and 4 residues, respectively, at their C-terminal ends (the whole β subunit contains 289 residues). The other 11 expressed mutants contained the following single mutations: A156V, K160N, Q234H, G237D, and L283F that was found seven times. These mutants are located in the characteristic class Ia α-helix-bundle-domain or in the C-terminal domain which was disordered in the crystal of T.thermophilus LeuRS (7).
Test of site-directed mutants of the β subunit by the 3HS
To complete the study of the tRNA binding site on the β subunit, we identified additional tRNA binding residues by means of site-directed mutagenesis combined with the 3HS selection. Based on multiple sequence alignment of LeuRSs, on the X-ray structure of LeuRS (7), and on the first results from the 3HS selection, we selected a set of six residues that we mutated in Ala in order to remove the contribution of their side chains to a hypothetical contact with the tRNA molecule (Figure 2). Three mutants (N152A, M159A, N163A) were constructed in the third long helix of the helix-bundle-domain (residues 740–761 in T.thermophilus LeuRS), the region where the previously isolated loss-of-binding mutations A156V and K160N are located. These three residues are highly conserved or invariant in the LeuRS sequences. In addition, the quasi-invariant residue R94 located in the close vicinity, but in the adjacent helix, was mutated in Ala. We also mutated two residues of the C-terminal domain, N236 and K238, both highly conserved and located close to the loss-of-binding mutants Q234H, G237D.
Figure 2.
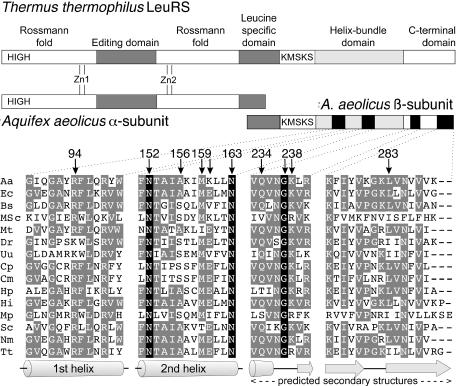
Schematic diagram of the domain structure of T.thermophilus and A.aeolicus LeuRSs and partial sequence alignment of the four β subunit peptides investigated. Amino acid sequences were aligned in the four peptidic regions here studied. Aa, A.aeolicus; Ec, E.coli; Bs, Bacillus subtilis; MSc, mitochondrial S.cerevisiae; Mt, Mycobacterium tuberculosis; Dr, Deinococcus radiodurans; Uu, Ureaplasma urealyticum; Cp, Chlamydia pneumoniae AR39; Cm, Chlamydia muridarum; Hp, Helicobacter pylori; Hi, Haemophilus influenzae; Mp, Mycoplasma pneumoniae; Sc, Streptomyces coelicolor; Nm, Neisseria meningitidis; Tt, T.thermophilus. Amino acids that are completely conserved in all sequences are boxed in black. Homologous residues are boxed in gray. The residues studied in this work with the yeast 3HS are marked with an arrow. The indicated secondary structure elements are from the T.thermophilus LeuRS 3D structure (7) and from the prediction of the GCG SeqWeb 2.0 software for the C-terminal domain.
All the six mutants were tested in 3HS. Coloration defects on X-Gal-containing medium, and measurement of β-galactosidase activity as well as drop tests on 3-AT-containing medium were determined in order to assess the effects of the mutations on the tRNA–protein interaction (Figure 3). Only mutants R94A, M159A and K238A induced phenotypes of loss-of-binding with white phenotypes, and drops of 3-AT tolerance. Mutants N152A and N163A only reduced the interaction slightly, with a light-blue phenotype, whereas N236A exhibited no effect at all.
Figure 3.
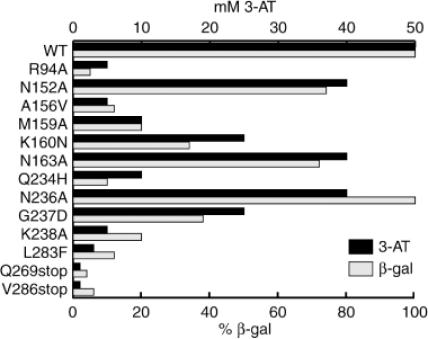
Measurement of the tRNA:β subunits interactions by the yeast 3HS. Growth in the presence of the 3-AT inhibitor was tested by drop test and the β-galactosidase activity was assayed on crude extracts from yeast (see Materials and Methods). The vertical axis shows the 3-AT concentrations and the percentage of the measured β-galactosidase activity compared to the activity measured with the wild-type β subunit.
tRNA affinity of the β subunits and αβ-heterodimers for tRNALeu
In order to correlate the in vivo results with the 3HS with changes in affinity of the β subunit of LeuRS for tRNALeu, we performed band-shift assays using transcript of ![]() . This excludes putative effects of the fusions of the protein and RNAs in the hybrids. Eight mutants (R94A, A156V, K160N, Q234H, G237D, L283F, V286stop and Q269stop), that displayed the most severe perturbation in 3HS were assayed by gel mobility shift (Figure 4). The mutated β subunits were overexpressed and purified to homogeneity. Remarkably, the thermostability of these eight proteins was not changed by the heating step of one hour at 75°C, suggesting that the tertiary structure of the mutants was preserved. The tRNA affinity values measured by gel-shift are shown in Figure 4B. They correlate perfectly with the β-galactosidase and 3-AT data obtained in vivo by the 3HS (Figure 3). The six single mutants showed a 2-fold decrease of the tRNA affinity whereas the two deletion mutants exhibited a significantly higher decrease (12-fold).
. This excludes putative effects of the fusions of the protein and RNAs in the hybrids. Eight mutants (R94A, A156V, K160N, Q234H, G237D, L283F, V286stop and Q269stop), that displayed the most severe perturbation in 3HS were assayed by gel mobility shift (Figure 4). The mutated β subunits were overexpressed and purified to homogeneity. Remarkably, the thermostability of these eight proteins was not changed by the heating step of one hour at 75°C, suggesting that the tertiary structure of the mutants was preserved. The tRNA affinity values measured by gel-shift are shown in Figure 4B. They correlate perfectly with the β-galactosidase and 3-AT data obtained in vivo by the 3HS (Figure 3). The six single mutants showed a 2-fold decrease of the tRNA affinity whereas the two deletion mutants exhibited a significantly higher decrease (12-fold).
Figure 4.
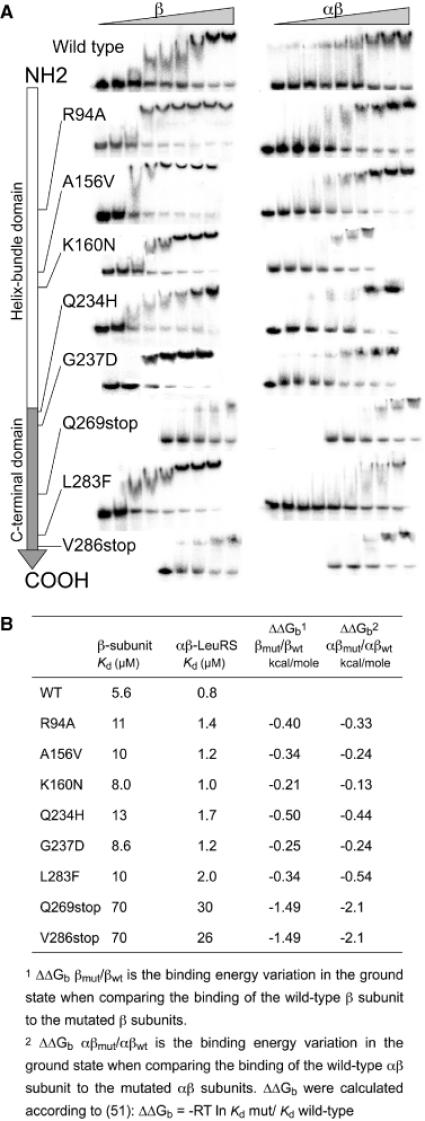
Measurement of the dissociation constant of LeuRS proteins for tRNALeu by gel-shift assay. (A) In vitro transcribed 32P-labelled A.aeolicus  was incubated with increasing concentrations of the β subunits or whole αβ-LeuRS (see Materials and Methods). Apparent dissociation constants (Kd) of the proteins were estimated from quantification of the free and retarded tRNA. Band shifts are displayed along the linear schematic representation of the β subunit (NH2- to the COOH-end). For clarity the protein concentrations were omitted from each picture. The large increases of Kd were shown by a shift of the gel picture towards the right side, which symbolizes the high values. (B) Kd values and binding energy variations for the β subunits and αβ-LeuRS proteins.
was incubated with increasing concentrations of the β subunits or whole αβ-LeuRS (see Materials and Methods). Apparent dissociation constants (Kd) of the proteins were estimated from quantification of the free and retarded tRNA. Band shifts are displayed along the linear schematic representation of the β subunit (NH2- to the COOH-end). For clarity the protein concentrations were omitted from each picture. The large increases of Kd were shown by a shift of the gel picture towards the right side, which symbolizes the high values. (B) Kd values and binding energy variations for the β subunits and αβ-LeuRS proteins.
Then, the eight mutants R94A, A156V, K160N, Q234H, G237D, L283F, V286stop and Q269stop, were co-expressed with the wild-type α-subunit in E.coli and the mutated heterodimers were purified to homogeneity. The αβ-LeuRSs were thermostable at 75°C for 1 h, like the β subunits displaying the same mutations. The Kd values of αβ-LeuRSs for ![]() were determined by gel-shift assay. Under these experimental conditions, the Kd of the wild-type αβ-LeuRS for its tRNA is 0.8 μM. Kd values from the mutated αβ-LeuRSs range from 1.2 to 30 μM (Figure 4). The resulting loss of tRNA binding energy is basically the same as for the β subunits alone (Figure 4), except for the deletion mutants V286stop and Q269stop which display a significant higher loss of binding energy when in the heterodimeric complex with the α subunit.
were determined by gel-shift assay. Under these experimental conditions, the Kd of the wild-type αβ-LeuRS for its tRNA is 0.8 μM. Kd values from the mutated αβ-LeuRSs range from 1.2 to 30 μM (Figure 4). The resulting loss of tRNA binding energy is basically the same as for the β subunits alone (Figure 4), except for the deletion mutants V286stop and Q269stop which display a significant higher loss of binding energy when in the heterodimeric complex with the α subunit.
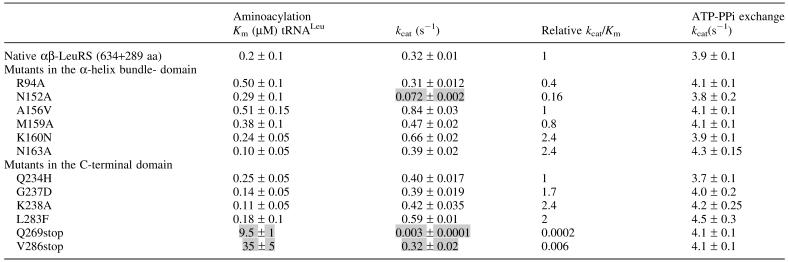
Kinetic analysis of the αβ-LeuRS proteins
Four additional αβ-heterodimers (N152A, M159A, N163A and K238A) were purified. The kinetic parameters of the 12 mutated αβ-LeuRSs for A.aeolicus tRNALeu were measured at the steady-state level. Both ATP-PPi exchange and aminoacylation activities were measured (Table 1). The ATP-PPi exchange activities were unaffected by the substitutions and deletions, excluding the participation of the C-terminal end of the β subunit to the leucine activation. Most of the mutated proteins that exhibited binding losses in the 3HS and gel-shift assays, showed moderate variations of their aminoacylation rates, except for N152A and Q269stop which showed a 5- and 100-fold reduction. Km values for tRNALeu were only increased for the deletion mutants V286stop and Q269stop. In non-saturating conditions the tRNA aminoacylation of the Q269stop mutant is almost undetectable. Thus, removal of 21 residues at the C-terminal end induces an almost complete decoupling of the two enzyme's activities, and renders the enzyme only capable to catalyze the leucyl-adenylate formation.
Table 1. Kinetic constants for A.aeolicus αβ-LeuRSs at 37°C.
Shaded boxes mark significant changes of the catalytic parameters.
DISCUSSION
The yeast 3HS has been developed for the research and for the study of interactions between RNA and protein (42,43). Here we report the first successful 3HS based on the interaction of tRNA and an aminoacyl-tRNA synthetase, more precisely between the A.aeolicus tRNALeu and the β subunit of its homologous LeuRS. Remarkably, this interaction occurs in vivo despite the fact that the dissociation constant measured in vitro for these two molecules is rather low (∼5.8 μM) when compared to typical Kd values measured for whole aminoacyl-tRNA synthetases with their cognate tRNA (∼1 μM and less). Despite this high dissociation constant, the β subunit and tRNALeu, in fusion with the HIS3-lacZ activation domain and in fusion with MS2 RNA respectively, interact in vivo in the 3HS and the binding strength was even higher than the positive control (IRE). This interaction was specific for the A.aeolicus tRNA, since E.coli tRNALeu that is recognized and charged by the whole αβ-LeuRS (41), failed to produce a functional 3HS. Thus, the E.coli tRNALeu can only give a productive complex in the presence of the α subunit that may provide additional binding energy. We previously demonstrated that in vitro, E.coli tRNAArg is able to compete and to displace a complex formed by the A.aeolicus β subunit and A.aeolicus tRNALeu (41). Nevertheless, we show here that tRNAArg does not form a complex with the β subunit of LeuRS, since no functional 3HS was generated in vivo, suggesting that the cellular environment required for three-hybrid formation is more stringent than the in vitro conditions used for complex formation.
Two special considerations about the hybrid RNA should be taken into account when the yeast 3HS is applied to a tRNA–protein system. The first is that tRNA processing enzymes present in the yeast cells can process the hybrid tRNA molecules. We only detected the expression of the reporter genes when the prokaryotic A.aeolicus tRNALeu was located at the 5′ end and not at the 3′ end of the hybrid RNA molecule. This difference might be attributed to the destruction of the tRNA-like molecule by the tRNA processing enzymes. A second consideration might be that the appending of the MS2 RNA to the acceptor stem of tRNA interferes with the tRNA–enzyme interaction. These complications suggest assaying the tRNA bait on both 5′ and 3′ ends of the MS2 RNA, but also assaying tRNA mutants deficient in processing as well as adding RNA spacers to prevent the steric hindrance.
As a direct application of the efficient 3HS, we selected mutations with loss-of-binding phenotypes and demonstrated that the yeast 3HS is sensitive enough to map the tRNA binding site on the β subunit of A.aeolicus LeuRS. Five point mutations and two C-terminal deletions that exhibited drastic decreases in the binding strength were selected from a randomly mutated library. On the other hand, based on other class Ia tRNA-aaRS complexes, and based on sequences homologies with LeuRSs from different organisms, six residues located in the potential tRNALeu binding site were altered by site directed mutagenesis. Severe effects were observed for three of them, and moderate or no effects for the three others. All together, the loss-of-binding effects observed for these mutants underline the strength of the 3HS to map and identify specific molecular interactions that are important to tRNA–enzyme complex formation.
We found that the mutated β subunits exhibited changes in the Kd range of 5.6 up to 70 μM. In this range, the yeast colony phenotype varied from the deep-blue (Kd = 5.6 μM) to the white (Kd = 70 μM). The variation in binding energies resulting from these variations in affinities ranges between −0.21 to −0.54 kcal/mol for the single substitution mutants [ΔΔGb βmut/βwt, Figure 4B, (51,52)]. These energy losses might correspond to the binding energy of a weak hydrogen bond. For the two deletion mutants, a −1.49 kcal/mol loss in binding energy was calculated, which suggests that stronger interactions have been lost. In the αβ heterodimers, the variations in binding energy are basically unchanged (ΔΔGb αβmut/αβwt, Figure 4B) except for the V286stop and Q269stop mutants which showed a significant decrease of the binding energy (−2.1 kcal/mol). This suggests that the assembly of the deleted β subunits with the native α subunit induces an additional negative impact on the tRNA binding, which might be related to some structural alteration of the β subunit itself or of the α subunit by a distal effect.
According to the moderate loss of binding energy that was observed for the single substitution mutants, the mutations cause only slightly altered kcat and Km values for the aminoacylation using the αβ constructions. With the two C-terminal deletion mutants, remarkable effects on the aminoacylation catalysis were observed. Both exhibited severe increases in the Km for tRNALeu, and whereas the shortest deletion, V286stop (4 residues), exhibited no change in its catalytic rate, a 100-fold decrease in the kcat for the Q269stop mutant (21 residues deleted) was measured.
LeuRS belongs to the class Ia synthetases that comprise ArgRS, CysRS, IleRS, LeuRS, MetRS and ValRS. LeuRS is more closely related to IleRS and ValRS, a subgroup characterized by the presence of large editing domain (CP domain), and C-terminal extensions appended to the idiosyncratic class Ia helix-bundle-domain. So far, three class Ia synthetases structures have been solved in complex with their tRNA, the Staphylococcus aureus IleRS (13), the T.thermophilus ValRS (9,26) and the S.cerevisiae ArgRS (8). The tRNAs found in these complexes interact by their D-loop side and approach the active site by the minor groove of their acceptor stem, according to the class I rules.
The helix-bundle-domain shared by the six class Ia synthetases is the major constituent of the tRNA anticodon recognition. The first and third α-helices of the bundle harbor-specific residues that are involved in tRNA–protein interactions (8,13,26). A 3D model of the β subunit interacting with tRNA has been previously proposed according to the 3D structure of T.thermophilus LeuRS and sequences homologies with the A.aeolicus LeuRS (Figure 5) (31). Unfortunately, the 63-residues long C-terminal domain could not be modeled since it is disordered in the crystal structure probably due to some flexibility or mobility property (7). If we consider the seven mutants that we have selected by the 3HS selection, only two of them can be localized on the 3D structure of the β subunit (A156V and K160N). Their homologs in the T.thermophilus enzyme are A746 and E750, and are located in the third long helix of the bundle. If we superimpose the helix-bundle-domain of LeuRS onto the equivalent domains of the Ile-, Val- and ArgRS complexed with their tRNA, it appears that A746 and E750 might interact with the D arm of tRNALeu at the level of the base pairs 13:22, or 14:21. In this study, we showed that the mutation of these two residues in the A.aeolicus enzyme produces exclusive decreases in the stability of the complex in its ground state, without any effect on the transition state of the aminoacylation reaction.
Figure 5.
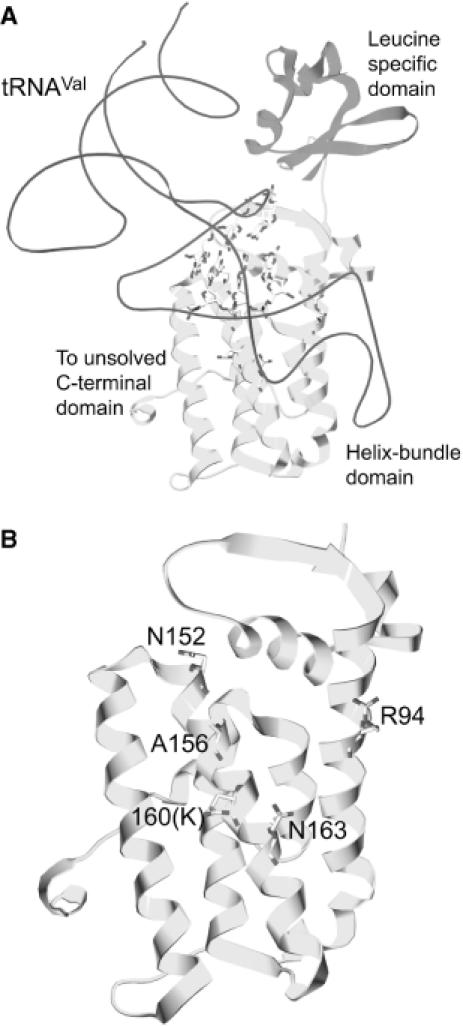
Partial representations of the LeuRS structure from T.thermophilus. (A) View of the leucine-specific domain (shown in magenta) and helix-bundle-domain (shown in yellow) of T.thermophilus LeuRS (7) corresponding to the β subunit of A.aeolicus LeuRS (31). T.thermophilus tRNAVal from the ValRS:tRNAVal complex (26) was docked after superimposition of the helix-bundle-domains of the T.thermophilus LeuRS and ValRS. Base pairs 11:24, 12:23, 13:22 and 14:21 of the D-arm are shown. The 63 last amino acid residues of the protein are missing in the structure (7). (B) View focused on the helix-bundle-domain of T.thermophilus LeuRS. Mutated residues are labeled according to the β subunit numbering. These residues are conserved in the two enzymes, except K160 of the A.aeolicus subunit that is E750 in T.thermophilus.
The five other mutants selected by 3HS are found in the unsolved C-terminal domain. The single substituted residues showed a moderate negative effect on the tRNA binding in its ground state. In addition, the 21-residues deletion mutant was impaired in the stabilization of the transition state of the aminoacylation reaction. This effect is similar to those observed for mutants of the C-terminal coiled-coil domain of ValRS, which interacts electrostatically with A20 and hydrophobically with the G19:C56 tertiary base pair, providing binding energy and tRNA structure stabilization (26). ArgRS, which is not really a ‘large class Ia synthetase’ in the sense it has no editing domain, uses instead of a C-terminal domain a specific N-terminal domain that recognizes the D-loop side of tRNAArg (8). Thus, at least for ValRS and ArgRS, the binding of the single stranded tRNA region called ‘variable pocket’ formed by the D- and T-loops is crucial (53,54). This binding may promote recognition but also structural stabilization of the tRNA L-shape during the aminoacylation reaction (26,55). Collectively, these data highlight the recruitment of new domains by these enzymes in order to recognize, stabilize or bind tRNA atoms located on its D-loop face. Concerning the tRNALeu binding on the A.aeolicus LeuRS no experimental data are presently available. In analogy to the E.coli tRNALeu, the discriminator base and a specific tertiary network of interactions located in the elbow region should be essential for the leucine identity (32–35,39). No identity elements have been found in the bacterial anticodon loop, in contrast to the S.cerevisiae tRNALeu (36). Since the tertiary structure of the tRNALeu elbow region is crucial for its identity, it is tempting to hypothesize that the C-terminal domain of LeuRS occupies the spatial location of the coiled-coil domain of ValRS that stabilizes the elbow structure of tRNAVal. At this location, it would check the tertiary structure features of the tRNA that control the leucine identity. We cannot rule out the possibility that the C-terminal domain is folded in some way that allows also the checking of the variable stem of the tRNA. Indeed, LeuRS from the archaea Haloferax volcanii measures the length of the variable stem and recognizes the nucleotides at position 47C, 47D, 47H (38). Previous studies on other synthetases that recognize the class II tRNA have shown that both SerRS and TyrRS recognize the shape of the variable stem of their cognate tRNA by an N- and C-terminal domain, respectively (56,57). Definitive answer to these questions would need the resolution of a complex between the A.aeolicus αβ-LeuRS with its tRNA.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr S. Barends for fruitful discussions and critical reading of the manuscript and to S. Jaeger for constant advice. This work was funded by the Natural Science Foundation of China (Grant 30170224, 30330180), the Chinese Academy of Sciences (Grant KSCX-2-2-04) and Shanghai Committee of Science and Technology (Grant 02DJ140567) and the exchange program between Chinese Academy of Sciences and Centre National de la Recherche Scientifique (#15129).
REFERENCES
- 1.Ibba M. and Söll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 2.Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebel J.-P., Giegé,R., Bonnet,J., Kern,D., Befort,N., Bollack,C., Fasiolo,F., Gangloff,J. and Dirheimer,G. (1973) Factors determining the specificity of the tRNA aminoacylation reaction. Non-absolute specificity of tRNA-aminoacyl-tRNA synthetase recognition and particular importance of the maximal velocity. Biochimie, 55, 547–557. [DOI] [PubMed] [Google Scholar]
- 4.Eriani G., Delarue,M., Poch,O., Gangloff,J. and Moras,D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature, 347, 203–206. [DOI] [PubMed] [Google Scholar]
- 5.Shimada A., Nureki,O., Goto,M., Takahashi,S. and Yokoyama,S. (2001) Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 98, 13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavarelli J., Delagoutte,B., Eriani,G., Gangloff,J. and Moras,D. (1998) l-arginine recognition by yeast arginyl-tRNA synthetase. EMBO J., 17, 5438–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cusack S., Yaremchuk,A. and Tukalo,M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J., 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delagoutte B., Moras,D. and Cavarelli,J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J., 19, 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukai S., Nureki,O., Sekine,S., Shimada,A., Tao,J., Vassylyev,D.G. and Yokoyama,S. (2000) Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell, 103, 793–803. [DOI] [PubMed] [Google Scholar]
- 10.Mechulam Y., Schmitt,E., Maveyraud,L., Zelwer,C., Nureki,O., Yokoyama,S., Konno,M. and Blanquet,S. (1999) Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol., 294, 1287–1297. [DOI] [PubMed] [Google Scholar]
- 11.Newberry K.J., Hou,Y.M. and Perona,J.J. (2002) Structural origins of amino acid selection without editing by cysteinyl-tRNA synthetase. EMBO J., 21, 2778–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nureki O., Vassylyev,D., Tateno,M., Shimada,A., Nakama,T., Fukai,S., Konno,M., Hendrickson,T., Schimmel,P. and Yokoyama,S. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- 13.Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an ile-tRNA synthetase structure with tRNAIle and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- 14.Sugiura I., Nureki,O., Ugaji-Yoshikawa,Y., Kuwabara,S., Shimada,A., Tateno,M., Lorber,B., Giege,R., Moras,D., Yokoyama,S. and Konno,M. (2000) The 2.0 Å crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Structure, 8, 197–208. [DOI] [PubMed] [Google Scholar]
- 15.Kise Y., Lee,S., Park,S., Fukai,S., Sengoku,T., Ishii,R., Yokoyama,S., Kim,S. and Nureki,O. (2004) A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat. Struct. Mol. Biol., 11, 149–156. [DOI] [PubMed] [Google Scholar]
- 16.Geslain R., Martin,F., Delagoutte,B., Cavarelli,J., Gangloff,J. and Eriani,G. (2000) In vivo selection of lethal mutations reveals two functional domains in arginyl-tRNA synthetase. RNA, 6, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geslain R., Bey,G., Cavarelli,J. and Eriani,G. (2003) Limited set of amino acid residues in a class Ia aminoacyl-tRNA synthetase is crucial for tRNA binding. Biochemistry, 42, 15092–15101. [DOI] [PubMed] [Google Scholar]
- 18.Shepard A., Shiba,K. and Schimmel,P. (1992) RNA binding determinant in some class I tRNA synthetases identified by alignment-guided mutagenesis. Proc. Natl Acad. Sci. USA, 89, 9964–9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H.Y., Pelka,H., Brunie,S. and Schulman,L.H. (1993) Two separate peptides in Escherichia coli methionyl-tRNA synthetase form the anticodon binding site for methionine. Biochemistry, 32, 10506–10511. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh G., Pelka,H. and Schulman,L.D.H. (1990) Identification of the anticodon recognition site of Escherichia coli methionyl-tRNA synthetases. Biochemistry, 29, 2220–2225. [DOI] [PubMed] [Google Scholar]
- 21.Meinnel T., Mechulam,Y., LeCorre,D., Panvert,M., Blanquet,S. and Fayat,G. (1991) Selection of suppressor methionyl-tRNA synthetases : mapping the tRNA anticodon binding site. Proc. Natl Acad. Sci. USA, 88, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt E., Meinnel,T., Panvert,M., Mechulam,Y. and Blanquet,S. (1993) Two acidic residues of Escherichia coli methionyl-tRNA synthetase act as negative discriminants towards the binding of non-cognate tRNA anticodons. J. Mol. Biol., 233, 615–628. [DOI] [PubMed] [Google Scholar]
- 23.Despons L., Senger,B., Fasiolo,F. and Walter,P. (1992) Binding of the yeast tRNAMet anticodon by the cognate methionyl-tRNA synthetase involves at least two independent peptide regions. J. Mol. Biol., 225, 897–907. [DOI] [PubMed] [Google Scholar]
- 24.Auld D.S. and Schimmel,P. (1995) Switching recognition of two tRNA synthetases with an amino acid swap in a designed peptide. Science, 267, 1994–1996. [DOI] [PubMed] [Google Scholar]
- 25.Auld D.S. and Schimmel,P. (1996) Single sequence of a helix-loop peptide confers functional anticodon recognition on two tRNA synthetases. EMBO J., 15, 1142–1148. [PMC free article] [PubMed] [Google Scholar]
- 26.Fukai S., Nureki,O., Sekine,S., Shimada,A., Vassylyev,D. and Yokoyama,S. (2003) Mechanism of molecular interactions for tRNAVal recognition by valyl-tRNA synthetase. RNA, 9, 100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander R.W. and Schimmel,P. (1999) Evidence for breaking domain-domain functional communication in a synthetase-tRNA complex. Biochemistry, 38, 16359–16365. [DOI] [PubMed] [Google Scholar]
- 28.Alexander R.W. and Schimmel,P. (2001) Domain-domain communication in aminoacyl-tRNA synthetases. Prog. Nucleic Acid Res. Mol. Biol., 69, 317–349. [DOI] [PubMed] [Google Scholar]
- 29.Lazard M., Agou,F., Kerjan,P. and Mirande,M. (2000) The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication. J. Mol. Biol., 302, 991–1004. [DOI] [PubMed] [Google Scholar]
- 30.Lincecum T.L., Tukalo,M., Yaremchuk,A., Mursinna,R.S., Williams,A.M., Sproat,B.S., Van Den Eynde,W., Link,A., Van Calenbergh, S., Grotli,M., Martinis,S.A. and Cusack,S. (2003) Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell, 11, 951–963. [DOI] [PubMed] [Google Scholar]
- 31.Zhao M.-W., Hao,R., Chen,J.-F., Martin,F., Eriani,G. and Wang,E.-D. (2003) Enzymes assembled from Aquifex aeolicus and Escherichia coli leucyl-tRNA synthetases. Biochemistry, 42, 7694–7700. [DOI] [PubMed] [Google Scholar]
- 32.Asahara H., Himeno,H., Tamura,K., Hasegawa,T., Watanabe,K. and Shimuzu,M. (1993) Recognition nucleotides of Escherichia coli transfer RNALeu and its elements facilitating discrimination from transfer RNASer and transfer RNATyr. J. Mol. Biol., 231, 219–229. [DOI] [PubMed] [Google Scholar]
- 33.Asahara H., Nameki,N. and Hasegawa,T. (1998) In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol., 283, 605–618. [DOI] [PubMed] [Google Scholar]
- 34.Tocchini-Valentini G., Sacks,M.E. and J., Abelson,J. (2000) tRNA leucine identity and recognition sets. J. Mol. Biol., 298, 779–793. [DOI] [PubMed] [Google Scholar]
- 35.Du X. and Wang,E.-D. (2003) Tertiary structure base pairs between D- and TC-loops of Escherichia coli tRNALeu play important roles in both aminoacylation and editing. Nucleic Acids Res., 31, 2865–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soma A., Kumagai,R., Nishikawa,K. and Himeno,H. (1996) The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNALeu. J. Mol. Biol., 263, 707–714. [DOI] [PubMed] [Google Scholar]
- 37.Soma A. and Himeno,H. (1998) Cross-species aminoacylation of tRNA with a long variable arm between Escherichia coli and Saccharomyces cerevisiae. Nucleic Acids Res., 26, 4374–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soma A., Uchiyama,K., Sakamoto,T., Maeda,M. and Himeno,H. (1999) Unique recognition style of tRNALeu by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol., 293, 1029–1038. [DOI] [PubMed] [Google Scholar]
- 39.Larkin D.C., Williams,A.M., Martinis,S.A. and Fox,G.E. (2002) Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res., 30, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouda M., Yokogawa,T., Asahara,H. and Nishikawa,K. (2002) Leucyl-tRNA synthetase from the extreme thermophile Aquifex aeolicus has a heterodimeric quaternary structure. FEBS Lett., 518, 139–143. [DOI] [PubMed] [Google Scholar]
- 41.Xu M.G., Chen,J.F., Martin,F., Zhao,M.W., Eriani,G. and Wang,E.D. (2002) Leucyl-tRNA synthetase consisting of two subunits from hyperthermophilic bacteria Aquifex aeolicus. J. Biol. Chem., 25, 41590–41596. [DOI] [PubMed] [Google Scholar]
- 42.Putz U., Skehel,P. and Kuhl,D. (1996) A tri-hybrid system for the analysis and detection of RNA-protein interactions. Nucleic Acids Res, 24, 4838–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SenGupta D.J., Zhang,B., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Wang,E.D. and Wang,Y.L. (1999) Analysis of RNA structure using chemical and enzymatic probing monitored by primer extension. Prot. Exp. Purif., 16, 355–358. [Google Scholar]
- 45.Schenk P.M., Baumann,S., Mattes,R. and H.,S.H. (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare Arg tRNAs. Biotechniques, 19, 196–200. [PubMed] [Google Scholar]
- 46.Kraemer B., Zhang,B., SenGupta,D., Fields,S. and Wickens,M. (2000) Using the yeast three-hybrid system to detect and analyze RNA-protein interactions. Meth. Enzymol., 328, 297–321. [DOI] [PubMed] [Google Scholar]
- 47.Martin F., Michel,F., Zenklusen,D., Müller,B. and Schumperli,D. (2000) Positive and negative mutant selection in the human histone hairpin-binding protein using the yeast three-hybrid system. Nucleic Acids Res., 28, 1594–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouda M., Yokogawa,T. and Nishikawa,K. (2002) The beta subunit of Aquifex aeolicus leucyl-tRNA synthetase is responsible for cognate tRNA recognition. Biochem. Biophys. Res. Commun., 297, 950–955. [DOI] [PubMed] [Google Scholar]
- 49.Rho S.B. and Martinis,S.A. (2000) The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA, 6, 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steiner-Mosonyi M., Leslie,D.M., Dehghani,H., Aitchison,J.D. and Mangroo,D. (2003) Utp8p is an essential intranuclear component of the nuclear tRNA export machinery of Saccharomyces cerevisiae. J. Biol. Chem., 278, 32236–32245. [DOI] [PubMed] [Google Scholar]
- 51.Carter P.J., Winter,G., Wilkinson,A.J. and Fersht,A.R. (1984) The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell, 38, 835–840. [DOI] [PubMed] [Google Scholar]
- 52.Eriani G. and Gangloff,J. (1999) Yeast aspartyl-tRNA synthetase residues interacting with tRNAAsp identity bases connectively contribute to tRNAAsp binding in the ground and transition-state complex and discriminate against non-cognate tRNAs. J. Mol. Biol., 291, 761–773. [DOI] [PubMed] [Google Scholar]
- 53.McClain W.H. and Foss,K. (1988) Changing the acceptor identity of a transfer RNA by altering nucleotides in a ‘variable pocket’. Science, 241, 1804–1807. [DOI] [PubMed] [Google Scholar]
- 54.Horowitz J., Chu,W., Derrick,W., Liu,J., Liu,M. and Yue,D. (1999) Synthetase recognition determinants of E. coli valine transfer RNA. Biochemistry, 38, 7737–7746. [DOI] [PubMed] [Google Scholar]
- 55.Geslain R., Martin,R., Camasses,A. and Eriani,G. (2003) A yeast knockout strain to discriminate between active and inactive tRNA molecules. Nucleic Acids Res., 31, 4729–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yaremchuk A., Kriklivyi,I., Tukalo,M. and Cusack,S. (2002) Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J., 21, 3829–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biou V., Yoremchuk,M., Tukalo,M. and Cusack,S. (1994) The 2,9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science, 263, 1404–1410. [DOI] [PubMed] [Google Scholar]