ABSTRACT
Male sex determination is initiated through the testis-determining factor SRY that promotes Sertoli cell differentiation and subsequent gonadal development. The basic helix-loop-helix (bHLH) gene Tcf21 was identified as one of the direct downstream targets of SRY. The current study was designed to identify the downstream targets of TCF21 and the potential cascade of bHLH genes that promote Sertoli cell differentiation. A modified ChIP-Chip comparative hybridization analysis identified 121 direct downstream binding targets for TCF21. The gene networks and cellular pathways potentially regulated by these TCF21 targets were identified. One of the main bHLH targets for TCF21 was the bHLH gene scleraxis (Scx). An embryonic ovarian gonadal cell culture was used to examine the functional role of Sry, Tcf21, and Scx to promote an in vitro sex reversal and induction of Sertoli cell differentiation. SRY and TCF21 were found to induce the initial stages of Sertoli cell differentiation, whereas SCX was found to induce the later stages of Sertoli cell differentiation associated with pubertal development using transferrin gene expression as a marker. Therefore, a cascade of SRY followed by TCF21 followed by SCX appears to promote, in part, Sertoli cell fate determination and subsequent differentiation. The current observations help elucidate the initial molecular events involved in the induction of Sertoli cell differentiation and testis development.
Keywords: basic helix-loop-helix, cell differentiation, ChIP-Chip, SRY, scleraxis, Scx, Sertoli cells, sex determination, testis development
The SRY target TCF21 induces a downstream cascade of bHLH genes and the induction of Sertoli cell differentiation.
INTRODUCTION
Male gonadal sex determination is induced by the expression of the testis-determining factor SRY (sex-determining region on the Y chromosome) in a subset of somatic cells that are induced to differentiate into Sertoli cells. Sertoli cells are somatic cells that orchestrate germ cell differentiation and nurture their development in the fetal, postnatal, pubertal, and adult spermatogenic stages. Sertoli cells are the first cell type known to differentiate within the gonad from the bipotential precursor cell lineage to initiate testis differentiation. In contrast, supporting cell precursors in XX female gonads differentiate into granulosa cells and surround the meiotic germ cells to form primordial follicles [1]. The initiation of testis cord formation occurs at the onset of gonadal sex determination, whereas ovarian follicle assembly occurs after birth in the rodent [2]. The current study was designed to investigate the molecular events involved in promoting Sertoli cell fate determination and subsequent differentiation.
During the differentiation of testicular Sertoli cells, SRY induces the expression of its autosomal counterpart Sox9 by acting synergistically with steroidogenic factor 1 (SF1) on its testis-specific enhancer region on the promoter [3, 4]. After reaching a threshold expression level, SOX9 acts on Sry to repress expression [5, 6]. Concurrently, Sox9 expression is maintained via a positive-feedback mechanism involving fibroblast growth factor 9 (FGF9) and prostaglandins [7, 8]. Loss of function mutations of Sry and Sox9 produce a male-to-female sex reversal phenotype in XY males, whereas the gain of function causes induction of testis development in XX females, suggesting SRY initiates and then SOX9 maintains testis development. Despite extensive research regarding the functions of SRY and SOX9 in mammals, downstream targets and genome-wide actions remain poorly understood. Recently, we used a genome-wide chromatin immunoprecipitation (ChIP) followed by a promoter tiling array chip (Chip) in a ChIP-Chip comparative hybridization approach to identify the in vivo downstream targets of SRY and SOX9 in the rat gonad [9]. This analysis identified 71 direct downstream binding targets for SRY and 109 binding targets for SOX9, with only five that overlap between the two. Recently, we also demonstrated that the growth factor neurotropin 3 (Ntf3) and the basic helix-loop-helix (bHLH) transcription factor Tcf21 (bHLHa23) are direct downstream targets of SRY [10, 11]. TCF21 was found to induce differentiation of Sertoli cells in vitro in rat Embryonic Day (E) 13 ovary primary cell cultures. Many cell differentiation events during early development involve a cascade of bHLH gene expression, such as muscle cell differentiation [12], neuronal differentiation [13–18], lung cell morphogenesis [19], and cardiac cell differentiation, [20, 21]. Therefore, the current study investigated the potential that a cascade of bHLH factors may be involved in Sertoli cell differentiation and testis development.
The bHLH proteins are characterized by the helix-loop-helix (HLH) domain, which mediates the interactions that form homo- and heterodimers between these proteins [22, 23]. They also contain a highly charged basic region upstream of the HLH domain, which functions as a specific DNA-binding domain that recognizes the bHLH consensus sequence known as an E-box (CANNTG). The bHLH protein heterodimers, which consist of a ubiquitous class of bHLH protein and more tissue-specific class of bHLH proteins, bind at this conserved E-box domain. The formation of these heterodimers can promote cell-specific gene expression that influences cellular differentiation and proliferation [23]. The bHLH proteins are negatively regulated by another class of HLH proteins termed the inhibitors of differentiation (Id). These Id proteins lack a basic region, which allows them to inhibit transcriptional activation with the bHLH proteins they bind [22]. The phylogenetic and unified nomenclature of the bHLH family of genes have been previously described [23].
A previous rat and mouse developmental microarray database that covers several stages of testis development was used to determine a potential cascade of bHLH expression [24, 25]. The corresponding early gonadal period of mouse E11.5 is E13 in the rat, with no testis cords detected. Tcf21 was found to be highly expressed in Sertoli cells during this rat developmental period, corresponding to the onset of testis differentiation [11]. Tcf21 has been shown to be a downstream-regulated target of SRY, and TCF21 promotes Sertoli cell differentiation [9, 11]. To investigate the cascade of bHLH genes associated with Sertoli cell differentiation, a genome-wide ChIP-Chip approach was used to identify downstream TCF21 binding targets. A major bHLH target previously shown to be associated with Sertoli cell differentiation was scleraxis (Scx) (bHLHa41) [26]. The sequential actions of SRY, TCF21, and SCX were found to induce, in part, fetal ovarian cell cultures to differentiate in vitro to express an adult Sertoli cell gene (i.e., transferrin). Observations suggest SRY induces a cascade of bHLH factors that promote Sertoli cell differentiation and testis development.
MATERIALS AND METHODS
Tissue Preparation
Harlan Sprague-Dawley rats (Harlan, Inc.) were used for the experiment. All rats were kept in a temperature-controlled environment (22°C) and given food and water ad libitum. Estrous cycles of female rats were monitored by cellular morphology from vaginal smears. Rats in early estrus were paired with males overnight, and mating was confirmed by sperm-positive smears, denoted as Day 0 of pregnancy. Pregnant rats were euthanized at E13 of pregnancy, and embryonic gonads were collected for ChIP. Sex was determined by PCR using primers specific for Sry on genomic DNA isolated from embryo tails as previously described [27]. All procedures were approved by the Washington State University Animal Care and Use Committee (IACUC approval 02568–26).
Immunohistochemistry
Immunolocalization of scleraxis was performed according to methods described previously [11]. Briefly, embryonic testis sections (E16) were deparaffinized, rehydrated through a graded ethanol series, boiled for 10 min in 10 mM sodium citrate buffer to expose the antigens, washed with 0.1% Triton-X solution, and then blocked with 10% serum of the species the secondary antibody was raised in for 60 min before incubation with 0.5 μg/ml of primary rabbit anti-SCX antibody (Life Technologies) and 5.0 μg/ml of anti-Müllerian hormone (AMH) antibody (R&D Systems) for 18 h. The sections were then washed with PBS and stained with diaminobenzidine according to the manufacturer's instructions (Vector Laboratories). Negative-control experiments were performed using a nonimmune immunoglobulin (Ig) G at 0.5 and 5.0 μg/ml concentrations (Sigma).
In Vivo ChIP Assay
Carrier ChIP assay was adopted from O'Neill et al. [28] and performed according to Bhandari et al. [11]. The testis was removed from the mesonephros. However, residual mesonephros cannot be eliminated, so some contamination may persist. Male gonads from 30 E13 (12- to 18-tail somite stage) rat embryos were used per array. Drosophila SL2 cells were used as a carrier. Densely grown cells (∼5 × 107 cells) were pelleted and washed three times in ice-cold PBS, then resuspended in NB buffer (15 mM Tris-HCL [pH 7.4], 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM ethyleneglycoltetra-acetic acid, 0.5 mM 2-mercaptoethanol, and 0.1 mM PMSF). Testis samples were mixed with SL2 cells and homogenized to isolate nuclei. After a minute of homogenization in a glass homogenizer (15 dounces), the lysate was put on ice for 3 min before repeating the homogenization and icing for a total of 20 repeats. Nuclei were pelleted, resuspended in NB buffer and 5% sucrose, then pelleted and resuspended again in digestion buffer (50 mM Tris-HCl [pH 7.4], 0.32 M sucrose, 4 mM MgCl2, 1 mM CaCl2, and 0.1 mM PMSF). Following micrococcal nuclease digestion, the digested samples were gently centrifuged (<1000 × g) and the supernatant set aside on ice. A fraction of the supernatant was used as input. The remaining supernatant was incubated with either nonimmune IgG or anti-TCF21 antibody at 4°C overnight. After incubation with preswollen protein A-Sepharose beads, the bead-bound immunoprecipitates were centrifuged gently (<1000 × g) and washed five times with TE buffer (50 mM Tris-HCl [pH 7.5], 10 mM ethylenediaminetetra-acetic acid, 5 mM sodium butyrate, and 50–150 mM NaCl). Immunoprecipitated DNA was eluted with elution buffer (TE plus 1% SDS) and extracted with a phenol/chloroform extraction. Final concentration of immunoprecipitated DNA varied from 200 to 500 ng/assay. Three different experiments with three different biological sample ChIPs were performed. Exactly 30 ng of immunoprecipitated DNA from each assay were amplified with a whole-genome amplification kit developed by Sigma (WGA2-50 RXN). At least five separate whole-genome amplifications were performed for each sample, and DNA was pooled. Pooled whole-genome amplified DNA was purified by using the Wizard SV40 PCR Clean-up System (A9281; Promega). Purified DNA was checked on the gel and sent to Roche NimbleGen for ChIP-chip hybridization, and a three-plex promoter array was used for competitive hybridizations. Confirmation of the selected candidate binding targets used a semiquantitative PCR method [9] as described below.
Analysis of ChIP-Chip Data
For ChIP-chip hybridization, Roche NimbleGen's Rat ChIP-Chip 3x720K RefSeq Promoter Array was used. The enrichment for each probe on the array was calculated as the log ratio of the intensities of hybridization for TCF21 ChIP DNA (Cy5) to control DNA from IgG ChIP control (Cy3). Arrays contained on average 4000 bp of promoter for each of 15 287 promoters in the rat genome, corresponding to 15 600 RefSeq transcripts (∼3880 bp upstream and 970 bp downstream from transcription start site).
The bioinformatics analysis of ChIP-Chip data was performed as previously described [29]. For each hybridization experiment, raw data from both the Cy3 and Cy5 channels were imported into R (http://www.R-project.org), checked for quality, and converted to MA values (M = Cy5 − Cy3; A = [Cy5 + Cy3]/2). The R codes that were used are available at http://skinner.wsu.edu/microarray.html.
Within each array, probes were separated into groups by guanine-cytosine (GC) content, and each group was separately normalized using the Loess normalization procedure [30]. After each array was normalized within array, the arrays were then normalized across arrays using the A-quantile normalization procedure [31]. Both Loess and A-quantile normalization were performed using the limma library [32]. Following normalization, each probe's normalized M (and then A) values were replaced with the median value of all probe normalized M (and then A) values across all arrays within a 600-bp window [33–35]. The average size of the DNA fragment was 600 bp, so the window correlated. Following normalization, the median intensity difference between Cy5 and Cy3 of a 600-bp window was determined. Significance was assigned to probe differences between experimental (TCF21) and IgG control by calculating the median value of the intensity differences as compared to a normal distribution scaled to the experimental mean and SD of M. Regions of interest were then determined by combining consecutive probes with P-values less than 1 × 10−3. A Z-score and P-value were computed from that distribution with the use of R code analysis [29]. The statistically significant peaks of hybridization were identified and the P-value associated with each peak presented. Each peak of interest was then annotated for the gene. Every promoter exceeding the intensity threshold was considered to be positive for TCF21 binding. The final list of TCF21 targets includes the promoter-proximal regions that made the threshold in an average for all three experiments. Hybridization signals for all the candidate promoters that were within the cutoff line (P ≤ 1 × 10−7) were plotted (average of the three experiments). The genes that were not in the list but seemed to be masked by IgG-negative signals were designated as questionable positives. Selected questionable positive promoters were manually chosen and confirmed by PCR.
Gene Network Analysis
To move beyond examining single-gene effects on a developmental process, gene network analysis can be employed to identify groups (e.g., modules) of genes for which expression is regulated in a coordinated and functionally interconnected manner (gene network). One important end product of gene coexpression network analysis is the construction of gene modules composed of highly interconnected genes. In the current study, a network for TCF21 downstream binding target genes was constructed separately using the Pathway Studio software (Ariadne Genomics, Inc) and previously published criteria for developmental network analysis [36].
Pathway Analysis
The pathway analysis of direct downstream target genes was performed according to the protocol previously described by Nilsson et al. [36]. The downstream binding targets of TCF21 and their interconnections were analyzed for KEGG (Kyoto Encyclopedia for Genes and Genome, Kyoto University, Japan) pathway enrichment using Pathway Express, a Web-based tool freely available as part of the Onto-Tools (http://vortex.cs.wayne.edu). The Partek software was used to identify gene functional categories. A program based on literature analysis (Pathway Studio) was used to evaluate a gene network of TCF21 targets.
Fetal Gonadal Cell Preparation and Culture
Cell preparation and culture were performed are previously described [11]. E13 embryos (12- to 18-tail somite stage) were collected from timed-pregnant females as described above. Gonads from E13 animals were dissected, and each pair of gonads from individual animals was placed into one well of a 24-well plate with 500 μl of Ham F-12 medium until embryos could be sexed as described above. The female gonads were then pooled and digested with trypsin (2.5%) and collagenase (1 mg/ml, type I) plus DNase (3 mg/ml) to disassociate the cells. All cells from the digested ovaries were then cultured on 100-mm plates in Ham F-12 with 10% bovine calf serum (Sigma). Cells initially multiplied well in culture and were split two times (1:2) as they reached confluence, at which point cell division slowed considerably. Cells were maintained in culture, with the medium changed every 3 days, until growth plaques were observed at approximately 1 mo. These growth plaques were then collected for further propagation, and frozen stocks were prepared for subsequent cell culture such that cells could be maintained at fewer than 12 subcultures.
Transfection
The E13 ovarian cell cultures between subcultures 8 and 12 were transfected using Lipofectamine 2000 (Invitrogen). For each well of a six-well plate, 2 μg of expression plasmid in 250 μl of Opti-Mem medium (Sry-pCMV-HA, Tcf21-pCMV-MYC, Scx-pCMV-HA; Invitrogen) were mixed with 10 μl of Lipofectamine 2000 in 250 μl of Opti-MEM medium and incubated for 20 min at room temperature. This 500-μl mix was added to each well containing approximately 90% confluent, cultured E13 ovarian cells in 2 ml of Ham F-12 without antibiotics and incubated at 32°C for 24 h. After 24 h, medium was aspirated from cells and replaced with 2 ml of Ham F-12 with 10% serum. Cells were cultured for 2–10 days and collected in TRIzol reagent (Sigma) for RNA extraction.
PCR and ChIP Confirmation
Total RNA was extracted from frozen cells using the TRIzol extraction method. A total of 2 μg of total RNA was used to generate cDNA using Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen) according to the manufacturer's instruction. At the end, cDNA was diluted to 20 ng/μl, and exactly 50 ng of cDNA were used for PCR. PCR was performed using the following conditions: 95°C for 5 min; followed by 25–35 cycles of 95°C for 30 sec to 1 min, 58°C for 30 sec, and 72°C for 40 sec; followed by 72°C for 7 min. PCR product was visualized in 2% agarose gel with ethidium bromide. Primers were designed for ChIP confirmation for selected statistically significant targets from the region of probe hybridization peaks and tested using WGA kit-amplified ChIP DNA in a PCR. Primers are listed in Supplemental Table S1 (all Supplemental Data are available online at www.biolreprod.org). The ChIP-PCR was performed with 25–30 cycles, so it was in the linear curve of the PCR and used as a semiquantitative procedure, as previously described [9].
RESULTS
Downstream Binding Targets of TCF21 During Male Gonadal Sex Determination
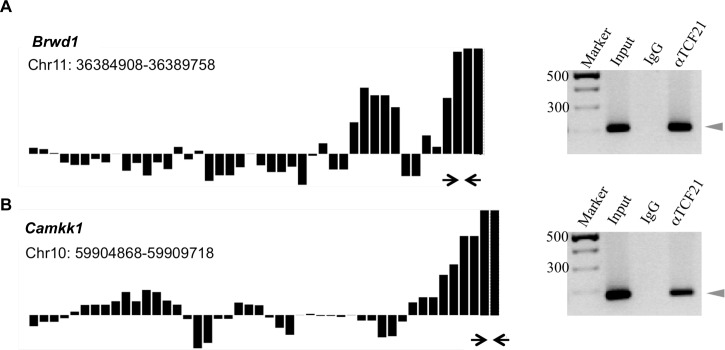
A bHLH protein, TCF21 was previously shown to be a direct downstream target of SRY to induce Sertoli cell differentiation [11]. The initial objective of the current study was to identify the genome-wide binding targets for TCF21, with a focus given to bHLH targets. Observations would extend previous observations involving the identification of the downstream targets of SRY and SOX9 [9] and help elucidate the molecular events involved in the induction of Sertoli cell differentiation and testis development. A modified ChIP followed by a Chip involving a comparative hybridization of a TCF21 antibody ChIP versus a control nonimmune IgG ChIP was used. This novel modified ChIP-Chip corrects for false-positive targets due to nonimmune IgG nonspecific binding, as previously described [9]. The rat Chip used contains approximately 4 kb of the majority of promoters in the genome. An R code was used to identify the positive-binding target genes, remove false positives, and determine statistical significance for the binding. The E13 rat male gonad was microdissected and identified with an embryonic tail clip and Sry PCR to confirm XY male genotype. The chromatin was isolated from this E13 male gonad and used in the ChIP. An antibody for TCF21 and corresponding nonimmune IgG were also used in the ChIP. The two different ChIP DNA samples were labeled with unique fluorescent colors and used in a competitive hybridization rat Chip analysis. A total of 1383 promoters were detected with P < 0.001, and a statistical cutoff of P < 1 × 10−7 was used to select potential targets to examine hybridization profiles and determine the downstream binding targets. The final analysis identified 121 downstream direct gene binding targets (Table 1). The functional categorization of these genes, chromosome location, and statistical significance are presented. The hybridization profiles for these gene targets are provided in Supplemental Figure S1. A minimum of three adjacent oligonucleotides on the tiling array had to have a statistically significant hybridization of P < 0.001 to be considered. Representative examples of direct binding targets are presented in Figure 1. A confirmation of the ChIP analysis of selected downstream targets was performed with a TCF21 antibody ChIP followed by PCR. The anti-TCF21 ChIP-PCR confirmation data are presented next to the hybridization profiles in Figure 1. All 121 targets had bHLH response elements (i.e., E-box) present, as indicated in Table 1.
TABLE 1.
TCF21 downstream binding promoter target regions.
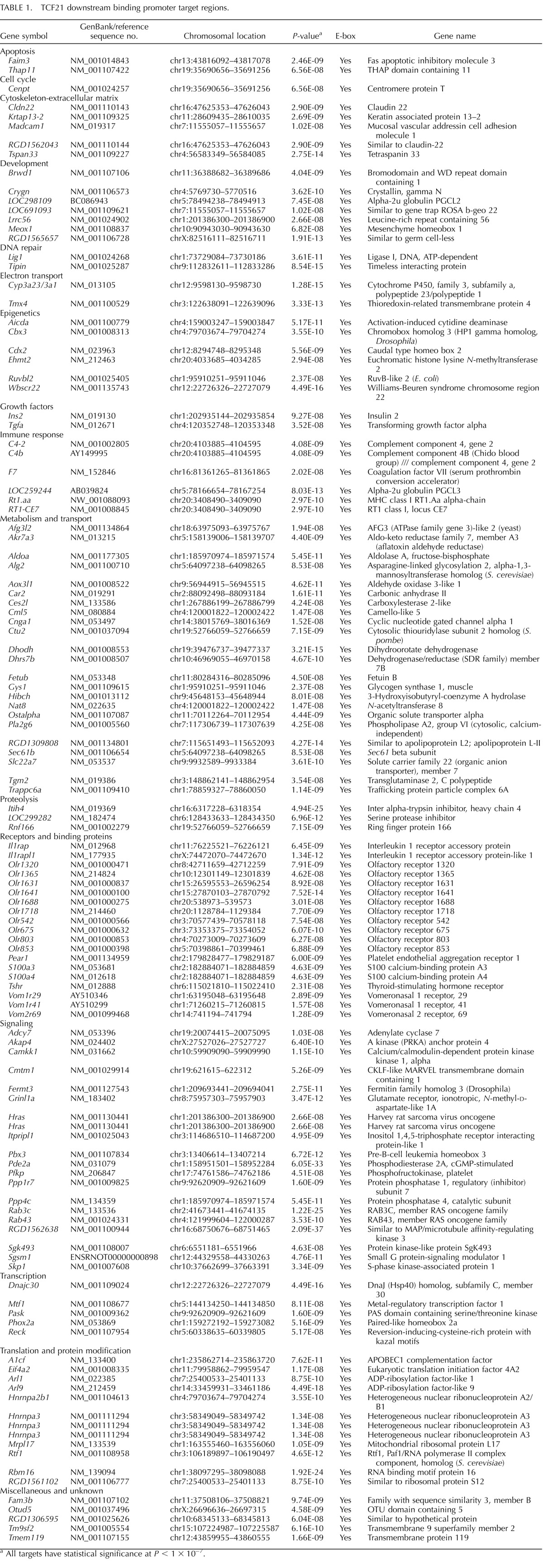
All targets have statistical significance at P < 1 × 10−7.
FIG. 1.
A and B) Hybridization signals of selected direct downstream binding target genes (Brwd1 and Camkk1) in the ChIP-Chip analysis and the PCR confirmation of target genes. PCR was performed with primers designed from the region of significant binding, indicated by arrows. The size marker (Marker), genomic DNA (Input), IgG ChIP (IgG), and TCF21 ChIP (aTCF21) are presented. The relative hybridization signal (black bar) for each oligonucleotide probe on the tiling array in the region of interest is presented. The positive bars represent TCF21 ChIP signal, and the negative bars represent the IgG ChIP signal. Data are representative of three different experiments.
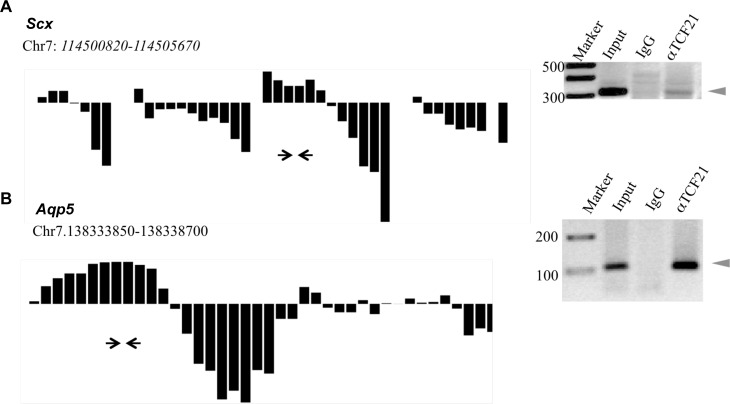
The ChIP-Chip comparative hybridization data profile identifies positive peaks representing the TCF21 ChIP pull-down, whereas negative peaks represent the nonimmune IgG ChIP (Fig. 1 and Supplemental Fig. S1). In the event a strong negative IgG peak is adjacent to a positive TCF21 peak, the positive target can be masked and considered to be questionable. A list of questionable binding targets is presented in Table S2. Representative profiles for these questionable targets are presented in Figure 2. A ChIP-PCR was performed to confirm the target and is presented. Clearly, the comparative hybridization allows the detection of nonspecific IgG binding (negative bars presented) and eliminates false positives in the ChIP-Chip assay. However, strong negative hybridization of the IgG can mask some positive targets (Fig. 2).
FIG. 2.
A and B) Hybridization signals of selected questionable positive TCF21 binding target genes in the ChIP-Chip analysis and the PCR confirmation of target genes. PCR was performed with primers designed from the region of significant binding, indicated by arrows. The DNA ladder (Marker), genomic DNA (Input), IgG ChIP (IgG), and TCF21 ChIP (aTCF21) are presented. The relative hybridization signal for each probe on the tiling array in the region of interest is presented. The positive bars represent TCF21 ChIP signal, and the negative bars represent the IgG ChIP signal. Data are representative of three different experiments.
The current study was designed to identify the binding targets for TCF21 and not to indicate direct transcriptional regulation during the E13 period. However, a previous experiment using the E13 ovarian cell culture overexpressed the TCF21 with a transient transfection of an expression construct [11]. A microarray of the cells identified genes that had significantly altered expression due to the overexpression of TCF21 [11]. A comparison of this regulated gene set with the TCF21 binding targets identified in the current study identified a number of binding targets that are transcriptionally regulated by TCF21. These statistically significant (P < 0.05) regulated TCF21 binding targets include Arl1, Rpl24, Stk3, Tipin, Pfkp, RGD1306839, Mcdcom1, Sl00a3, Phox2a, F7, Akap4, and Itih4. Therefore, more than 10% of the binding targets identified were previously shown to be regulated by TCF21 [11]. Because TCF21 may also have a role in gene repression and may regulate genes at later stages of development, not all genes are anticipated to be regulated at E13. Observations confirm a transcriptional role for TCF21 at some of the binding targets identified.
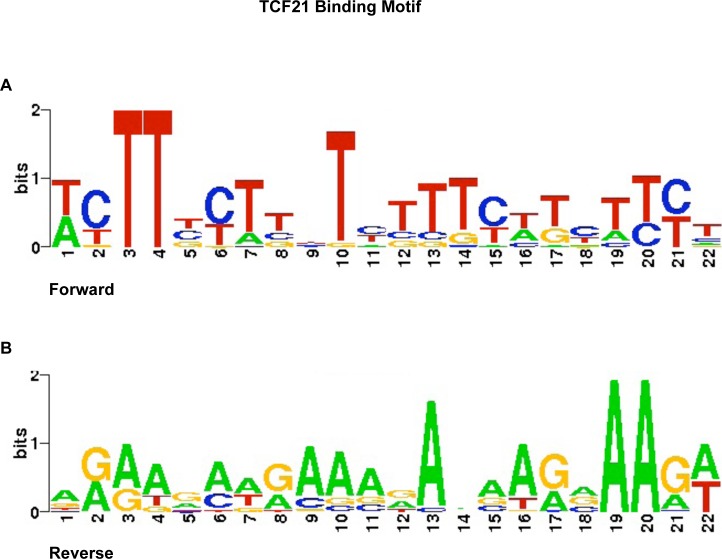
Analysis of the TCF21 DNA-binding site for the 121 downstream targets identified a consensus binding motif (Fig. 3). The TCF21 DNA-binding motif identified contained an E-box consensus sequence and flanking sequence on both sides. The largest nucleotide shown in Figure 3 represents the most prominent nucleotide present, but others are possible, providing flexibility in TCF21 binding. Although a more specific TCF21 binding site was identified, the core E-box sequence was present. A predominant E-box sequence present was CANNTC, which previouslywas shown to be used by other bHLH factors [37, 38].
FIG. 3.
Binding motif identified to be highly conserved in DNA sequences identified in TCF21 binding target gene promoters. Analysis was done using MEME software with TOMTOM follow up. Approximately 500–800 bp of all direct downstream target gene promoters were aligned and used for analysis. The core sequence for TCF21 binding was found to be CANNTC, which is common for many bHLH factors. A) The 22-bp core sequence for TCF21 binding. B) The reverse complementary sequence for TCF21 binding motif.
TCF21 Downstream Gene Target Pathway and Gene Network Analysis
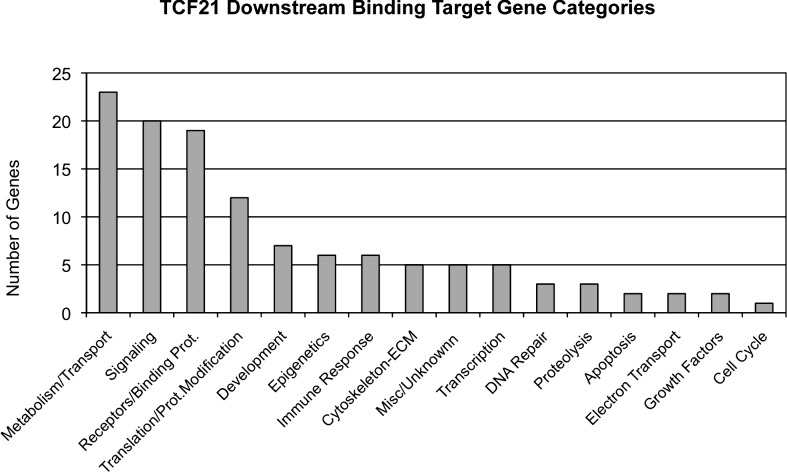
The TCF21 downstream binding target gene functional categories are presented in Table 1 and Figure 4. Predominant gene categories include metabolism/transport, signaling, receptive binding proteins, and translation/promoter modification and development (Fig. 4). Further analysis of the TCF21 gene target list for specific pathways and cellular processes is presented in Table 2. The pathways containing more than three TCF21 target genes were considered. The total number of genes in the pathways and impact of the correlated genes is also presented in Table 2. The pathway with the highest number of genes present was the olfactory transduction pathway (Supplemental Fig, S2). Other predominant developmentally relevant pathways include the cell adhesion molecule, chemokine signaling pathway, and mitogen-activated protein kinase pathway (Table 2).
FIG. 4.
TCF21 binding target gene functional categories. Total numbers of target genes associated with a specific category are presented on the y-axis and gene functional categories on the x-axis.
TABLE 2.
Number of regulated TCF21 downstream target genes in the pathways as analyzed by KEGG, with total number of genes in pathway indicated.
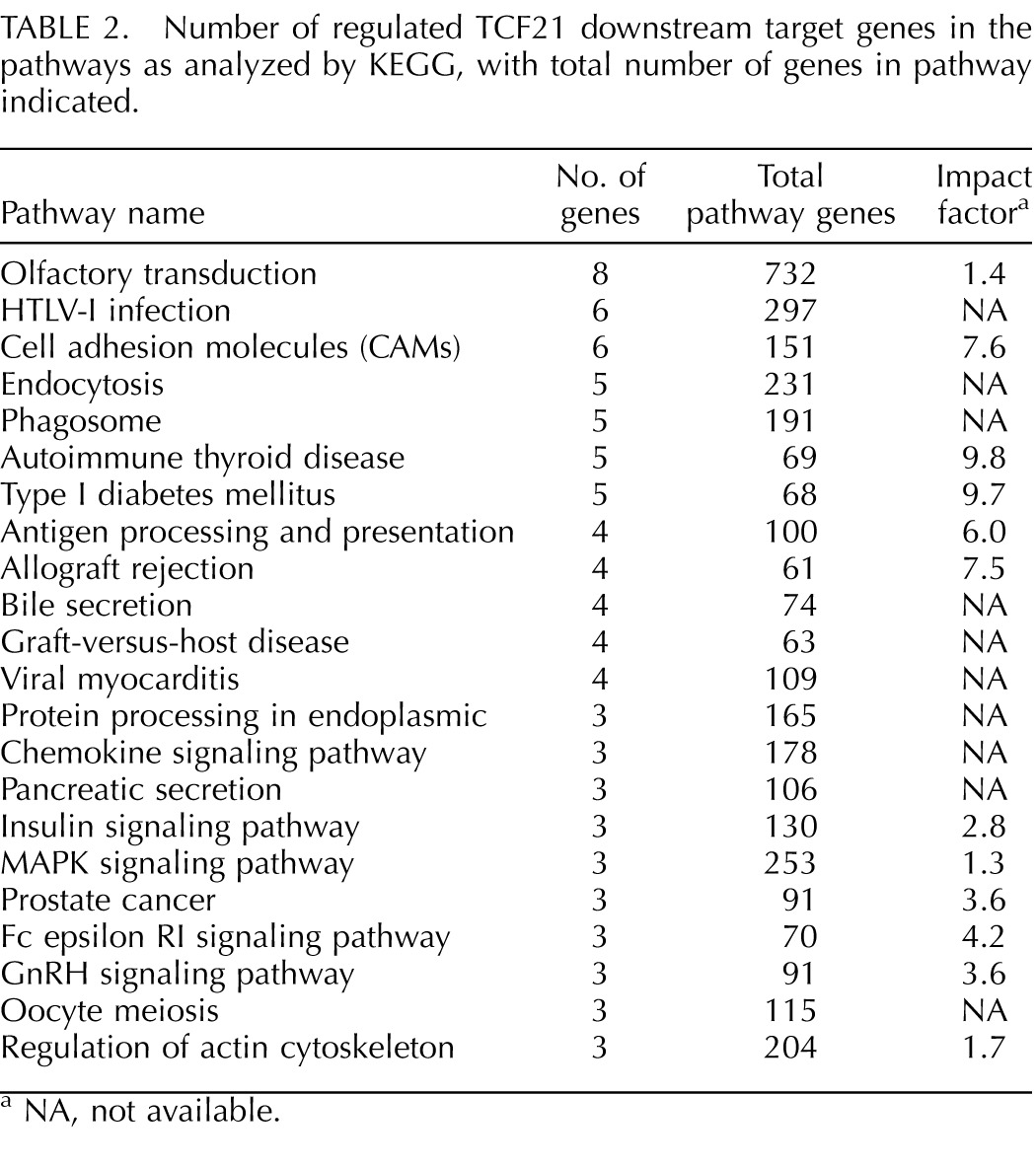
NA, not available.
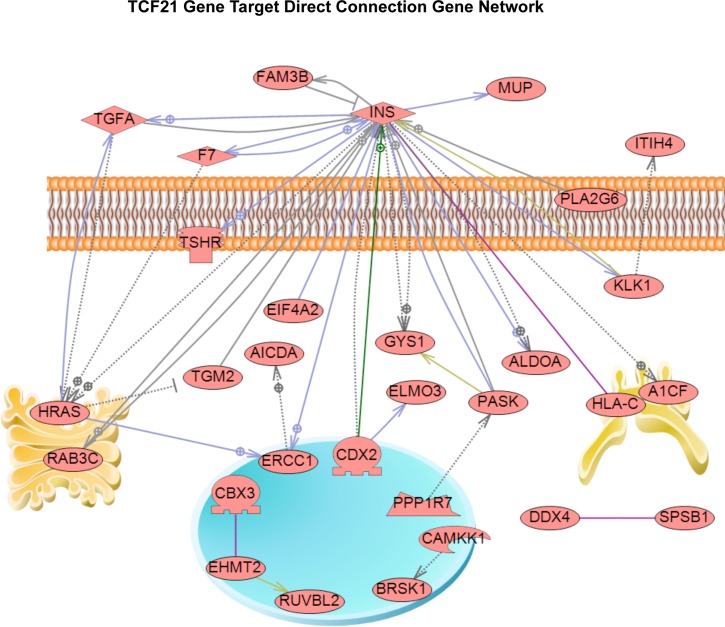
A gene network analysis was performed by considering all the TCF21 binding target genes and the use of a literature-based bioinformatics tool to assess gene connectivity (Pathway Studio). A direct connection gene network for the TCF21 target genes is presented in Figure 5. Central to this gene network is the insulin signaling system along with a number of signaling and transcriptional genes. This potential regulatory network of TCF21 downstream binding target genes was identified and speculated to be involved in the downstream regulation of Sertoli cell differentiation and testis development.
FIG. 5.
Gene network of TCF21 downstream binding target genes. Cell membrane and organelles have been graphically presented. Direct interconnected genes are shown as solid lines with positive or negative relationships and indirectly connected genes as dotted lines.
TCF21 Downstream bHLH Target Genes
Tcf21 (bHLHa23) is a member of the bHLH family of transcription factors [23]. It has been previously shown that the bHLH proteins heterodimerize to functionally induce transcription of target genes, and as stated, some bHLH genes should be associated with TCF21. The current study concentrated on TCF21 gene targets belonging to the bHLH transcription factor family. Manual screening of genes was performed to reveal bHLH gene promoters beyond a statistical cutoff of P < 1 × 10−7. A total of six bHLH genes were found within the cutoff of P < 1 × 10−4. During the comparative hybridization, the signals generated by negative-control IgG often masked the signal generated by anti-TCF21 antibody for several bHLH genes. Such genes were tested by PCR for presence in the enriched ChIP-DNA and termed questionable positive downstream targets (Fig. 2 and Supplemental Table S2). Four additional bHLH genes were also questionable targets. A total of 10 bHLH genes were found to be direct binding targets of TCF21 (Table 3).
TABLE 3.
Basic helix-loop-helix genes that are identified as downstream targets (both direct and questionable) of TCF21 in the rat testis during sex determination.
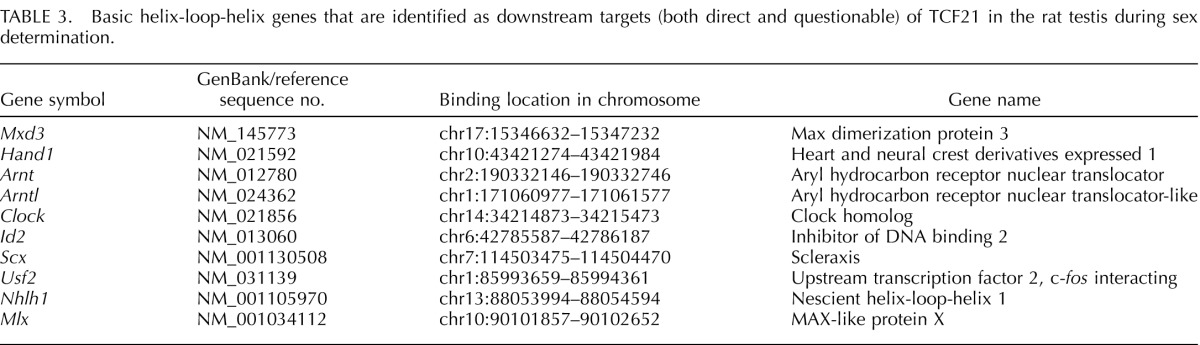
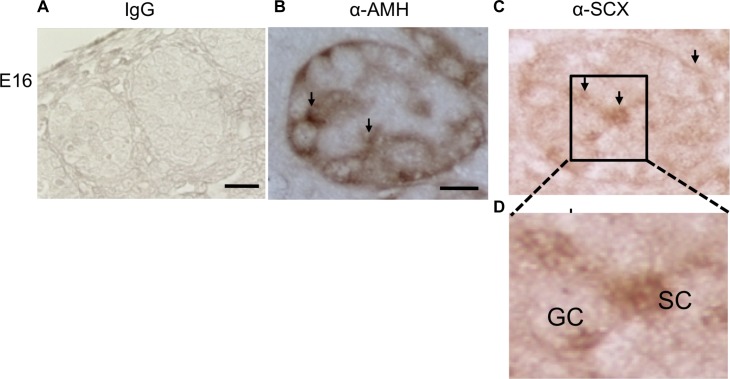
The expression profiles of the bHLH candidates indicated scleraxis had the highest level of expression following the expression of Tcf21 [23, 24]. The bHLH family transcription factor scleraxis (Scx) (bHLHa41) was one of the downstream targets in the questionable positive group. SCX has previously been shown to be localized to Sertoli cells and to promote Sertoli cell differentiation in neonatal and prepubertal rats [26]. No transferrin expression occurs during fetal development, and the only cell type that expresses transferrin is the Sertoli cell. Therefore, transferrin gene expression is a marker of pubertal and adult Sertoli differentiation [26]. The presence of Scx in TCF21 ChIP-enriched DNA was confirmed by ChIP-PCR (Fig. 2). Localization of SCX to Sertoli cells in E13 rat testis was confirmed by immunohistochemistry (Fig. 6). No SCX was detected in the interstitial or precursor Leydig cells at E13, only in Sertoli cells, but SCX has been shown to localize to Leydig cells in the E16 testis [26]. A control of AMH is presented to confirm Sertoli localization. Previously, SCX was localized to postpubertal Sertoli cells [26]. Therefore, scleraxis was selected as the primary TCF21 bHLH target for further analysis.
FIG. 6.
Immunohistochemical localization of scleraxis protein in the E13 testis. A) Negative-control staining with nonimmune IgG. B) Localization of Sertoli cells identified by anti-AMH antibody. C and D) Localization of scleraxis identified by antibody against scleraxis (higher-magnification view in D). Data are representative of three different experiments. Bar = 50 μm.
Functional Analysis of the Role of TCF21 and SCX in the Induction of Sertoli Cell Differentiation
The functional role of TCF21 and SCX was investigated with an E13 fetal gonad stem cell-like culture system as previously described [11]. The objective was to transiently overexpress expression constructs for SRY, TCF21, or SCX in the fetal E13 ovarian cells to induce an in vitro sex reversal and promote Sertoli cell differentiation. Previously, this was accomplished with the overexpression of SRY or TCF21 to induce expression of the Sertoli cell-specific marker gene Amh [11]. AMH is a marker of perinatal and prepubertal Sertoli cell differentiation that is lost at the onset of puberty. A pubertal and adult marker of Sertoli cell differentiation that was selected was transferrin gene expression [39]. Transferrin is specifically expressed in Sertoli cells in the pubertal and adult testis. This fetal ovarian cell culture was used to investigate the ability of TCF21 and SCX to induce the cascade of events leading to Sertoli cell differentiated gene expression. This experiment was not designed to suggest a normal developmental process but, rather, simply the ability of Tcf21 and Scx to induce the cascade of events leading to the induction of a more adult state of Sertoli cell differentiation, similar to other stem cell-type experiments.
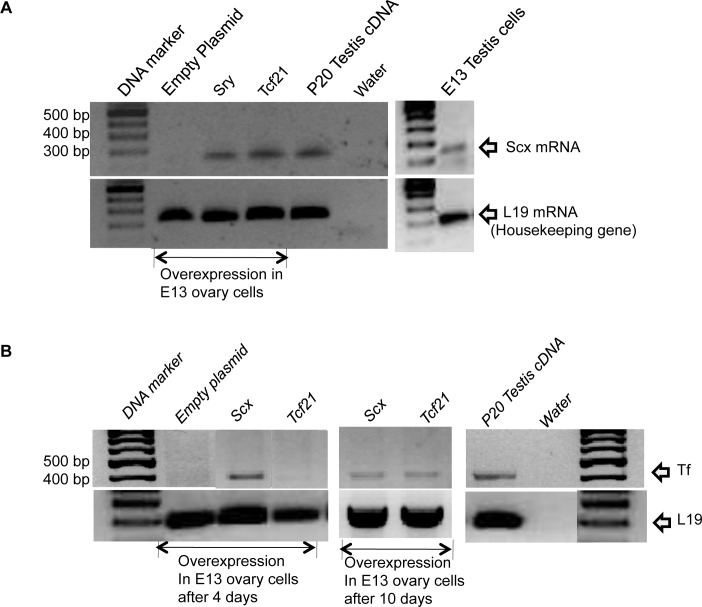
The ability of TCF21 to induce Scx expression was investigated with the in vitro E13 ovary culture system. The cell culture of E13 rat ovaries was transfected with a Tcf21 expression construct, and then the expression of Scx was tested by PCR. Preliminary studies indicated that E13 testis cell cultures express Scx but that ovarian cell cultures do not. After 48 h of culture, TCF21 induced the expression of Scx in ovarian cell cultures in vitro (Fig. 7A). In addition to TCF21 inducing Scx expression, SRY also induced Scx expression as an upstream gene to TCF21. The Postnatal Day (P) 20 testis was used as a positive control and had Scx expression [26] (Fig. 7A). Observations suggest that TCF21 induces the expression of Scx that correlates with the promotion of Sertoli cell differentiation. This supports the observation that Scx is a downstream target for TCF21.
FIG. 7.
E13 gonadal cell culture gene expression analysis. A) In vitro induction of scleraxis (Scx) expression in primary E13 ovarian cell cultures. PCR was performed with Scx primers, and DNA bands represent scleraxis. DNA ladder (Marker), overexpressed empty expression plasmid (Empty plasmid), SRY expression plasmid (Sry), Tcf21 expression plasmid (Tcf21), P20 testis cDNA, and water are presented. L19 was used as an internal control. B) In vitro induction of mature Sertoli cell marker transferrin (Tf) gene expression in E13 primary cell cultures with transfected Scx, Tcf21, and empty expression constructs. PCR bands indicate Tf and L19 control for 4- and 10-day culture durations. Data are representative of three or more different experiments.
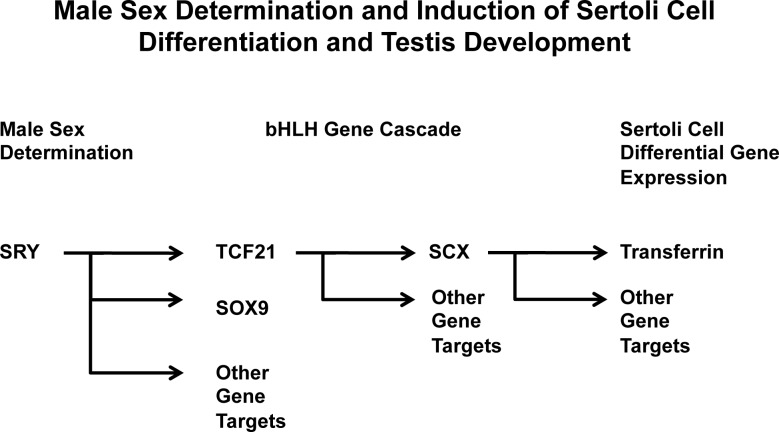
Induction of Scx by TCF21 suggested a cascade of bHLH genes may be involved in the induction of Sertoli cell differentiation. Previous studies suggested that TCF21 can induce Sertoli cell differentiation in vitro by causing sex reversal of fetal ovarian cell cultures [11]. The final experiment of the current study examined if SCX can induce the expression of an adult Sertoli cell differentiated marker in vitro. Scx was overexpressed in the fetal ovarian cell culture, and the presence of transferrin was examined by PCR. Transferrin is a cell-specific marker gene of mature Sertoli cell function and differentiation [40] and is not expressed prepubertally. After 4 days of cell culture, transferrin expression was seen in the cells overexpressing SCX in vitro (8/10 experiments) (Fig. 7B). Interestingly, TCF21 did not consistently promote transferrin expression within this 4-day culture duration (5/10 experiments) but did consistently promote transferrin expression after 10 days of culture (8/10 experiments) (Fig. 7B). Therefore, an intermediate step, such as SCX expression, appears to be required. The P20 testis was used as a positive control for transferrin expression. These observations suggest that SCX can induce further maturation of a more adult Sertoli cell differentiation state in vitro. Because TCF21 was not able to consistently promote transferrin expression within the 4-day culture duration used but was able to promote SCX expression, a cascade of SRY-induced TCF21 followed by TCF21-induced SCX appears to be required to promote, in part, Sertoli cell differentiation (Fig. 8). However, a large cascade of multiple factors likely is required in the in vivo differentiation of Sertoli cells.
FIG. 8.
Schematic diagram of the hypothesized cascade of bHLH transcription factors involved in Sertoli cell differentiation and gonadal sex determination.
DISCUSSION
Sertoli cell differentiation is one of the earliest events in sex determination and testis morphogenesis, which is initiated by expression of the male sex-determining gene Sry. During gonadal sex determination and morphogenesis, precursor Sertoli cells aggregate with primordial germ cells and then become surrounded by peritubular myoid cells to form cord-like structures. The cords in the adult testis develop into seminiferous tubules and support the process of spermatogenesis. As Jost proposed in his theory of sex determination in the 1940s, chromosomal or genetic sex is followed by gonadal sex determination, which is induced by a testis-determining factor on the Y chromosome to promote phenotypic sex through endocrine processes [41]. The sex-determining region on the Y chromosome (SRY) was identified in 1990 as the testis-determining factor [42, 43] that induces Sertoli cell differentiation to then promote testis development and male sex determination. Although SRY is known to act as Jost's testis-determining factor (TDF) and induce Sertoli differentiation, the downstream targets have only recently been elucidated [9]. Because Sertoli cell differentiation is essential for male gonadal sex determination and adult male fertility, the downstream targets of SRY and the cascade of molecular events involved are important to understand. The current study was designed to further investigate the SRY-induced molecular events that lead to Sertoli cell differentiation and male gonadal sex determination.
The initial downstream target for SRY identified was Sox9 [5, 6]. SRY acts in concert with SF1 to regulate Sox9 expression [3]. Recently, we identified the neurotrophin 3 (Ntf3) growth factor as a downstream target of SRY [10]. A genome-wide analysis of the SRY direct downstream binding targets identified 71 different genes and a large number of atypical binding targets not involving apparent direct DNA binding [9]. The downstream binding targets of SOX9 were found to be distinct from SRY, with negligible overlap [9]. Observations identified gene networks and cellular pathways involved in the SRY-induced Sertoli cell differentiation and testis determination.
One of the previously identified primary downstream SRY targets was the bHLH protein TCF21 (bHLHa23) [11]. Tcf21 was also detected in the genome-wide analysis of SRY target genes [9]. Tcf21 is a bHLH gene involved in the development of a number of tissues, such as the kidney, lungs, and heart [12–19, 21, 44]. The bHLH transcription factor family is critical for cellular differentiation and tissue development for a wide number of organ systems [12–18, 21, 44]. Although previously shown to be primarily expressed at E13 in Sertoli cells [11], potential Tcf21 expression in other cell types needs to be considered as a limitation to any data interpretations. Often, a cascade of bHLH genes orchestrate cellular differentiation and development [45, 46]. An example involves the myod and myogenin cascade for muscle development [12]. Tissue-specific bHLH factors exist for muscle (myod), neurogenesis (Neurogenin) [47–49], and heart (Hand) [50]. In addition to the cell-specific focus of bHLH genes, more widely expressed bHLH proteins integrate with other bHLH proteins to promote tissue-specific development [12–19, 44, 45]. A phylogenetic analysis of the bHLH family of genes in several different species identified groups of bHLH genes with functional similarities [23]. The Tcf21 gene clusters with a number of different genes (e.g., Musc, Tcf23, Hand, Atoh, Twist, NeuroG, NeuroD, Lyl, Tal, etc.) and appears to be highly conserved between species. Because the TCF21 bHLH protein is involved in cellular differentiation and a direct downstream target of SRY, the current study was designed to investigate the downstream targets of TCF21, with a focus on bHLH targets. An objective was to determine if a cascade of bHLH genes has a role in the induction and promotion of Sertoli cell differentiation.
A modified ChIP followed by a promoter tiling microarray (Chip) was performed to identify the downstream targets of TCF21. A ChIP-Chip involving a competitive hybridization of the antibody to TCF21-ChIP compared to the nonimmune IgG-ChIP was used to eliminate false-positive sites due to nonspecific binding of IgG. The comparative hybridization in the ChIP-Chip allows the direct comparison not possible in next-generation sequencing, ChIP-Seq, that requires different experiments to be compared. The hybridization profiles clearly identified TCF21-specific binding sites (position peaks) (Fig. 2 and Supplemental Fig. S1). This novel competitive hybridization clearly indicates nonspecific IgG binding is a critical issue to address in ChIP-Chip analysis. A limitation to this analysis is that nonspecific IgG-binding peaks can mask true-positive TCF21 binding sites, but the elimination of the false-positive binding sites is a significant advance to previous ChIP-Chip analysis and is not possible in ChIP-Seq analysis. Therefore, the number of TCF21 binding targets identified likely will be a subset of a larger number of targets. In addition, a Chip was used with approximately 4 kb of upstream promoter, such that more distal binding sites would not be detected. Therefore, the ChIP-Chip analysis used will identify targets within 4 kb of the promoter without a strong nonspecific binding of IgG. This TCF21 ChIP-Chip analysis used a stringent statistical cut off of P < 1 × 10−7, which may not have considered less significant binding sites. Therefore, the analysis used likely is a subset of TCF21 targets but includes the most highly significant detectable binding targets.
The TCF21 ChIP-Chip analysis identified 121 direct binding target genes for the fetal male gonad at the rat E13 stage of development. This is a period when SRY expression is high and testis development has been initiated (corresponding to the mouse E10.5–11.5 stage of development). Our previous studies have demonstrated the expression of Tcf21 and Scx at this stage of gonadal development in rat and mouse microarray studies [24, 25] and protein expression studies [11]. A recent study using a transgenic mouse line and fluorescent automated cell sorting to sort gonadal cell populations and examine gene expression with microarrays [51] did not detect Tcf21 expression at mouse E11.5 or Tcf21 or Scx at E12.5. This information, which is in conflict with the previous literature and the current study, remains to be elucidated, but may be an element of the mouse-versus-rat model and of the analysis used [51]. In the current study, the TCF21 target gene set was analyzed with bioinformatics protocols to identify potential common cellular pathways and processes. A number of major pathways were identified, with the most predominant being the olfactory transduction pathway, a G protein-coupled receptor family of genes (Supplemental Fig. S2). The functional role of this pathway in testis development is unclear, but evidence suggests a nonolfactory role. A gene network analysis of the TCF21 targets also identified a network of interconnected genes (Fig. 5). The insulin signaling was central to this network, with signal transduction and transcriptional regulation components. These cellular pathways and gene networks now can be further investigated to help elucidate potential roles in Sertoli cell differentiation and testis development. The combination of the previous gene networks and binding targets for SRY and SOX9 [9] with the current TCF21 targets provide a genome-wide set of molecular events in male gonadal sex determination. A comparison of the SRY [9] and TCF21 target genes indicated that 12 genes overlapped (i.e., Tmen95, Afg312, Arfgef2, Gata1, Il1rep11, Olr853, Pdcd6ip, Phox2a, RGD1562638, Rpl24, Rtf1, Sec1, and Timm86).
The analysis of TCF21 downstream targets identified 10 different bHLH genes. All these bHLH targets now need to be considered in regards to Sertoli cell and/or testis development. One gene that had a high level of fetal testis gene expression [24] and that had previously been shown by our laboratory to be associated with Sertoli cell differentiation was scleraxis [26]. Scleraxis (Scx) (bHLHa41) is involved in connective tissue development and cartilage [52, 53]. Scx clusters with Tcf15 (bHLHa40) in a phylogenic manner [23] and appears to have functions in a variety of tissues [54–57]. In adult Sertoli cells, Scx is expressed during pubertal and adult periods and can influence differential functions, such as transferrin expression [26]. Because Scx is a direct target for TCF21, is primarily localized in Sertoli cells at the E13 stage of development (Fig. 6), and has a role in pubertal and adult Sertoli cell differentiation [26], the functional role of Scx in a cascade of events associated with SRY and TCF21 was investigated.
Previously, a fetal gonadal cell culture system was established to examine the ability of SRY and TCF21 to promote an in vitro sex reversal and induction of Sertoli cell differentiation in vitro [11]. In the current study, TCF21 was found to induce Scx expression in the fetal ovarian cells, providing support for its role as a direct target for TCF21. The overexpression of SCX was found to induce the expression of transferrin gene expression in the fetal ovarian cell culture, suggesting an in vitro sex reversal and induction of mouse adult Sertoli cell differentiation. Transferrin gene expression is not detected in ovaries at this stage of development; it is only expressed in pubertal and adult Sertoli cells in the testis [24–26]. Interestingly, TCF21 alone did not have the ability to consistently promote transferrin gene expression (Fig. 7B) within the 4-day culture duration, but an extended 10-day duration of TCF21 overexpression was found to more consistently promote transferrin gene expression. This suggests an intermediate, such as SCX, in the TCF21 induction of transferrin expression. Therefore, the hypothesis is that a sequential cascade of SRY acting on Tcf21, which then acts on Scx, is part of a cascade that can induce Sertoli cell differentiation through fetal development to a more adult differentiated state (Fig. 8). The assumption is that the ultimate molecular in vivo cascade of events involved in Sertoli cell differentiation and testis development will be more complex and involve many of the other gene networks and bHLH factors identified. The current gonadal cell culture experiment was designed to determine the potential that TCF21 and SCX can promote, in a stem cell-like culture system, the differentiation of Sertoli cells, not to suggest the in vivo timing or developmental programming. A similar approach was recently made to differentiate Sertoli cells in vitro from a stem cell-like culture [58]. Observations demonstrate that TCF21 and SCX can promote fetal gonadal ovarian cells to sex reverse and induce Sertoli cells to become differentiated. Clearly, in vivo, a more complex cascade of molecular events will be involved over the developmental period of normal male testis development. Future studies need to examine more thoroughly the expression profiles and cascade of molecular events and to utilize conditional knockouts for Tcf21 and Scx in Sertoli cells to investigate this phenomenon further. The current mechanism proposed provides a framework for these further investigations. Observations support a role for bHLH in Sertoli cell differentiation and provide further insights regarding the molecular processes in male sex determination and testis development.
ACKNOWLEDGMENT
We thank Ms. Tiffany Hylkema for assistance in PCR confirmation of downstream targets, Dr. Marina Savenkova for gene family and network data analysis, Dr. Mohan Manikkam and Ms. Rebecca Tracey for time-pregnant animals, Dr. Eric Nilsson for critical review of the manuscript, and Ms. Heather Johnson for assistance in preparation of the manuscript.
Footnotes
Supported by an NICHD National Institutes of Health grant to M.K.S.
REFERENCES
- DeFalco T, Capel B. Gonad morphogenesis in vertebrates: divergent means to a convergent end. Annu Rev Cell Dev Biol 2009; 25: 457 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Regulation of primordial follicle assembly and development. Hum Reprod Update 2005; 11: 461 471. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet 2009; 25: 19 29. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008; 453: 930 934. [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet 1996; 14: 62 68. [DOI] [PubMed] [Google Scholar]
- Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol 2004; 274: 271 279. [DOI] [PubMed] [Google Scholar]
- Kim Y, Bingham N, Sekido R, Parker KL, Lovell-Badge R, Capel B. Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc Natl Acad Sci U S A 2007; 104: 16558 16563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J Biol Chem 2007; 282: 10553 10560. [DOI] [PubMed] [Google Scholar]
- Bhandari R, Haque Md M, Skinner M. Global genome analysis of the downstream binding targets of testis-determining factor SRY and SOX9. PLoS ONE 2012; 7: e43380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement TM, Bhandari RK, Sadler-Riggleman I, Skinner MK. Sry directly regulates the neurotrophin-3 promoter during male sex determination and testis development in rats. Biol Reprod 2011; 85: 227 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Sadler-Riggleman I, Clement TM, Skinner M. Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex-determining gene SRY. PLoS ONE 2011; 6: e19935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies MK, Bate M. Ruiz Gomez M. Myogenesis: a view from Drosophila. Cell 1998; 93: 921 927. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C, Jan LY, Jan YN. Cell interactions and gene interactions in peripheral neurogenesis. Genes Dev 1993; 7: 723 733. [DOI] [PubMed] [Google Scholar]
- Modolell J, Campuzano S. The achaete-scute complex as an integrating device. Int J Dev Biol 1998; 42: 275 282. [PubMed] [Google Scholar]
- Goulding SE. zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron 2000; 25: 69 78. [DOI] [PubMed] [Google Scholar]
- Huang ML, Hsu CH, Chien CT. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron 2000; 25: 57 67. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 1993; 73: 1307 1321. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell 1996; 87: 43 52. [DOI] [PubMed] [Google Scholar]
- Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development 1999; 126: 5771 5783. [DOI] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 1996; 86: 47 57. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 1997; 16: 154 160. [DOI] [PubMed] [Google Scholar]
- Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol 1998; 8: 58 65. [PubMed] [Google Scholar]
- Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 2010; 80: 1 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement TM, Anway MD, Uzumcu M, Skinner MK. Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes. Reproduction 2007; 134: 455 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod 2005; 72: 492 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir T, Sadler-Riggleman I, Skinner MK. Role of the basic helix-loop-helix transcription factor, scleraxis, in the regulation of Sertoli cell function and differentiation. Mol Endocrinol 2005; 19: 2164 2174. [DOI] [PubMed] [Google Scholar]
- Levine E, Cupp AS, Miyashiro L, Skinner MK. Role of transforming growth factor-alpha and the epidermal growth factor receptor in embryonic rat testis development. Biol Reprod 2000; 62: 477 490. [DOI] [PubMed] [Google Scholar]
- O'Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet 2006; 38: 835 841. [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner M. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE 2010; 5: e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods 2003; 31: 265 273. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19: 185 193. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, VJ Carey, Huber W, Irizarry RA, Dudoit S. (eds.), Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005: 397 420. [Google Scholar]
- Tukey J. Exploratory Data Analysis. Reading, MA: Addison-Wesley; 1977. [Google Scholar]
- Hardle W, Steiger W., Algorithm AS. 296: optimal median smoothing. J R Stat Soc Ser C Appl Stat 1995; 44 2: 258 264. [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 2007; 8: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson EE, Savenkova MI, Schindler R, Zhang B, Schadt EE, Skinner MK. Gene bionetwork analysis of ovarian primordial follicle development. PLoS ONE 2010; 5: e11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ray SK, Yang XQ, Luntz-Leybman V, Chiu IM. A splice variant of E2-2 basic helix-loop-helix protein represses the brain-specific fibroblast growth factor 1 promoter through the binding to an imperfect E-box. J Biol Chem 1998; 273: 19269 19276. [DOI] [PubMed] [Google Scholar]
- Lei H, Fukushige T, Niu W, Sarov M, Reinke V, Krause M. A widespread distribution of genomic CeMyoD binding sites revealed and cross validated by ChIP-Chip and ChIP-Seq techniques. PLoS ONE 2010; 5: e15898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Griswold MD. Sertoli cells synthesize and secrete transferrin-like protein. J Biol Chem 1980; 255: 9523 9525. [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK. E-box and cyclic adenosine monophosphate response elements are both required for follicle-stimulating hormone-induced transferrin promoter activation in Sertoli cells. Endocrinology 1999; 140: 1262 1271. [DOI] [PubMed] [Google Scholar]
- Jost A, Price D, Edwards G. Hormonal factors in the sex differentiation of the mammalian fetus. Phil Trans R Soc Lond B Biol Sci 1970; 259: 119 131. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990; 346: 245 250. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990; 346: 240 244. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 1989; 58: 537 544. [DOI] [PubMed] [Google Scholar]
- Henke RM, Savage TK, Meredith DM, Glasgow SM, Hori K, Dumas J, MacDonald RJ, Johnson JE. Neurog2 is a direct downstream target of the Ptf1a-Rbpj transcription complex in dorsal spinal cord. Development 2009; 136: 2945 2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Hermeking H. The c-MYC-AP4-p21 cascade. Cell Cycle 2009; 8: 982 989. [DOI] [PubMed] [Google Scholar]
- Brunet JF, Ghysen A. Deconstructing cell determination: proneural genes and neuronal identity. Bioessays 1999; 21: 313 318. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Anderson JP, Dodou E, Kelly KK, Greene SB, North BJ, Cripps RM, Black BL. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol 2002; 249: 174 190. [DOI] [PubMed] [Google Scholar]
- Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev 2008; 18: 411 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli BA, VanDusen NJ, Morikawa Y, Conway SJ, Cserjesi P, Vincentz JW, Firulli AB. Hand2 loss-of-function in Hand1-expressing cells reveals distinct roles in epicardial and coronary vessel development. Circ Res 2011; 108: 940 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet 2012; 8: e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Ligon KL, Lyons GE, Copeland NG, Gilbert DJ, Jenkins NA, Olson EN. Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 1995; 121: 1099 1110. [DOI] [PubMed] [Google Scholar]
- Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 2007; 134: 2697 2708. [DOI] [PubMed] [Google Scholar]
- Alberton P, Popov C, Pragert M, Kohler J, Shukunami C, Schieker M, Docheva D. Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev 2011; 21: 846 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bresee CS, Jiang H, He W, Ren T, Schweitzer R, Brigande JV. Scleraxis is required for differentiation of the stapedius and tensor tympani tendons of the middle ear. J Assoc Res Otolaryngol 2011; 12: 407 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumatsu T, Shukunami C, Amemiya-Kudo M, Shimano H, Ozaki T. Scleraxis and E47 cooperatively regulate the Sox9-dependent transcription. Int J Biochem Cell Biol 2010; 42: 148 156. [DOI] [PubMed] [Google Scholar]
- Levay AK, Peacock JD, Lu Y, Koch M, Hinton RB, Jr, , Kadler KE, Lincoln J. Scleraxis is required for cell lineage differentiation and extracellular matrix remodeling during murine heart valve formation in vivo. Circ Res 2008; 103: 948 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Itskovich E, Hu YC, Cheng AW, Ganz K, Sarkar S, Fu D, Welstead GG, Page DC, Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell 2012; 11: 373 386. [DOI] [PMC free article] [PubMed] [Google Scholar]