ABSTRACT
Spermatozoa are highly specialized cells that, when mature, are capable of navigating the female reproductive tract and fertilizing an oocyte. The sperm cell is thought to be largely quiescent in terms of transcriptional and translational activity. As a result, once it has left the male reproductive tract, the sperm cell is essentially operating with a static population of proteins. It therefore is theoretically possible to understand the protein networks contained in a sperm cell and to deduce its cellular function capabilities. To this end, we performed a proteomic analysis of mouse sperm isolated from the cauda epididymis and confidently identified 2850 proteins, which to our knowledge is the most comprehensive sperm proteome for any species reported to date. These proteins comprise many complete cellular pathways, including those for energy production via glycolysis, beta-oxidation and oxidative phosphorylation, protein folding and transport, and cell signaling systems. This proteome should prove a useful tool for assembly and testing of protein networks important for sperm function.
Keywords: mouse, proteome, proteomics, sperm
The mouse cauda epididymal sperm proteome comprises many cellular pathways, including those for energy production, protein folding and transport, and signaling and should prove useful for determining protein networks important for sperm function.
INTRODUCTION
The spermatozoon is a highly specialized cell that contains molecular systems supporting its primary functions of navigating the female reproductive tract and fertilizing an oocyte. Mouse sperm obtained from the cauda epididymis are capable of capacitation, hyperactivated motility, and fertilization of oocytes in vitro [1]. Thus, cauda epididymal sperm appear to possess all the necessary functionality of mature sperm. The sperm is thought to be transcriptionally quiescent; thus, its population of proteins is much less dynamic than a somatic cell, changing only by acquisition or loss of proteins (i.e., sperm maturation proteins or decapacitation factors, respectively) [2].
The purpose of the present study was to comprehensively identify the proteins in mouse cauda epididymal sperm, to determine their site of origin, and to begin the process of organizing the sperm proteome into a set of cellular systems that account for the function of the mature murine sperm. We hypothesize that the sperm, being a highly specialized cell, will have complete systems required for its biological functions, such as energy metabolism and signaling, but will be lacking in certain systems that are not essential, like DNA replication and transcription. Given that sperm must transit the epididymis to acquire functional maturation, we also hypothesize that a subset of necessary sperm proteins, which are associated with cauda sperm, will originate in the epididymis.
To this end, we performed a deep mass spectrometry (MS)-based proteomic analysis of mouse cauda epididymal sperm and compared the resulting set of proteins identified with high confidence to transcription profiles of the testis (http://public.wsu.edu/∼griswold/microarray.html) and epididymis (http://www.biolreprod.org/content/73/3/404.long) [3, 4]. Our results show that sperm possess many complete cellular systems that are consistent with their specialized function.
MATERIALS AND METHODS
Isolation of Cauda Sperm
Adult male mice (age, 12 wk) were used for cauda sperm isolation. Mice were maintained and handled in accordance with the Institutional Animal Care and Use policies at Washington State University. Mice were fed ad libitum and housed in a room with a controlled photoperiod (12L:12D). Mice were euthanized by CO2 asphyxiation, and their epididymides were removed and dissected free of fat. Radial slits were made in each of the cauda epididymides. The epididymides were then placed in siliconized/low-retention microcentrifuge tubes containing 1× PBS and agitated on an orbital shaker for 10 min to facilitate the swim out of the sperm. The tubes were then placed upright on a bench top, and the epididymal tissues were allowed to settle for 10 min. The sperm suspension was then removed, and an aliquot was taken to ensure purity (∼99%) and for counting purposes. The sperm suspension was placed in a new siliconized/low-retention microcentrifuge tube and pelleted at 16 000 × g for 10 min, after which the pellet was frozen at −80°C until proteomic analysis occurred.
Proteomics Sample Preparation
The sperm pellet was resuspended in 50 mM ammonium bicarbonate buffer (pH 7.4) and subjected twice to a freeze (in liquid nitrogen)-thaw cycle, followed by five 30-sec bursts of sonication with cooling on ice. Guanidine hydrochloride was added to the sample to reach a final concentration of 6 M. The sample was incubated with 10 mM dithiothreitol for 60 min at 37°C and then with 40 mM iodoacetamide for 60 min at 37°C in the dark. Samples were diluted 10-fold with 50 mM ammonium bicarbonate buffer (pH 7.4) and supplemented with 2 mM CaCl2. Sequencing-grade modified trypsin (Promega) was then added to the samples at an enzyme:protein ratio of 1:50 (w/w). After 3 h of initial incubation, the sample was diluted 2-fold further with 50 mM ammonium bicarbonate buffer (pH 7.4), and another aliquot of trypsin (enzyme:protein ratio, 1:50) was added for incubation overnight. The digests were then acidified to a final concentration of 0.1% trifluoroacetic acid before purification over C18 Solid-Phase Extraction (SPE) columns (Supelco). The eluate was dried under vacuum to remove organic solvents and then resuspended in water. Final peptide concentrations were determined by BCA Protein Assay (bicinchoninic acid; Thermo Scientific). Samples were stored at −80°C until fractionation.
High-pH Reversed-Phase Liquid Chromatography Fractionation
High-pH reversed-phase (RP) liquid chromatography (LC) fractionation was performed as described previously [5] on an Agilent 1200 Series high-performance liquid chromatography (HPLC) system at a flow rate of 0.5 ml/min using an XBridge C18 column (inner diameter, 4.6 mm; length, 250 mm; particle size, 5 μm; Waters) equipped with a guard column (inner diameter, 4.6 mm; length, 20 mm; Waters) [5]. Solvent A consisted of 10 mM ammonium formate (pH 10), whereas solvent B consisted of 10 mM ammonium formate and 90% acetonitrile (pH 10). The separation gradient was set up as follows: from 0% to 5% solvent B in 10 min, from 5% to 35% solvent B in 60 min, from 35% to 70% solvent B in 15 min, and held at 70% solvent B for another 10 min. The fraction collection started at the beginning, and a total of 96 fractions were collected. The 96 fractions were dried in a Speed-Vac (Thermo Scientific Savant) and resuspended in water before being concatenated into 24 samples in a rolling fashion—namely, pooling fractions 1, 25, 49, and 73; 2, 26, 50, and 74; 3, 27, 51, and 75; and so on. The samples were stored at −80°C until analysis by high-resolution RPLC coupled to tandem MS (MS/MS).
LC-MS/MS Analysis
The 24 concatenated fractions from the high-pH RPLC fractionation were analyzed using an in-house, automated, four-column capillary RP-HPLC system coupled to an LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific). The LC columns (inner diameter, 75 μm; length, 65 cm) were packed in-house with 3-μm Jupiter C18 bonded particles (Phenomenex). Peptides were loaded and separated using an exponential gradient starting with 100% mobile-phase solvent A (0.2% formic acid in water), which was gradually increased to 60% solvent B (0.2% formic acid in 100% acetonitrile) over 100 min. Each MS scan (m/z 400-2000) was measured with a resolution setting of 30 000 and followed by data-dependent MS/MS of the 10 most intense ions in the ion trap. The normalized collision energy for collision-induced dissociation was set at 35%, and the dynamic exclusion was enabled such that the MS/MS spectrum of a precursor ion, once acquired, was excluded from future MS/MS acquisitions for 60 sec.
Proteomics Data Analysis
The MS/MS spectra acquired from the offline two-dimensional (2D) LC-MS/MS analysis were preprocessed using in-house tools DeconMSn and DtaRefinery to assign the correct monoisotopic peak and to remove the systematic errors in mass measurement accuracy, respectively [6, 7]. SEQUEST (Thermo Fisher Scientific) MS/MS search engine was then used to match the MS/MS spectra against a concatenated database, including the Mus musculus UniProt (version 2010-05-05) protein sequences and their reversed counterparts. Partial trypsin cleavage rule was required for all the considered peptides. Static modification of cysteine alkylated with iodoacetamide and dynamic modification of methionine oxidation were considered. The distribution of mass deviation (from the theoretical masses) was determined to have an SD (σ) of 1.65 ppm. Data filtering criteria including a parent-ion mass error of smaller than 3σ, and others based on the cross-correlation score (Xcorr) and delta correlation (ΔCn) values, along with tryptic cleavage and charge states, were then developed using the decoy database approach and applied for filtering the raw data to limit false-positive identifications to less than 1% at the peptide level [8]. The resulting lists of peptides were further processed by ProteinProphet software (http://proteinprophet.sourceforge.net) to remove redundancy in protein identification [9].
Comparison of Sperm Proteome to Transcriptome
Comparisons between the sperm proteome and the readily available Affymetrix mouse testis and mouse epididymis transcriptomes were conducted. The mouse testis and epididymis transcriptomes databases are available online (http://public.wsu.edu/∼griswold/microarray.html and http://www.biolreprod.org/content/73/3/404.long, respectively). Any transcripts that were flagged as present in the Affymetrix data and had raw expression values of greater than 50 were used in comparison studies. Data were compared using gene symbols; data points to which gene symbols were not assigned were omitted from analysis.
Functional Cluster and Pathway Analysis
The mouse sperm proteome was analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; version 6.7; http://david.abcc.ncifcrf.gov) [10, 11]. The UniProt identifiers (UniProt_ID) for the 2850 protein database were entered into the DAVID functional annotation program. Overall, 2839 of the proteins were assigned DAVID IDs for analysis. The list was subjected to functional annotation clustering using GOTERM_FAT Gene Ontology (GO) terms and a classification stringency of medium. Pathways were analyzed using the KEGG PATHWAY database (http://www.genome.jp/kegg/pathway.html) available in DAVID. Illustrations of complete molecular pathways in sperm (Supplemental Figs. S1–S12; all Supplemental Data are available online at www.biolreprod.org) were generated using the KEGG PATHWAY software and are used with permission (http://www.genome.jp/kegg/kegg1.html). To determine which proteins in the cauda sperm proteome may be important for normal reproductive function in the male, the proteome was screened against the subset of mutant mice with a male reproductive phenotype in The Jackson Laboratory mutant mouse database (http://www.informatics.jax.org/).
RESULTS
Mouse Cauda Epididymal Sperm Proteome
A total of 66 561 tandem mass spectra acquired from 2D LC-MS/MS analysis of the trypsinized sample of cauda epididymal mouse sperm proteins yielded confident peptide identifications (Table 1). These spectra corresponded to 29 212 unique peptide species, from which 4004 proteins were identified. For this analysis, we set detection criteria of at least two unique peptides for high-confidence identification of protein. In all, 2850 proteins were identified with at least two unique peptides. The list of proteins identified with two or more unique peptides was subjected to the analysis reported herein and is referred to as the cauda sperm proteome. The entire list of 2850 proteins comprising the cauda sperm proteome, with number of unique peptides and total spectral count, is shown in Supplemental Table S1. The peptide identifications, with all the associated spectra and charge state, database searching scores, and mass error, are shown in Supplemental Table S2. To our knowledge, this proteome is the deepest proteome coverage reported to date in the literature for any species of sperm. The proteins identified cover between 75% and 100% of previously reported murine sperm proteins identified by proteomic analysis, including 82% overlap with the next deepest proteome of 858 sperm proteins reported by Baker et al. [12] in 2008 (Supplemental Table S3). When compared to the recently published proteome of mouse testicular germ cells, in which 2116 proteins were identified, 52% of the germ cell proteins were detected in our mouse cauda proteome [13].
TABLE 1.
Characteristics of the mouse sperm proteome.

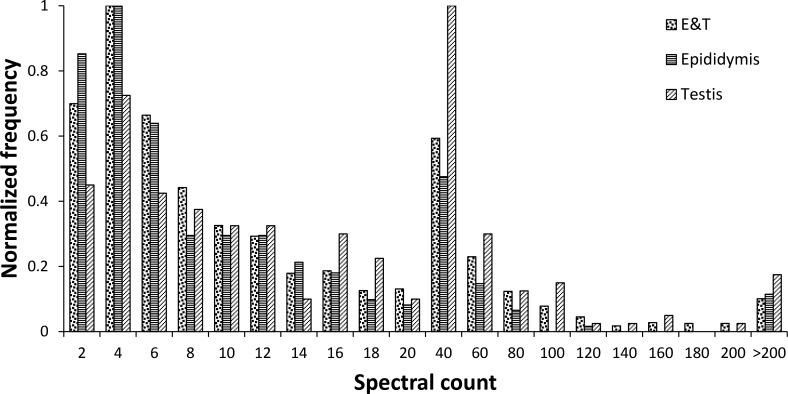
The cauda sperm proteome was compared to transcription array data for mouse testis and epididymis [3, 4] (Fig. 1). The majority of transcripts encoding the cauda sperm proteins (2727/2850; 96%) were accounted for in at least one of the two transcription array databases using a minimum signal of 50 from the Affymetrix array data. In all, 46 transcripts with a signal of less than 50 on the Affymetrix arrays corresponded to mRNA-encoding proteins in the cauda sperm proteome. These 46 proteins were indeed detected at lower concentrations in the proteome, identified with an average of 12 spectra per protein, compared to an average of 28 spectra per protein for which the Affymetrix array data showed a signal of greater than 50 (Supplemental Table S1). In addition, 77 proteins were in the cauda sperm proteome that had no expression data, because they were not tiled on the Affymetrix arrays. The majority of the cauda sperm proteins (2182/2727; 80%) were expressed in both the testis and the epididymis. A total of 245 proteins were expressed only in the testis and may represent a population of proteins enriched in germ cell-specific proteins; 300 proteins were expressed only in the epididymis and may represent a pool of proteins enriched in sperm maturation proteins. Interestingly, those proteins that were expressed specifically in either the testis or epididymis had a similarly broad distribution of spectral count (i.e., relative protein abundance) compared to those expressed in both the testis and epididymis (Fig. 2). This peptide spectral count distribution appears to reflect the true endogenous abundance distribution of these proteins and suggests that the levels measured are not biased by analytical measurements, which are typically concentration sensitive. The site of gene transcription data for each protein in the cauda sperm proteome is included in Supplemental Table S1.
FIG. 1.
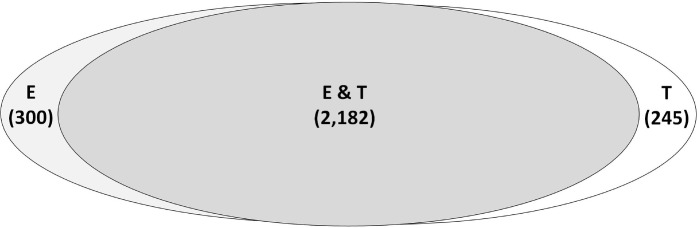
Venn diagram illustrating the site of transcription of cauda proteins as determined by transcription array analysis. E, 300 transcripts detected only in the epididymis; T, 245 transcripts detected only in the testis; E & T, 2182 transcripts detected in both the testis and the epididymis. For a list of each set of transcripts, see Supplemental Table S1.
FIG. 2.
Relative protein abundance for proteins expressed in both epididymis and testis, only in epididymis, and only in testis. The frequencies in the protein spectral count bins were normalized to a zero-to-one scale in each case.
Cluster Analysis of the Cauda Sperm Proteome
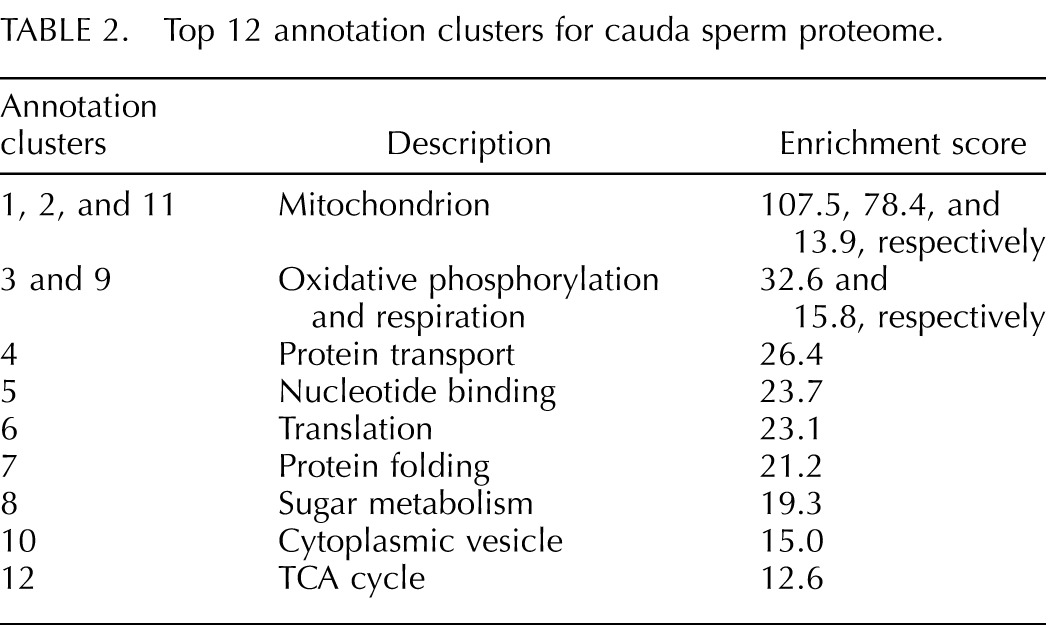
Cluster analysis of the cauda sperm proteome was conducted to determine if proteins involved in particular biological or cellular functions, or those located in particular cellular compartments, are enriched in the proteomic data set. The data represent a statistically significant clustering of proteins associated with related GO annotation terms and is interpreted here as an indication of the importance of certain systems within the cell. Cluster analysis of the cauda sperm proteome using the DAVID program produced 527 protein clusters from 1647 proteins [10, 11]. Of these annotation clusters, 176 were considered to be statistically significant (enrichment score, >1.3). Table 2 lists the top 12 annotation clusters in the cauda sperm proteome. Annotation clusters 1, 2, and 11 all represent proteins clustered in association with the mitochondrion or a subcomponent of the mitochondrion. Annotation clusters 3 and 9 cover proteins involved in oxidative phosphorylation and cellular respiration and are consistent with the significant clustering of mitochondrial proteins. Annotation clusters 8 and 12 cover proteins involved with sugar metabolism and the tricarboxylic acid (TCA) cycle and add to the emphasis on energy metabolism in sperm. Annotation clusters 6 and 7 are associated with protein translation and folding, and cluster 4 includes proteins involved with localization and transport of proteins. Proteins with nucleotide-binding activity and proteins located in, or associated with, cytoplasmic vesicles comprise the other two clusters (clusters 5 and 10). These results are consistent with a high priority for energy production in sperm, robust transport and signaling systems, and a possible role for protein translation in posttesticular germ cells.
TABLE 2.
Top 12 annotation clusters for cauda sperm proteome.
Metabolic Pathways
To further characterize the energy production potential of the cauda sperm, the cauda sperm proteome was analyzed for the presence of complete metabolic pathways using the KEGG PATHWAY. Within the proteome, 1143 (40%) of the proteins were assigned to pathways. Analysis of pathways relative to sugar metabolism confirmed that the cauda sperm proteome contained all the enzymes of the glycolytic pathway and the TCA cycle (Supplemental Figs. S1 and S2). The sperm-specific forms of glycolytic enzymes phosphoglycerate kinase (Pgk2), glyceraldehyde-3-phosphate dehydrogenase (Gapdhs), and lactate dehydrogenase (Ldhc) were the predominant forms detected (579, 700, and 1177 spectral counts, respectively). The somatic forms of each of these proteins were also present in the proteome, albeit at lower abundance (spectral counts detected reflecting relative protein abundance: Pgk1, 158; Gapdh, 160; Ldha, 278; Ldhb, 108). The majority of the protein subunits in the five enzyme complexes of the oxidative phosphorylation pathway were also present in the proteome (Supplemental Fig. S3). The undetected subunits were either small (<15 kDa) or part of the membrane-spanning portion of the a complex (i.e., ATP synthase subunits a and c). Because sperm mitochondria are known to maintain a membrane potential, the undetected subunits likely are present [14]. The pathway proteins required for metabolism of fructose and mannose were also detected, as were all enzymes of the pentose phosphate pathway and pyruvate metabolism (Supplemental Figs. S4 and S5). Two detected proteins are members of the solute carrier family 2 glucose transporters (Slc2a3/GLUT3 and Slc2a5/GLUT5), indicating that sperm can uptake both glucose and fructose, which is consistent with the ability of sperm to metabolize these two sugars as primary energy sources [14, 15]. The cauda sperm proteome is deficient in galactokinase, which suggests that mouse cauda sperm are unable to metabolize galactose.
The cauda sperm proteome contains all the proteins required for β-oxidation of lipids for the production of energy (Supplemental Fig. S6). The proteome also contains the complete enzymatic pathway necessary to synthesize fatty acids, which is expected given the important role of lipid metabolism in sperm maturation (Supplemental Fig. S7). The proteome also possesses the proteins required to metabolize most amino acids and to transfer the nitrogen of ammonia to glutamate for safe elimination of nitrogen from the cell. Analysis of nucleotide synthesis pathways revealed that mouse cauda sperm contain the enzymes needed to synthesize purines but not those required for pyrimidines. Components of several other synthetic pathways are also present in the mouse cauda sperm proteome, including all enzymes necessary for the synthesis of glutathione from precursor amino acids and for the interconversion between glutathione and glutathione disulfide (Supplemental Fig. S8).
Sperm Signaling Proteins
Signaling pathways are very important for posttesticular sperm function, including capacitation, and for fertilization. Cluster analysis of the cauda sperm proteome showed that small GTP-binding proteins were significantly enriched in cauda sperm (Table 3). Analysis of proteins associated with the GO term for “small GTPase-mediated signal transduction” (GO:0007264) revealed 66 sperm proteins in this category, and these are one of two statistically significant clusters of proteins involved with signal transduction (Table 3). This group of proteins includes 23 RAB proteins, most of which are completely unstudied in sperm. As expected, the cauda sperm proteome also included several proteins involved with G protein-coupled receptor signaling pathways, including the proteins of the adenylate cyclase/protein kinase A pathway.
TABLE 3.
Signaling pathway proteins.
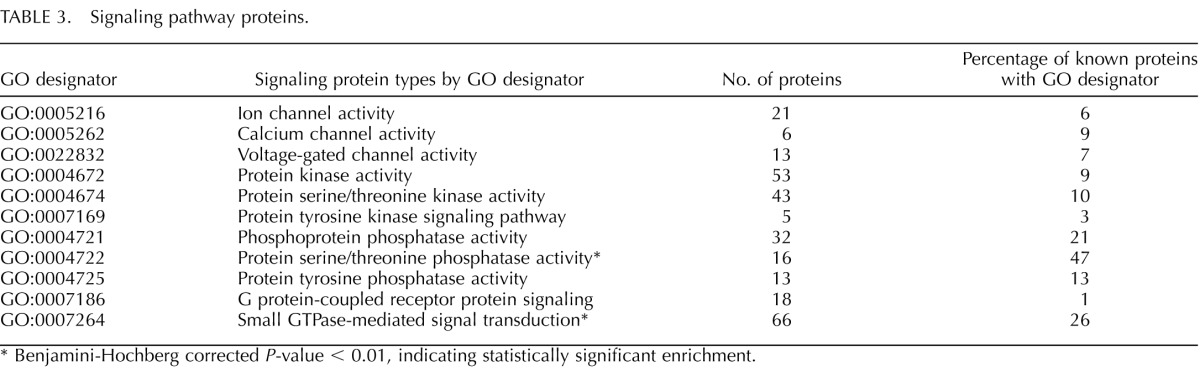
Benjamini-Hochberg corrected P-value < 0.01, indicating statistically significant enrichment.
Also within the mouse cauda sperm proteome were 21 ion channel proteins, including ATP-driven transport proteins. Six calcium channels were found, including CatSper subunits 1–4, β, and γ, and two forms of the inositol 1,4,5-triphosphate receptor (1 and 2). The ion channel list also included several voltage-dependent ion channels. Transient receptor potential cation (TRPC) channels were not detected in the cauda sperm proteome, suggesting that the proteome is incomplete for channel proteins, because these channels have been described in mouse sperm [16, 17]. Proteins necessary for generating membrane potentials (Na+/K+ ATPases) and calcium gradients (Ca2+ ATPases) were observed. These ion pumps and channels are also part of the cluster of transport proteins in sperm. Protein kinases and phosphatases were also well-represented signaling proteins in the proteome, including a statistically significant clustering of proteins with serine/threonine phosphatase activity (Table 3).
Translation-Related Pathways
The cluster analysis shown in Table 2 suggests that protein translation and folding are represented processes in sperm. When subjected to KEGG PATHWAY analysis, the cauda sperm proteome was found to contain 67 ribosomal proteins (85% of total), along with cytoplasmic tRNA synthases for all 20 amino acids with the exception of threonine, for which only one unique peptide was detected (Supplemental Figs. S9 and S10). Consistent with the dogma that sperm are transcriptionally quiescent, proteins involved with transcription, most notably RNA polymerase II, were largely lacking in the cauda sperm proteome, as were proteins involved with RNA splicing. Together, these data suggest that mouse sperm may be able to synthesize proteins from existing mRNAs within the cell. The mouse cauda sperm proteome was found to be rich in proteolytic enzymes, including proteins required for a functional proteasome and ubiquitin-conjugation system (Supplemental Figs. S11 and S12). In addition, 93 other proteases were present, and 36 protease inhibitors were identified.
Reproduction-Associated Proteins
The mouse cauda sperm proteome includes 35% of mouse proteins annotated for involvement with the process of spermatogenesis and 41% of proteins annotated for involvement with fertilization (Table 4). The cauda sperm proteome also contained the majority of proteins known to be associated with the flagellum and acrosome. All the proteins associated with reproductive GO terms were clustered in the cauda sperm proteome with strong statistical significance.
TABLE 4.
Proteins associated with spermatogenesis or germ cell function.
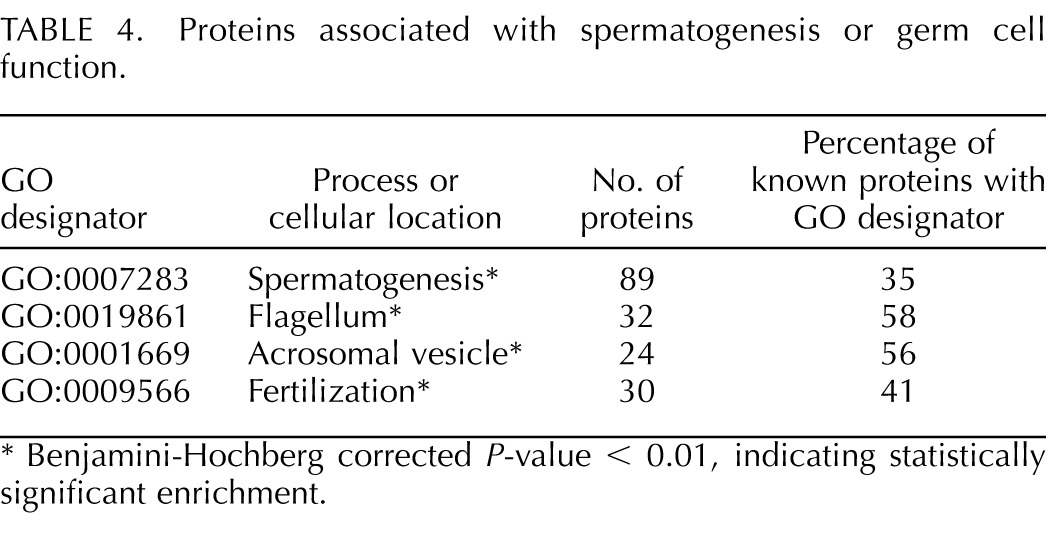
Benjamini-Hochberg corrected P-value < 0.01, indicating statistically significant enrichment.
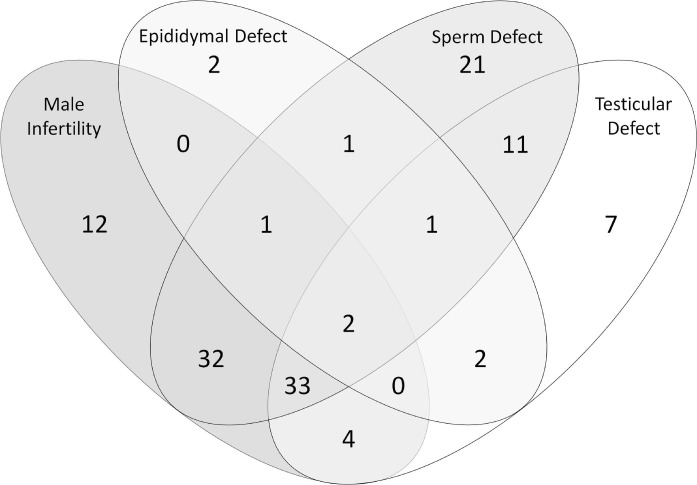
When screened against The Jackson Laboratory mutant mouse database, 137 proteins were identified that have genes required for normal male reproductive function. When these genes are knocked out or mutated, 87 produce infertile male mice, 110 a sperm defect, 60 a testicular defect, and 9 an epididymal defect. The Venn diagram in Figure 3 illustrates the overlap in the effects of mutations in these genes. Of these genes, 36 are expressed exclusively in the testis, 7 exclusively in the epididymis, and 78 in both tissues. The source of expression for all proteins in the cauda sperm proteome is detailed in the Supplemental Table S1.
FIG. 3.
Cauda sperm proteins associated with defects in male fertility or a functional or morphological defect in the epididymis, testis, or sperm. Data were obtained from The Jackson Laboratory mouse knockout database (http://www.informatics.jax.org/). Note that not all mutants were assessed in all four categories. For an interactive version of this figure with links to a list of each protein set, see Supplemental Table S4.
DISCUSSION
The present results build upon, and substantially extend, the mouse sperm proteome published by Baker et al. [12] in 2008 that cataloged 858 proteins from mouse sperm. The Baker et al. proteome also uses detection of two unique peptides as criteria for inclusion in the list. Our proteome contains 703 (82%) of these proteins and extends the list of known mouse sperm proteins by 1984 proteins, to a total of 2850 proteins; this is well in excess of the 2000–2300 proteins estimated to be in human sperm [18, 19]. All the proteins identified in the cauda sperm proteome are annotated in the UniProt database. In 2004, Nass and Strauss [20] published a report on the future of contraceptive development that included a recommendation that protein networks in sperm be determined. The mouse sperm proteome is now likely sufficient to begin deciphering protein networks that can be interrogated for contraceptive development.
The requirement of detecting two unique peptides for inclusion in the cauda sperm proteome dramatically diminishes the likelihood that the database contains misidentified proteins. There may be minor contamination from sources such as epididymal epithelial cells, epididymal fluid, and serum; however, the swim out method for preparation of cauda sperm used in the present study allows isolation of a very pure population of sperm. The cauda sperm population was judged to be greater than 99% pure based on inspection of cell smears from the sample preparation. However, we did detect albumin in the sperm proteome, suggesting that the cauda sperm came into contact with serum proteins either in the caudal fluid or during isolation of the cells. The lack of highly expressed epididymal gene products suggests the contamination of the sperm sample with other cellular proteins appears to be minimal. For example, lipocalin 9 has the second-highest level of expression in the epididymis (expression level, 17 194) and is not present in the sperm proteome. Likewise, CRIP1, Ly6E, GPR64, and lipocalin 8, all in the most abundant 0.2% of epididymal transcripts, are absent from the mouse sperm proteome. Lipocalin 5, also among the most abundant epididymal transcripts, has been shown to associate with sperm and is abundant in the mouse sperm proteome [21]. These observations are consistent with highly pure epididymal sperm used for the proteomic analysis.
As expected, proteins involved with cellular energy metabolism were found to be dominant in the mouse sperm proteome (Table3). Glycolysis, the TCA cycle, and oxidative phosphorylation pathways have all been shown to function in sperm [14, 22, 23]. The cauda sperm proteome contains every enzyme necessary for both glycolysis and the TCA cycle, and 80% of the enzymes, or enzyme complex subunits, of the mitochondrial electron-transport system were detected. A total of 599 mitochondrial proteins were detected in the cauda sperm proteome, which is consistent with the importance of this organelle in sperm function. The sperm-specific forms of Pgk2, Gapdhs, and Ldhc predominated, as measured by the total spectral counts of the peptides detected, compared to the somatic forms of these proteins. Studies using gene knockout models have demonstrated the requirement for the sperm-specific forms of these glycolytic enzymes and suggest that the levels, or possibly the cellular distribution, of the somatic form of these enzymes is not sufficient to support normal sperm function [24–26].
Other pathways of carbohydrate metabolism were also completely intact in cauda epididymal sperm. These included all the enzymes of the pentose phosphate pathway, which has been implicated in sperm-egg fusion and regulation of oxygen free radical production [27, 28]. The facilitated glucose transporters, Slc2a3 and Slc2a5, were both identified, which is consistent with the ability of sperm to actively take up glucose and fructose [27, 29]. Interestingly, solute carrier protein 2a8 (Slc2a8; also known as GLUT8), which has been implicated in knockout mice to be important for glucose uptake in sperm, was not among the 40 Slc family proteins identified in the cauda sperm proteome [30, 31].
A complete enzymatic pathway for saturated lipid synthesis and breakdown was detected in the cauda proteome. In addition, lipid desaturase enzymes were detected in the proteome. Sperm membrane lipids are known to undergo substantial change during epididymal transit, including, in human sperm, an increase in the concentration of unsaturated fatty acids [32–35]. Very little published data on the ability of sperm to synthesize lipids de novo are available.
Sperm utilize significant amounts of ATP to drive movement of the flagellum and other cellular processes and, as noted above, have robust representation of proteins involved in glycolysis, the TCA cycle, and oxidative phosphorylation for the purpose of regenerating ATP from precursor forms of adenosine. Consistent with the need for purines, the cauda sperm proteome contains the necessary enzymes to synthesize adenosine de novo, although to our knowledge, no reports of such synthesis have appeared in the literature. All but one enzyme involved in synthesizing guanine are also present (xanthine oxidase was detected only at the one-peptide level in cauda sperm). This agrees with the energy production needs of sperm (ATP synthesis) and the large number of GTP-binding proteins present in sperm (see below). In contrast, sperm are missing several key enzymes required for the synthesis of pyrimidines, including the trifunctional enzyme carbamoyl-phosphate synthetase 2. Given the allosteric regulation of both biosynthetic pathways, sperm would have to maintain adequate levels of pyrimidines to facilitate de novo synthesis of purines, if such synthesis occurs in sperm. Also missing from the proteome are all DNA polymerases and all but one subunit of RNA polymerase II. This is concurrent with the lack of requirement for DNA or RNA synthesis.
Although cauda epididymal sperm are thought to be translationally quiescent, the majority of proteins needed for successful translation of mRNA into protein were detected in the proteome. In all, 67 of 79 cytoplasmic ribosomal proteins were detected, as were all the enzymes for activating tRNAs for each amino acid except threonine. These data suggest that functional translational machinery may exist in cauda sperm. Several reports of protein synthesis in sperm have appeared in the literature, although each of these studies implicates mitochondrial rather than cytoplasmic ribosomes [36–39]. Interestingly, although ribosomal proteins were well represented in the proteome, many were detected with a low number of unique peptides, indicating that the abundance of these proteins is relatively low. This implies that translation, if it occurs as a normal part of sperm function or maturation, is not a robust feature of these cells.
The cauda sperm proteome contained 63 proteins annotated as involved with protein folding. This list includes a number of known chaperone proteins, such as 10 members of the Dnaj family, as well as members of the heat shock protein 60, 70, and 90 as well as endoplasmin, calreticulin, calmegin, and calnexin families. In addition to the standard role these proteins play in protein folding, transport, and degradation, molecular chaperones have been postulated to play a role in sperm-egg interactions [40].
Mouse cauda epididymal sperm contain 468 proteins that carry the GO functional tag of transport proteins. In addition to proteins that transport small molecules and ions within cells and across membranes, 237 of these proteins are involved with the transport and localization of proteins. The proteins involved with protein localization included RABs, dynein and kinesin molecular motors, adaptor protein (AP) complexes, and filamins. Both axonemal and cytoplasmic forms of dynein proteins were found. The axonemal dyneins are involved in producing flagellar motion, whereas the cytoplasmic dyneins act as molecular motors, moving membrane proteins, vesicles, and other cargo in the cell [41, 42]. In all, 18 of the 23 RAB proteins in the proteome are known to have regulatory roles in trafficking of membrane-bound vesicles [43]. The most abundant dyneins in the sperm proteome were the axonemal type. All three kinesin proteins detected appear to be heavy subunits containing the dynein motor domain. Kinesin KIF9, implicated in cell shape change and podocyte formation, was the most abundant (27 unique peptides detected) kinesin protein found in cauda sperm [44]. Proteins of the AP complex 1 and the AP complex 2 were also present. AP-1 proteins are implicated in acrosome formation, whereas AP-2 proteins are utilized in formation of endocytic vesicles [45, 46]. Filamin A and B have well-established roles in the formation of actin networks that communicate with various membrane proteins [47]. Filamins, to our knowledge, have not been described in sperm previously.
Nucleotide-binding proteins were a major cluster, due in large part to the number of proteins (n = 335) that bind ATP. Also detected were 148 GTP-binding proteins, including 67 small GTPases involved in signal transduction. More than 40 of these GTP-binding proteins were RAS-related small G proteins, 20 of which were RABs. A set of eight large G proteins was detected as well. The involvement of large G proteins in sperm signaling is well documented [48]. However, fewer reports have appeared concerning the role of small G-signaling proteins in sperm. RAB3A is the primary small G protein studied in sperm and has been implicated in the induction of the acrosome reaction [49].
In addition to large and small G proteins, many other signaling proteins were identified in the cauda proteome. Ion channels found included voltage-dependent anion channels 1–3, CatSper 1–4 and the beta and gamma subunits, the IP3 receptor, and potassium channels. All these channels have been implicated in sperm signaling [50, 51]. Not detected were the TRPC channels, which also have been implicated in murine sperm function [17]. Integral membrane proteins, such as ion channels, are difficult to efficiently extract and are often underrepresented in proteomic analyses. This may be the case in the cauda sperm proteome. Other intracellular signaling molecules detected included five members of the testis-specific serine/threonine kinase family, cAMP-dependent kinase, five tyrosine kinases, and a large number of phosphatases.
As expected, the cauda sperm proteome contains a significant cluster of proteins with the associated GO terms related to spermatogenesis, the flagellum and acrosome, and fertilization. Of the proteins associated with the flagellum identifier, 58% were found in the proteome. This relatively low percentage representation of flagellar proteins may be due to poor efficiency of extracting these proteins from sperm. Of course, not all proteins associated with spermatogenesis, the acrosome, or fertilization would be expected to be part of the mature epididymal sperm.
Crossing the cauda proteome with the database of gene knockout mice phenotypes at The Jackson Laboratory produced a list of 131 proteins (4.6% of the proteome) that corresponded with a documented male reproductive phenotype when the encoding gene was knocked out. Using the known epididymal and testis transcriptomes and comparing them to this proteome, we have observed that of these genes, 36 are expressed only in the testis, 9 are specifically expressed in the epididymis, and 78 are expressed in both tissues. Seventy-nine percent of the knockout mice with a male reproductive phenotype had a documented defect in the sperm, and 65% were infertile. More than 80% of the infertile animals had a demonstrated sperm defect, suggesting that the majority of genetic mutations leading to infertile male mice cause sperm dysfunction rather than azoospermia. Only eight knockout mice had an observable defect in the epididymis, and only three of these mice were infertile, indicating that genetic causes of infertility that affect the epididymis are less common than those affecting the testis. As the number of knockout animals grows, the percentage with a reproductive phenotype will certainly increase, given the large number of genes in the sperm proteome that are not germ cell specific.
In summary, we have produced a deep proteome of mouse cauda epididymal sperm that illustrates many complete, or nearly complete, metabolic and cellular system pathways. It would appear from this proteome that the sperm is capable of a broad range of cellular processes, many of which have not been experimentally explored. This proteome should extend the ability of reproductive biologists to assemble hypothetically sound protein networks and systems for experimental validation. These proposed networks will facilitate the work of finding new pathways that can be explored for possible contraceptive interventions or diagnostics for sperm function.
Footnotes
Portions of this research were supported by grants from the National Center for Research Resources ( 5 P41 RR018522-10) and the National Institute of General Medical Sciences ( 8 P41 GM103493-10) from the National Institutes of Health (to R.D.S.) and by funding from Washington State University (to K.P.R.). The proteomics experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy (DOE) and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
These authors contributed equally to this work.
REFERENCES
- Chang MC. The meaning of sperm capacitation. A historical perspective. J Androl 1984; 5: 45 50. [DOI] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. Structural and transcriptional features of the mouse spermatid genome. J Cell Biol 1975; 65: 258 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod 2005; 73: 404 413. [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319 330. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 2011; 11: 2019 2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayampurath AM, Jaitly N, Purvine SO, Monroe ME, Auberry KJ, Adkins JN, Smith RD. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics 2008; 24: 1021 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petyuk VA, Mayampurath AM, Monroe ME, Polpitiya AD, Purvine SO, Anderson GA, Camp DG, Smith RD. DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Mol Cell Proteomics 2010; 9: 486 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W-J, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res 2004; 4: 53 62. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003; 75: 4646 4658. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44 57. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Hetherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 2008; 8: 1720 1730. [DOI] [PubMed] [Google Scholar]
- Guo X, Shen J, Xia Z, Zhang R, Zhang P, Zhao C, Xing J, Chen L, Chen W, Lin M, Huo R, Su B, et al. Proteomic analysis of proteins involved in spermiogenesis in mouse. J Proteome Res 2010; 9: 1246 1256. [DOI] [PubMed] [Google Scholar]
- Mukai C, Okuno M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod 2004; 71: 540 547. [DOI] [PubMed] [Google Scholar]
- Cao W, Aghajanian HK, Haig-Ladewig LA, Gerton GL. Sorbitol can fuel mouse sperm motility and protein tyrosine phosphorylation via sorbitol dehydrogenase. Biol Reprod 2009; 80: 124 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel MK, Marrero H, Birnbaumer L, Lemos JR, Florman HM. Trp2 regulates entry of Ca2+ into mouse sperm triggered by egg ZP3. Nat Cell Biol 2001; 3: 499 502. [DOI] [PubMed] [Google Scholar]
- Stamboulian S, Moutin MJ, Treves S, Pochon N, Grunwald D, Zorzato F, De Waard M, Ronjat M, Arnoult C. Junctate, an inositol 1,4,5-triphosphate receptor associated protein, is present in rodent sperm and binds TRPC2 and TRPC5 but not TRPC1 channels. Dev Biol 2005; 286: 326 337. [DOI] [PubMed] [Google Scholar]
- Baker MA, Reeves G, Hetherington L, Muller J, Baur I, Aitken RJ. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin Appl 2007; 1: 524 532. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Wooters J, Kopf GS, Qiu Y, Roberts KP. Analysis of the human sperm proteome. Ann N Y Acad Sci 2005; 1061: 190 202. [DOI] [PubMed] [Google Scholar]
- Nass SJ, Strauss JF. New Frontiers in Contraceptive Research: A Blueprint For Action. Washington, DC: National Academies Press; 2004. [PubMed] [Google Scholar]
- Rankin TL, Tsuruta KJ, Holland MK, Griswold MD, Orgebin-Crist MC. Isolation, immunolocalization, and sperm-association of three proteins of 18, 25, and 29 kilodaltons secreted by the mouse epididymis. Biol Reprod 1992; 46: 747 766. [DOI] [PubMed] [Google Scholar]
- Fraser LR, Quinn PJ. A glycolytic product is obligatory for initiation of the sperm acrosome reaction and whiplash motility required for fertilization in the mouse. J Reprod Fertil 1981; 61: 25 35. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Takahashi T, Iguchi N, Kitamura K, Miyagawa Y, Tsujimura A, Matsumiya K, Okuyama A, Nishimune Y. Ketone bodies could support the motility but not the acrosome reaction of mouse sperm. Int J Androl 2004; 27: 172 177. [DOI] [PubMed] [Google Scholar]
- Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O'Brien DA. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod 2010; 82: 136 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci U S A 2004; 101: 16501 16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odet F, Gabel SA, Williams J, London RE, Goldberg E, Eddy EM. Lactate dehydrogenase C (LDHC) and energy metabolism in mouse sperm. Biol Reprod 2011; 85: 556 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urner F, Sakkas D. A possible role for the pentose phosphate pathway of spermatozoa in gamete fusion in the mouse. Biol Reprod 1999; 60: 733 739. [DOI] [PubMed] [Google Scholar]
- Urner F, Sakkas D. Involvement of the pentose phosphate pathway and redox regulation in fertilization in the mouse. Mol Reprod Dev 2005; 70: 494 503. [DOI] [PubMed] [Google Scholar]
- Angulo C, Rauch MC, Droppelmann A, Reyes AM, Slebe JC, Delgado-Lopez F, Guaiquil VH, Vera JC. Concha II. Hexose transporter expression and function in mammalian spermatozoa: cellular localization and transport of hexoses and vitamin C. J Cell Biochem 1998; 71: 189 203. [PubMed] [Google Scholar]
- Gawlik V, Schmidt S, Scheepers A, Wennemuth G, Augustin R, Aumuller G, Moser M, Al-Hasani H, Kluge R, Joost HG, Schurmann A. Targeted disruption of Slc2a8 (GLUT8) reduces motility and mitochondrial potential of spermatozoa. Mol Membr Biol 2008; 25: 224 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann A, Axer H, Scheepers A, Doege H, Joost HG. The glucose transport facilitator GLUT8 is predominantly associated with the acrosomal region of mature spermatozoa. Cell Tissue Res 2002; 307: 237 242. [DOI] [PubMed] [Google Scholar]
- Evans RW, Setchell BP. Lipid changes in boar spermatozoa during epididymal maturation with some observations on the flow and composition of boar rete testis fluid. J Reprod Fertil 1979; 57: 189 196. [DOI] [PubMed] [Google Scholar]
- Haidl G, Opper C. Changes in lipids and membrane anisotropy in human spermatozoa during epididymal maturation. Hum Reprod 1997; 12: 2720 2723. [DOI] [PubMed] [Google Scholar]
- Hall JC, Hadley J, Doman T. Correlation between changes in rat sperm membrane lipids, protein, and the membrane physical state during epididymal maturation. J Androl 1991; 12: 76 87. [PubMed] [Google Scholar]
- Rejraji H, Sion B, Prensier G, Carreras M, Motta C, Frenoux JM, Vericel E, Grizard G, Vernet P, Drevet JR. Lipid remodeling of murine epididymosomes and spermatozoa during epididymal maturation. Biol Reprod 2006; 74: 1104 1113. [DOI] [PubMed] [Google Scholar]
- Bragg PW, Handel MA. Protein synthesis in mouse spermatozoa. Biol Reprod 1979; 20: 333 337. [DOI] [PubMed] [Google Scholar]
- Gur Y, Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev 2006; 20: 411 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur Y, Breitbart H. Protein synthesis in sperm: dialog between mitochondria and cytoplasm. Mol Cell Endocrinol 2008; 282: 45 55. [DOI] [PubMed] [Google Scholar]
- Zhao C, Guo XJ, Shi ZH, Wang FQ, Huang XY, Huo R, Zhu H, Wang XR, Liu JY, Zhou ZM, Sha JH. Role of translation by mitochondrial-type ribosomes during sperm capacitation: an analysis based on a proteomic approach. Proteomics 2009; 9: 1385 1399. [DOI] [PubMed] [Google Scholar]
- Nixon B, Asquith KL, Aitken RJ. The role of molecular chaperones in mouse sperm-egg interactions. Mol Cell Endocrinol 2005; 240: 1 10. [DOI] [PubMed] [Google Scholar]
- Allan VJ. Cytoplasmic dynein. Biochem Soc Trans 2011; 39: 1169 1178. [DOI] [PubMed] [Google Scholar]
- Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol Hum Reprod 2011; 17: 524 538. [DOI] [PubMed] [Google Scholar]
- Horgan CP, McCaffrey MW., GTPases Rab. and microtubule motors. Biochem Soc Trans 2011; 39: 1202 1206. [DOI] [PubMed] [Google Scholar]
- Cornfine S, Himmel M, Kopp P, El Azzouzi K, Wiesner C, Kruger M, Rudel T, Linder S. The kinesin KIF9 and reggie/flotillin proteins regulate matrix degradation by macrophage podosomes. Mol Biol Cell 2011; 22: 202 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM. Lack of acrosome formation in Hrb-deficient mice. Science 2001; 294: 1531 1533. [DOI] [PubMed] [Google Scholar]
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol 2009; 10: 583 596. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr 2011; 5: 160 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser LR, Adeoya-Osiguwa SA, Baxendale RW. First messenger regulation of capacitation via G protein-coupled mechanisms: a tale of serendipity and discovery. Mol Hum Reprod 2003; 9: 739 748. [DOI] [PubMed] [Google Scholar]
- Lopez CI, Belmonte SA, De Blas GA, Mayorga LS. Membrane-permeant Rab3A triggers acrosomal exocytosis in living human sperm. FASEB J 2007; 21: 4121 4130. [DOI] [PubMed] [Google Scholar]
- Lishko PV, Kirichok Y, Ren D, Navarro B, Chung JJ, Clapham DE. The control of male fertility by spermatozoan ion channels. Annu Rev Physiol 2012; 74: 453 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez SS. Control of hyperactivation in sperm. Hum Reprod Update 2008; 14: 647 657. [DOI] [PubMed] [Google Scholar]