Abstract
Rationale: Microbiome studies typically focus on bacteria, but fungal species are common in many body sites and can have profound effects on the host. Wide gaps exist in the understanding of the fungal microbiome (mycobiome) and its relationship to lung disease.
Objectives: To characterize the mycobiome at different respiratory tract levels in persons with and without HIV infection and in HIV-infected individuals with chronic obstructive pulmonary disease (COPD).
Methods: Oral washes (OW), induced sputa (IS), and bronchoalveolar lavages (BAL) were collected from 56 participants. We performed 18S and internal transcribed spacer sequencing and used the neutral model to identify fungal species that are likely residents of the lung. We used ubiquity–ubiquity plots, random forest, logistic regression, and metastats to compare fungal communities by HIV status and presence of COPD.
Measurements and Main Results: Mycobiomes of OW, IS, and BAL shared common organisms, but each also had distinct members. Candida was dominant in OW and IS, but BAL had 39 fungal species that were disproportionately more abundant than in the OW. Fungal communities in BAL differed significantly by HIV status and by COPD, with Pneumocystis jirovecii significantly overrepresented in both groups. Other fungal species were also identified as differing in HIV and COPD.
Conclusions: This study systematically examined the respiratory tract mycobiome in a relatively large group. By identifying Pneumocystis and other fungal species as overrepresented in the lung in HIV and in COPD, it is the first to determine alterations in fungal communities associated with lung dysfunction and/or HIV, highlighting the clinical relevance of these findings.
Clinical trial registered with www.clinicaltrials.gov (NCT00870857).
Keywords: microbiome, fungi, bronchoalveolar lavage, COPD, HIV
At a Glance Commentary
Scientific Knowledge on the Subject
Culture-independent techniques have been instrumental in defining the microbiome in health and disease. Studies of the lung microbiome are at an early stage, and most attention has focused on bacterial communities. The mycobiome, referring primarily to the fungal biota in a habitat, is also an important component of the human microbiome, but little work has been performed to determine fungal communities in health and disease. Wide gaps exist in the understanding of the mycobiome and its relationship to lung disease.
What This Study Adds to the Field
We performed culture-independent examination of oral wash, induced sputum, and bronchoalveolar lavage for fungi in a multicenter cohort of healthy control subjects and HIV-infected individuals with and without chronic obstructive pulmonary disease. Each specimen type (oral, sputum, bronchoscopy) had overlapping communities but also contained unique fungal species. Pneumocystis jirovecii was found in greater abundance in the lungs of HIV-infected individuals and in those with HIV infection and chronic obstructive pulmonary disease. This study is the largest to date to systematically investigate the fungal microbiome of the respiratory tract and to explore the association of fungal communities to lung and systemic disease.
Culture-independent techniques have been instrumental in defining the bacterial microbiome in health and disease. Fungi are important constituents of the mucosa and respiratory tract, but fewer culture-independent datasets regarding these organisms are available. Whether there are resident fungal communities in the respiratory tract of most individuals is unknown. The lung has a microbiome that shifts in health and disease (1–3). Complex microbial communities affect human physiology, shape the immune response (4), and may play an important role in lung diseases (1, 2). Studies of the lung microbiome are at an early stage, and most attention has focused on bacterial communities; however, the mycobiome—referring primarily to the fungal biota in a habitat—is also an important component of the human microbiome (5). Findings regarding the lung mycobiome have been reported for a small number of individuals after lung transplantation and in individuals with cystic fibrosis (1, 2). Whether a distinct lung mycobiome exists in immunocompetent hosts without accompanying lung disease is currently unknown. Alterations associated with immunosuppression or lung diseases have also not been explored in depth.
Pulmonary disease, particularly chronic obstructive pulmonary disease (COPD), remains a significant problem in HIV-infected individuals despite availability of antiretroviral therapy (ART). Causes of COPD in HIV are poorly understood. Although smoking is clearly important, it is not the only risk. We see pulmonary function abnormalities in never smokers and in animal models without smoke exposure, and HIV is an independent risk factor for COPD (6–10). Although there may be common pathways involved in COPD in HIV-infected and HIV-uninfected individuals, underlying triggers of these pathways may differ. It is likely, given the multiple microorganisms already implicated in COPD pathogenesis in the HIV-uninfected population, that microbial populations may be related to development of COPD in this immunocompromised population. Fungal communities in the respiratory tract of HIV-infected individuals with and without COPD have not been examined using next-generation sequencing techniques. Given the increased susceptibility to fungal infection seen in HIV, it is possible that alterations in these communities are related to pulmonary disease.
We performed a study of the fungal communities in the mouth and the lung in a cohort of healthy individuals with normal lung function and in HIV-infected individuals with and without COPD to determine normal constituents of the respiratory tract mycobiome and how it is altered in immunosuppression and in chronic lung disease. Some of the results of this study have been previously reported in the form of unpublished abstracts (11, 12).
Methods
Study Participants
Fifty-six participants were recruited at the University of Pittsburgh, the University of California San Francisco, and the University of California Los Angeles (6, 13). Subjects were outpatients not experiencing an acute change in respiratory symptoms in the prior 4 weeks and had not received antibiotics in the previous 6 months. Additional details of participant recruitment and characteristics are provided in the online supplement. All participants signed written informed consent, and the institutional review boards of the University of Pittsburgh, the University of California San Francisco, and the University of California Los Angeles approved the study.
Sample Collection
Oral wash (OW), induced sputum (IS), and bronchoalveolar lavage (BAL) samples were collected from each subject, per protocols. Additional details of the procedures are provided in the online supplement. Clinical and laboratory data were collected, including CD4 cell count within 6 months of BAL, use of ART, and history of cigarette or marijuana smoking.
Pulmonary Function Testing
Prebronchodilator and post-bronchodilator (after 400 μg albuterol) spirometry was performed by trained personnel according to American Thoracic Society guidelines (14, 15). The third National Health and Nutrition Examination Survey equations were used for spirometry reference values (14). Participants also performed single-breath determination of carbon monoxide uptake in the lungs to measure diffusing capacity of the lung for carbon monoxide (DlCO) per ATS/ERS standards (16). DlCO was adjusted for hemoglobin and carboxyhemoglobin, and reference values for DlCO used Neas and Schwartz's equations (17).
DNA Extraction
Fungal genomic DNA from OW, IS, and BAL samples, and bronchoscopic control samples were extracted using the cetyl trimethylammonium bromide method (18) without the liquid nitrogen step.
Polymerase Chain Reaction Amplification
Primer sequences and the polymerase chain reaction (PCR) protocol for amplifying the 18S and internal transcribed spacer (ITS) of the rRNA gene were adapted from Dollive and coworkers (19), using barcoded primer pair 18S_0067a_deg/NSR399, and ITS1F/ITS2. Additional details of the method for amplicon purification are provided in the online supplement. Both 18S and ITS amplicons were sequenced on the Ion PGM Sequencer (Life Technologies, Carlsbad, CA) using the 400-bp protocol. Sequencing data were deposited in Sequence Read Archive (accession number: SRP040237).
Sequence Processing
The QIIME pipeline (20) was used in demultiplexing and trimming raw sequences. Additional detail is provided in the online supplement. Because Pneumocystis was the most overrepresented fungus in HIV-infected subjects with or without COPD, we confirmed its presence in BAL samples using nested PCR as previously described (21).
Statistical Analysis
Participant characteristics were summarized using means or medians. Clinical characteristics were compared for HIV-infected and HIV-uninfected individuals using t tests, rank sum, or chi-square in R as appropriate. We defined COPD as a post-bronchodilator FEV1/FVC less than 70%, or diffusing capacity of the lung for carbon monoxide less than 60% predicted. These criteria were chosen based on the Global Initiative for Obstructive Lung Diseases definition and on our previous work demonstrating that an impaired diffusing capacity of the lung for carbon monoxide is the most common COPD phenotype in HIV (9, 10). CD4 cell count was dichotomized as above and below 500 cells per microliter. Marijuana use was defined as any self-reported use in the previous 12 months.
18S data were used for operational taxonomic unit (OTU)-type analysis. The Principle Coordinate Analysis plot using the unweighted UniFrac distance metric was applied to visually display clustering of samples using QIIME (20, 22). Ubiquity-ubiquity plots were generated in Corbata, and quantification of the differences between two biomes was performed using the “abundance-weighted” Kolmogorov-Smirnov statistic in Corbata (23).
The ITS data were used for species-level analysis. The neutral model was applied to separate OW, IS, and BAL populations, and 95% binomial confidence intervals were constructed as the selection criteria based on the Wilson method with the binom package in R (24). To identify species important in disease status prediction (HIV, COPD) and associated with specific clinical risk factors (low CD4 cell count, marijuana use), two machine-learning classifiers (random forest and logistic regression with elastic Net) were applied separately in R packages randomForest and glmnet (25, 26). Metastats, an R-based open-source software, was used to compare divergence between different populations and to pick significantly overrepresented fungi characterized by a q value below 0.05 (27). Only species identified by all three tests were kept for downstream analysis. If logistic regression failed to identify any species because of unbalanced group sizes, only species identified by both random forest and metastats were kept for downstream analysis. Additional detail is provided in the online supplement.
Results
Subjects
Fifty-six participants were recruited from three centers (Table 1). Thirty-two were HIV infected, and 24 were HIV uninfected. HIV-infected and HIV-uninfected participants were similar in age, sex, and ethnicity. Of those with HIV, 13 had a CD4 cell count lower than 500 cells per microliter, but were otherwise similar in their demographics to those with CD4 cell counts greater than 500 cells per microliter. Among the 32 HIV-infected individuals, 10 had COPD. The HIV-infected individuals with and without COPD were similar in age, sex, and ethnicity (Table 1).
Table 1.
Participant Sociodemographic, Clinical, and Lung Function Characteristics (n = 56)
| Characteristic | HIV Uninfected | HIV Infected | P Value | HIV Infected, without COPD | HIV Infected with COPD | P Value |
|---|---|---|---|---|---|---|
| n = 24 | n = 32 | n = 22 | n = 10 | |||
| Age, mean (SD) | 51.8 (9.8) | 51.8 (8.0) | NS | 49.8 (7.0) | 55.0 (9.2) | NS |
| Female, n (%) | 5 (20.8) | 4 (12.5) | NS | 4 (18.2) | 1 (10.0) | NS |
| Race, n (%) | ||||||
| White | 15 (62.5) | 19 (59.4) | NS | 12 (54.5) | 6 (60.0) | NS |
| Black | 8 (33.3) | 13 (40.6) | NS | 9 (40.9) | 4 (40.0) | NS |
| Hispanic, n (%) | 0 (0) | 1 (3.13) | NS | 1 (4.55) | 0 | NS |
| Cumulative pack-years, median (range) | 0 (0–54.3) | 5.5 (0–75.0) | NS | 5.49 (0–33.8) | 7.5 (0–75) | NS |
| Marijuana use in last year, n (%) | 5 (20.8) | 15 (46.9) | NS | 9 (40.9) | 6 (60.0) | NS |
| ART use, n (%) | NA | 29 (90.6) | — | 20 (90.1) | 9 (90.0) | NS |
| Current CD4, cells/μl, median (range) | NA | 599 (133–1,175) | — | 533 (208–1,175) | 641.5 (133–1,091) | NS |
| CD4 < 500 cells/μl, n (%) | NA | 13 (40.6) | — | 9 (40.9) | 4 (40.0) | NS |
| Current HIV RNA viral load, copies/ml, median (range) | NA | <50 (<50–88,246) | — | <50 (<50–8,770) | 102 (<50–88,246) | NS |
| Post-bronchodilator FEV1/FVC, mean (SD) | 0.77 (0.10) | 0.78 (0.10) | NS | 0.82 (0.04) | 0.70 (0.13) | 0.0115 |
| DlCO percent predicted, mean (SD) | 0.82 (0.13) | 0.71 (0.13) | 0.0026 | 0.75 (0.09) | 0.62 (0.16) | 0.0376 |
Definition of abbreviations: ART = antiretroviral therapy; COPD = chronic obstructive pulmonary disease; DlCO = diffusing capacity of the lung for carbon monoxide; NS = not significant.
Participant characteristics were summarized using means or medians, and compared using t tests, rank sum, or chi-square in R as appropriate.
Distinct Mycobiome in the Lung
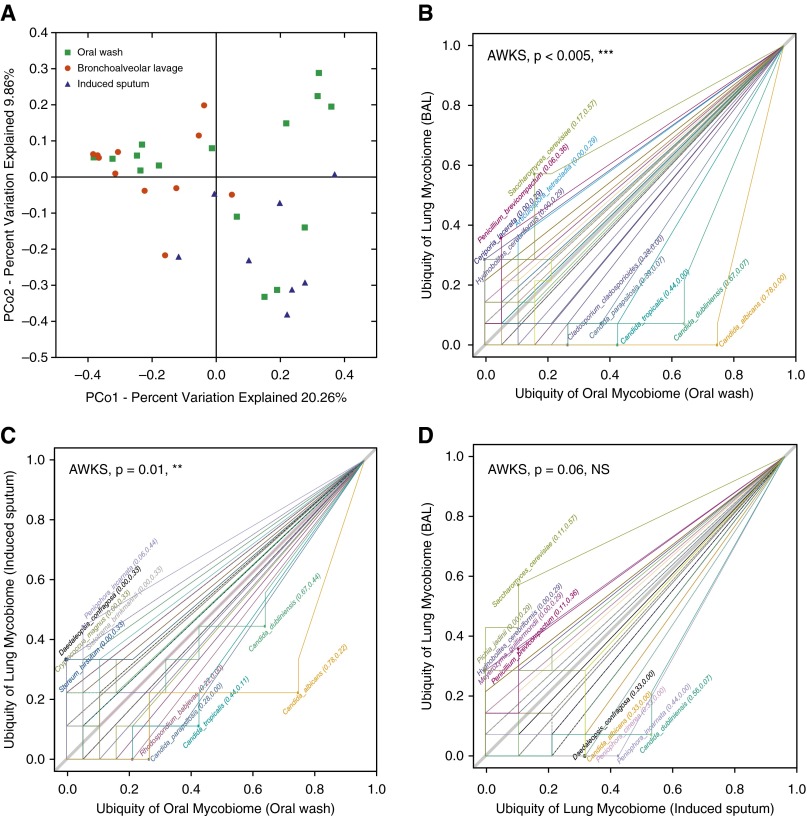
To characterize members of the fungal community in the normal lung, we compared 18S and ITS rRNA sequence data from OW, IS, and BAL from HIV-uninfected individuals with normal lung function. In Principle Coordinate Analysis of 18S data, BAL samples clustered together with OW; IS samples overlapped in part with OW samples, but not with BAL (Figure 1A). A similar distribution pattern was seen with the entire cohort including HIV-infected and HIV-uninfected individuals with or without normal lung function (see Figure E1 in the online supplement).
Figure 1.
Principal coordinate analysis plot and ubiquity–ubiquity plots (U–U plot) of the mycobiome of oral wash (OW), induced sputum (IS), and bronchoalveolar lavage (BAL) in HIV-uninfected individuals with normal lung function. (A) Principal coordinate analysis plot showing the clustering trend of OW (n = 15), IS (n = 8), and BAL (n = 11) samples (18S data). PCo = principal coordinate. (B–D) U–U plot generated using Corbata (21), comparing the ubiquity of every fungal species present in two biomes of either OW (n = 18), IS (n = 9), or BAL (n = 16) (internal transcribed spacer data). The top five taxa in every biome were labeled, and the P value showing the significance of difference between two biomes was generated in one-sided “abundance-weighted” Kolmogorov-Smirnov (AWKS) in Corbata (21). (B) U–U plot comparing the oral mycobiome (OW) with the lung mycobiome (BAL). (C) U–U plot comparing the oral mycobiome (OW) with the lung mycobiome (IS). (D) U–U plot comparing the lung mycobiome (IS) with the lung mycobiome (BAL). NS = not significant. **P ≤ 0.01; ***P ≤ 0.005.
These clustering patterns point toward shared and unique taxa in each respiratory tract level. To identify which species were likely core taxonomic members of the OW, IS, and BAL, we plotted prevalence of specific taxa comparing the ITS data of the OW with the BAL (Figure 1B) or with the IS (Figure 1C) using ubiquity–ubiquity plots (23). The genus Candida, represented by more than 90% of the ITS reads, was clearly dominant in OW compared with BAL (Figure 1B). There was also a higher prevalence of Candida in IS as compared with BAL (Figure 1D). Conversely, there were fungal species that were predominant in BAL as compared with OW (Figure 1B), including Ceriporia lacerata, Saccharomyces cerevisiae, and Penicillium brevicompactum. These organisms have been reported to cause opportunistic lung infection (28–30), demonstrating that they may be present in the lungs as commensals. None of these species were detected in the corresponding bronchoscopic control samples, indicating that environmental contamination is an unlikely explanation for their overrepresentation in BAL.
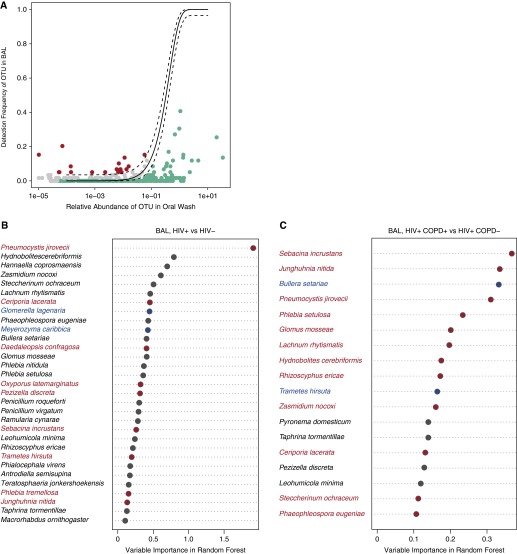
Overlap of BAL and OW communities in healthy hosts suggests that some lung microbes originate from the mouth, leading to shared communities between the two environments. To clarify which fungal species are likely to be true residents of the lung, we applied the neutral model of community ecology to compare fungal composition of OW and BAL using the ITS data in our entire cohort. This model examines whether distribution of organisms in the lung results from dispersal from the mouth or from active environmental selection in the lungs (3, 31). We identified organisms overrepresented in BAL (Figure 2A) and IS (Figure 3A) by using 95% binomial confidence intervals as the criteria. Although most OTUs were shared between oral and lung communities (falling between the 95% confidence intervals in the neutral model), 39 fungal species were disproportionately more abundant (falling outside the upper confidence interval) in the BAL and 203 species in the IS, as compared with the OW (see Table E1).
Figure 2.
Neutral model comparing the lung community (bronchoalveolar lavage [BAL]) with the oral community (oral wash) in the entire cohort and comparing lung communities in HIV infection and in chronic obstructive pulmonary disease (COPD). (A) In the neutral model plot, the solid line represents the frequency predicted in the model and dashed lines are 95% binomial confidence intervals. Although most operational taxonomic units (OTUs) were shared between the oral and lung community (found between the 95% confidence intervals in the neutral model; gray in the plots), 39 fungal species fell outside the upper confidence interval (red) and were considered to have a fitness advantage in the lung (those OTUs that were too close to the axis were omitted for plotting, but a detailed taxonomic list is provided in Table E1). The fungal species that fell below the lower confidence interval (green) did not have a fitness advantage in the lung. (B) Overrepresented species in BAL from HIV-infected and HIV-uninfected individuals. (C) Overrepresented species in BAL from HIV-infected individuals with and without COPD. The intersection of the results from three statistical methods (random forest, logistic regression, and metastats) (adjusted for multiple t tests) was colored in the plot. Only random forest and metastats results are colored for COPD species in C because logistic regression failed to identify any species. Red-labeled species are overrepresented in the disease group compared with the control group, and the blue-labeled species are found to be dominant in the control group. The ranking by importance in the group classification is based on the output from the random forest classifier (internal transcribed spacer data, n = 53 oral wash, 44 BAL).
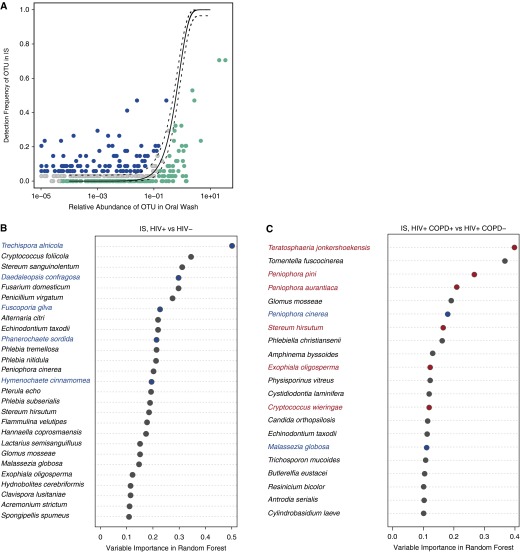
Figure 3.
Neutral model comparing the lung communities in induced sputum (IS) with the oral community (oral wash) and comparing lung communities in HIV infection and in chronic obstructive pulmonary disease (COPD) using IS. (A) In the neutral model plot, the solid line represents the frequency predicted in the model and dashed lines are 95% binomial confidence intervals. Although most operational taxonomic units (OTUs) were shared between the oral and lung community (found between the 95% confidence intervals in the neutral model; gray in the plots), 225 fungal species fell outside the upper confidence interval (blue) and were considered to have a fitness advantage in the lung (those OTUs that were too close to the axis were omitted for plotting, but a detailed taxonomic list is provided in Table E1). The fungal species that fell below the lower confidence interval (green) did not have a fitness advantage in the lung. (B) Overrepresented species in IS from HIV-infected and HIV-uninfected individuals. (C) Overrepresented species in IS from HIV-infected individuals with and without COPD. The intersection of the results from three statistical methods (random forest, logistic regression, and metastats) (adjusted for multiple t tests) was colored in the plot. Only random forest and metastats results are colored for COPD species in C because logistic regression failed to identify any species. Red-labeled species are overrepresented in the disease group compared with the control group, and the blue-labeled species are found to be dominant in the control group. The ranking by importance in the group classification is based on the output from the random forest classifier (internal transcribed spacer data, n = 53 oral wash, 30 IS).
Correlation of the Lung Mycobiome with HIV Infection, Lung Function, and Marijuana Use
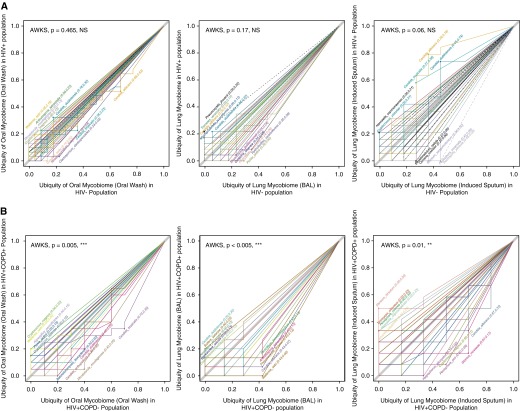
We then performed similar analyses to identify taxa representative of disease groups. First, we determined the impact of HIV infection on the mycobiome (Figure 4A) and the impact of a low CD4 cell count in HIV-infected individuals (defined as CD4 count <500 cells per microliter). Using our three statistical methods (random forest, logistic regression, and metastats), nine species were overrepresented in BAL of HIV-infected individuals as compared with HIV-uninfected individuals (Pneumocystis jirovecii, Junghuhnia nitida, Phlebia tremellosa, Oxyporus latemarginatus, Sebacina incrustans, Ceriporia lacerata, Pezizella discrete, Trametes hirsute, and Daedaleopsis confragosa) (Figure 2B). We also looked at the impact of degree of immunosuppression within the HIV-infected individuals. Two species were associated with a CD4 less than 500 cells per microliter, compared with those with CD4 cell counts greater than 500 cells per microliter: Zasmidium nocoxi and Teratosphaeria jonkershoekensis (see Figure E2A). For all comparisons, species identified were not found in bronchoscopic control samples, and we confirmed presence of P. jirovecii in all positive samples using nested PCR.
Figure 4.
Ubiquity–ubiquity plots (U–U plot) comparing the ubiquity of every fungal species present in two disease groups. The top five taxa in every biome were labeled, and the P value showing the significance of difference between two biomes was generated in one-sided “abundance-weighted” Kolmogorov-Smirnov (AWKS) in Corbata (21). (A) U–U plots comparing HIV-infected individuals with HIV-uninfected individuals in oral wash (n = 53), induced sputum (n = 30), and bronchoalveolar lavage (BAL) (n = 44) samples. (B) U–U plots comparing HIV-infected individuals with or without chronic obstructive pulmonary disease (COPD) in oral wash (n = 31), induced sputum (n = 19), and BAL (n = 26) samples (internal transcribed spacer data). NS = not significant. **P ≤ 0.01; ***P ≤ 0.005.
We then compared mycobiome composition in HIV-infected individuals with and without COPD (Figure 4B). Because cumulative pack-years smoking is a well-established risk factor for COPD and showed a nonsignificant trend to be higher in HIV-infected individuals with COPD, we excluded its direct effects on mycobiome composition by running statistical methods twice with and without the features of pack-years included, and only kept those fungal species that were selected in both models as those that were associated with COPD in HIV infection exclusively. Logistic regression failed to identify any species associated with COPD in the HIV-infected cohort as compared with HIV-infected individuals with normal lung function. Random forest and metastats overlapped in the identification of 12 species (S. incrustans, J. nitida, P. jirovecii, Phlebia setulosa, Glomus mosseae, Lachnum rhytismatis, Hydnobolites cerebriformis, Rhizoscyphus ericae, Z. nocoxi, C. lacerata, Steccherinum ochraceum, and Phaeophleospora eugeniae) (Figure 2C). As in the analyses of HIV infection, species of interest were not detected in bronchoscopic control samples, and Pneumocystis was confirmed by PCR.
Because marijuana is smoked without a filter and might contain pathogens that could enter the lung, we examined the mycobiome in those who reported smoking marijuana within the past year versus those without recent marijuana exposure. In marijuana smokers, with and without HIV infection, four species were overrepresented as compared with individuals who had not smoked marijuana in the past year: L. rhytismatis, P. discreta, Phialocephala virens, and Taphrina tormentillae (see Figure E2B). The latter two species are known plant pathogens (32, 33).
We also compared the IS communities by HIV, CD4, COPD, or marijuana smoking status. Although there were 225 fungal species found to be residents of the IS as compared with the oral community (Figure 3A), none were overrepresented in HIV-infected individuals as compared with HIV-uninfected individuals (Figure 3B). Lung function, however, did seem to be associated with six fungal species, based on the two successful statistical analyses used both with and without adjustment for pack-years (Figure 3C). Among these species, Exophiala oligosperma has been reported in immunocompromised patients with respiratory disorders (34). Low CD4 cell count (see Figure E2C) had no associated species, whereas marijuana use (see Figure E2D) had two associated species (Clavispoa lusitaniae and E. oligosperma).
Discussion
We investigated the mycobiome at various levels of the respiratory tract in healthy individuals and in those with HIV infection and with COPD. Although there was significant overlap between oral and lung fungal communities, we found distinct differences between the mouth and lung fungal populations in both healthy subjects and in the cohort overall. Candida was the most common species in the mouth with other species, such as Saccharomyces, overrepresented in the lung. We also found significant alterations in the fungal lung populations in those who were HIV infected compared with HIV uninfected, and in HIV-infected individuals with low CD4 cell counts. HIV-infected individuals with COPD had different fungal lung populations compared with HIV-infected individuals with normal lung function; those who smoked marijuana in the past year differed from those who had not reported marijuana use, regardless of HIV status.
Bacterial communities in the lung largely resemble those in the mouth, likely from microaspiration (3, 35). We also found that fungal communities in the lung resembled those in the mouth, which may also occur from microaspiration that occurs even in an immunocompetent host without accompanying lung disease. The overlap seen in this study could be caused, at least in part, by contamination during bronchoscopy when the instrument passes through the oral cavity to the bronchi, or by detection of lung organisms in the mouth that arise via coughing or exhalation. The neutral model clearly indicates that there are also organisms overrepresented in the lung compared with the mouth, and detection of organisms, such as P. jirovecii, a known lung pathogen not part of the oral flora, provides strong evidence that a mycobiome specific to the lung does indeed exist.
HIV infection has profound effects on systemic and lung immunity. Alterations in inflammation and immune function persist despite effective ART and may impact the mycobiome. In bacterial studies of BAL, certain species, such as Tropheryma whipplei, are overrepresented in HIV-infected individuals compared with HIV-uninfected control subjects (36). In our mycobiome study, two fungal species that were overrepresented in HIV were known lung pathogens associated with immunosuppression (P. jirovecii and C. lacerata) (28, 37). Pneumocystis is a leading cause of pneumonia in HIV-infected and other immunocompromised individuals (38), and low levels of the organism have previously been detected in the respiratory tract of HIV-infected individuals using targeted PCR (39). Other species that were overrepresented in the lungs in HIV infection or associated with a low CD4 cell count have been either poorly studied or reported strictly as plant pathogens.
Chronic lung diseases, such as COPD, have long been postulated to have an infectious component. COPD is an important cause of respiratory impairment in HIV-infected persons, and HIV infection is an independent risk factor for COPD (6, 13, 40). Pulmonary infections could stimulate replication of HIV in the lungs, augment pulmonary inflammation, and cause lung destruction by release of proteases, thereby leading to or worsening COPD (41). Previous studies examined the bacterial lung microbiome in HIV-uninfected individuals with advanced COPD and found profound changes in the microbiome associated with lung dysfunction (42, 43). No previous studies have examined the role of the microbiome or mycobiome in HIV-associated COPD. We postulated that the respiratory mycobiome in COPD may play a role in HIV-associated COPD given the increased susceptibility to fungal infection seen even in treated HIV.
The primary fungus enriched in the lungs of individuals with HIV and COPD was Pneumocystis. Pneumocystis detection was previously reported in both HIV-infected and HIV-uninfected individuals with COPD in studies using PCR (21, 39, 44). Pneumocystis also results in COPD-like changes in rodent and nonhuman primate models (8, 45). Pneumocystis colonization has been postulated to contribute to COPD by stimulating inflammation and release of matrix metalloproteases, which can damage the lung (8, 39). The current findings, using novel technology and analysis methods, provide additional support of the role of Pneumocystis in HIV-associated COPD.
Our findings also suggest that Pneumocystis is not the sole fungus involved in HIV-associated COPD, but that fungal communities are altered. Other fungal species overrepresented in the BAL of HIV-infected individuals with or without abnormal lung function have not yet been reported to be associated with human disease. Some of these fungi, including half of those associated with smoking marijuana, are regarded as purely plant pathogens, but hold potential as opportunistic microbes. C. lacerata, for example, now known to be an opportunist basidiomycete causing pneumonia, was once regarded solely as an agent of white rot on wood (28), and plant pathogenic fungi, such as Aspergillus flavus, can result in a life-threatening infection in animals, including humans (46).
The overrepresented fungal species in our cohort may have clinical significance that is not yet realized, but implications of these findings for clinicians are not yet established. It is possible in the future that these types of culture-independent techniques could be widely applied both in pneumonia and in chronic diseases, such as COPD, but further study of the impact of these techniques on patient care is needed. Those individuals who smoke marijuana may be at increased risk of colonization with plant pathogens, but effects on lung health are not currently known.
The use of IS has been proposed as an alternative to the more invasive BAL, but no studies have directly compared the results of IS with either BAL or OW in the mycobiome. Interestingly, we found that IS shared little similarity with BAL. Even when we performed this analysis with our entire cohort including HIV-infected and HIV-uninfected individuals with and without COPD, we observe the same distribution pattern. These data demonstrate that, although there is some overlap with both the OW and the BAL, IS has a distinct community structure. IS and BAL are collected using different methods and may reflect different features of the lower airways. Sputum induction results in a sample from a larger region of the lung as compared with BAL, which only samples a subsegment. In addition, IS may include material from larger airways than BAL and may contain a higher biomass because it is not diluted by saline. IS may also contain a higher proportion of contamination from the mouth. The question of which sample is most appropriate for use depends on the question to be answered, but the samples do not seem to be interchangeable.
We detected fewer fungal reads and fewer fungal genera in the 18S data compared with the ITS data. The eukaryotic rRNA gene consists of 18S, 5.8S, and 28S ribosomal subunits, separated by two ITSs, named ITS1 and ITS2. Both 18S and ITS can be used for sequencing of fungi (5). The 18S rRNA gene is more conserved across different fungal species and is thus used for determination of phylogenetic distance between OTUs, whereas the ITS region is more diverse across species and used for assignments at lower taxonomic levels (19, 47). We performed amplification of both genes to maximize strengths of each method. The 18S rRNA gene gave higher PCR-positive rates, but fewer fungal reads compared with the ITS, because the 18S primers cross-react with host rRNA even though they were designed with mismatches at the 3′ end (19). Consequently, approximately 90% of the raw 18S sequencing reads were human. Considering the high conservation of the 18S rRNA gene in Eukaryota, this high proportion is not surprising. Human 18S reads thus reduced the sequencing depth of fungal 18S reads.
There are several limitations to our study. We could not determine the causal role of alterations in lung fungal communities in development of HIV-associated COPD, but prior animal studies have shown that Pneumocystis colonization results in COPD-like changes in both an HIV-like nonhuman primate model and in an immunocompetent, smoke-exposed rodent model (8, 45). Differences between the populations could have affected our results, although we took steps to minimize these differences. For example, we excluded individuals with other chronic comorbidities or who were currently using antibiotics, and adjusted for smoking history. Also, the data on smoking status and marijuana use are based on patient self-report, which can have a recall bias or self-report bias. Although it would be interesting to compare results of culture techniques with the current findings, we were unable to do so. In addition, many organisms, such as Pneumocystis, have no established culture system and can only be identified through sequencing, thus we are unable to confirm viability of the organisms detected. We also were unable to compare bacterial and fungal communities in the lung because of the low number of fungi common to both HIV-infected and HIV-uninfected individuals. Finally, although we took steps to minimize contamination in sample collection and to exclude contaminants in analyses, it is possible that some of the species detected represent environmental contamination, although they did not match control samples.
This study is the first to systematically analyze the respiratory mycobiome in a large cohort and to determine changes that occur with immunosuppression and associated lung disease. Our results suggest that complex fungal communities exist at different levels of the respiratory tract and that the lung has unique fungal members. Changes in specific fungal species are seen in individuals with HIV infection, particularly in those with COPD demonstrating a potential role in pathogenesis of lung disease in immunocompromised populations.
Acknowledgments
Acknowledgment
The authors thank the following individuals for their work on this study: Sofya Tokman, Elizabeth Auld, and Alisa Malki (San Francisco, CA), Meghan Fitzpatrick, M. Patricia George, Matthew Gingo, and Robert Hoffman (Pittsburgh, PA).
Data in this article were collected by the Multicenter AIDS Cohort Study (MACS) with centers at Los Angeles (U01-AI35040): University of California Los Angeles, Schools of Public Health and Medicine: Roger Detels (primary investigator [PI]), Otoniel Martínez-Maza (co-PI), Aaron Aronow, Robert Bolan, Elizabeth Breen, Anthony Butch, Beth Jamieson, Eric N. Miller, John Oishi, Harry Vinters, Dorothy Wiley, Mallory Witt, Otto Yang, Stephen Young, and Zuo Feng Zhang; Pittsburgh (U01-AI35041): University of Pittsburgh, Graduate School of Public Health: Charles R. Rinaldo (PI), Lawrence A. Kingsley (co-PI), James T. Becker, Ross D. Cranston, Jeremy J. Martinson, John W. Mellors, Anthony J. Silvestre, and Ronald D. Stall; and the Data Coordinating Center (UM1-AI35043): The Johns Hopkins University Bloomberg School of Public Health: Lisa P. Jacobson (PI), Alvaro Munoz (co-PI), Alison Abraham, Keri Althoff, Christopher Cox, Jennifer Deal, Gypsyamber D'Souza, Priya Duggal, Janet Schollenberger, Eric C. Seaberg, Sol Su, and Pamela Surkan. The MACS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional cofunding from the National Cancer Institute (NCI). MACS data collection is also supported by UL1-TR000424 (JHU CTSA). Website located at http://www.statepi.jhsph.edu/macs/macs.html. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH).
Data in this article were collected by the Women's Interagency HIV Study (WIHS) Collaborative Study Group with a center at The Connie Wofsy Study Consortium of Northern California (PI, Ruth Greenblatt). The WIHS is funded by the National Institute of Allergy and Infectious Diseases (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (UO1-HD-32632). The study is cofunded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI grant number UL1 RR024131). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Supported by National Heart, Lung, and Blood Institute–National Institutes of Health grants U01HL098962 (E.G. and A.M.), R01 HL090339 (A.M.), and K24 HL087713 (L.H.); National Center for Advancing Translational Sciences–National Institutes of Health grant UL1TR000124 (J.C.D. and E.C.K.); and the University of Pittsburgh CTSI (UL1 RR024153). L.C. is a visiting scholar from Tsinghua University and supported in part by the China Scholarship Council.
Author Contributions: Conception and design, E.C.K., L.H., L.K., A.M., and E.G. Acquisition of data, L.C., L.L., L.T., M.B.R., A.F., C.K., D.C., N.L., S.F., S.S., and J.C.D. Analysis and interpretation of data, L.C., L.T., N.L., A.M., and E.G. Drafting or revising the article, L.C., L.T., A.M., and E.G. Final approval of the manuscript, L.C., L.L., L.T., M.B.R., A.F., C.K., D.C., L.K., N.L., R.M.G., S.F., S.S., J.C.D., E.C.K., L.H., A.M., and E.G.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201409-1583OC on January 20, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, Bushman FD, Collman RG. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One. 2012;7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, et al. Lung HIV Microbiome Project. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huttenhower C, Gevers D Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crothers K, McGinnis K, Kleerup E, Wongtrakool C, Hoo GS, Kim J, Sharafkhaneh A, Huang L, Luo Z, Thompson B, et al. HIV infection is associated with reduced pulmonary diffusing capacity. J Acquir Immune Defic Syndr. 2013;64:271–278. doi: 10.1097/QAI.0b013e3182a9215a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzpatrick M, Crothers K, Morris A. Future directions: lung aging, inflammation, and human immunodeficiency virus. Clin Chest Med. 2013;34:325–331. doi: 10.1016/j.ccm.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shipley TW, Kling HM, Morris A, Patil S, Kristoff J, Guyach SE, Murphy JE, Shao X, Sciurba FC, Rogers RM, et al. Persistent Pneumocystis colonization leads to the development of chronic obstructive pulmonary disease in a nonhuman primate model of AIDS. J Infect Dis. 2010;202:302–312. doi: 10.1086/653485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gingo MR, George MP, Kessinger CJ, Lucht L, Rissler B, Weinman R, Slivka WA, McMahon DK, Wenzel SE, Sciurba FC, et al. Pulmonary function abnormalities in HIV-infected patients during the current antiretroviral therapy era. Am J Respir Crit Care Med. 2010;182:790–796. doi: 10.1164/rccm.200912-1858OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingo MR, He J, Wittman C, Fuhrman C, Leader JK, Kessinger C, Lucht L, Slivka WA, Zhang Y, McMahon DK, et al. Contributors to diffusion impairment in HIV-infected persons. Eur Respir J. 2014;43:195–203. doi: 10.1183/09031936.00157712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui L, Fitch A, Lucht L, Saira K, Rogers M, Kessinger C, Kingsley LA, Wang J, Leo N, Morris A, et al. Characterization of the lung mycobiome in individuals with HIV and chronic obstructive pulmonary disease. Presented at the Pittsburgh International Lung Conference. October 17–18, 2013, Pittsburgh, PA. [Google Scholar]
- 12.Cui L, Rogers M, Lucht L, Fitch A, Saira K, Kessinger C, Huang L, Kleerup E, Greenblatt RM, Kingsley LA, et al. Characterization of the oral fungal microbiota in patients with HIV and chronic obstructive pulmonary disease. Presented at the Thomas L Petty Aspen Lung Conference. June 5–8, 2013, Aspen, CO. [Google Scholar]
- 13.Fitzpatrick ME, Gingo MR, Kessinger C, Lucht L, Kleerup E, Greenblatt RM, Claman D, Ponath C, Fong S, Huang L, et al. HIV infection is associated with diffusing capacity impairment in women. J Acquir Immune Defic Syndr. 2013;64:284–288. doi: 10.1097/QAI.0b013e3182a9213a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 15.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Crapo R, Enright P, van der Grinten CP, Gustafsson P, et al. ATS/ERS Task Force. Standardisation of spirometry. Eur Respir J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 16.Macintyre N, Crapo RO, Viegi G, Johnson DC, van der Grinten CP, Brusasco V, Burgos F, Casaburi R, Coates A, Enright P, et al. Standardisation of the single-breath determination of carbon monoxide uptake in the lung. Eur Respir J. 2005;26:720–735. doi: 10.1183/09031936.05.00034905. [DOI] [PubMed] [Google Scholar]
- 17.Neas LM, Schwartz J. The determinants of pulmonary diffusing capacity in a national sample of U.S. adults. Am J Respir Crit Care Med. 1996;153:656–664. doi: 10.1164/ajrccm.153.2.8564114. [DOI] [PubMed] [Google Scholar]
- 18.Zolan ME, Pukkila PJ. Inheritance of DNA methylation in Coprinus cinereus. Mol Cell Biol. 1986;6:195–200. doi: 10.1128/mcb.6.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, Nabel CS, Hill DA, Artis D, Bachman MA, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13:R60. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris A, Kingsley LA, Groner G, Lebedeva IP, Beard CB, Norris KA. Prevalence and clinical predictors of Pneumocystis colonization among HIV-infected men. AIDS. 2004;18:793–798. doi: 10.1097/00002030-200403260-00011. [DOI] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li K, Bihan M, Methé BA. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One. 2013;8:e63139. doi: 10.1371/journal.pone.0063139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorai-Raj S. Binomial confidence intervals for several parameterizations. R package 2014 version 1.1-1.
- 25.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 26.Liaw A, Wiener M. Classification and regression by randomforest. R News. 2002;2:18–22. [Google Scholar]
- 27.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLOS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chowdhary A, Agarwal K, Kathuria S, Singh PK, Roy P, Gaur SN, de Hoog GS, Meis JF. Clinical significance of filamentous basidiomycetes illustrated by isolates of the novel opportunist Ceriporia lacerata from the human respiratory tract. J Clin Microbiol. 2013;51:585–590. doi: 10.1128/JCM.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Cámara R, Pinilla I, Muñoz E, Buendía B, Steegmann JL, Fernández-Rañada JM. Penicillium brevicompactum as the cause of a necrotic lung ball in an allogeneic bone marrow transplant recipient. Bone Marrow Transplant. 1996;18:1189–1193. [PubMed] [Google Scholar]
- 30.Tawfik OW, Papasian CJ, Dixon AY, Potter LM. Saccharomyces cerevisiae pneumonia in a patient with acquired immune deficiency syndrome. J Clin Microbiol. 1989;27:1689–1691. doi: 10.1128/jcm.27.7.1689-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbell SP. Neutral theory in community ecology and the hypothesis of functional equivalence. Funct Ecol. 2005;19:166–172. [Google Scholar]
- 32.Bacigalova K, Petrydesova J, Mulenko W, Kozlowska M. A Taphrina on Potentilla erecta new in Slovakia. Biologia. 2014;69:163–167. [Google Scholar]
- 33.Swart WJ, Oelofse RM, Labuschagne MT. Susceptibility of South African cactus pear varieties to four fungi commonly associated with disease symptoms. J Prof Assoc Cactus Devel. 2003;5:86–97. [Google Scholar]
- 34.Badali H, Hedayati MT, Bahoosh M, Kasir A, Ghasemi M, Motahari J, Meis JF, De Hoog GS. Exophiala oligosperma involved in a refractory chronic rhinosinusitis. Eur Rev Med Pharmacol Sci. 2011;15:319–323. [PubMed] [Google Scholar]
- 35.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, et al. Lung HIV Microbiome Project. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am J Respir Crit Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottlieb MS, Schroff R, Schanker HM, Weisman JD, Fan PT, Wolf RA, Saxon A. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N Engl J Med. 1981;305:1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan JE, Benson C, Holmes KK, Brooks JT, Pau A, Masur H Centers for Disease Control and Prevention (CDC); National Institutes of Health; HIV Medicine Association of the Infectious Diseases Society of America. Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recomm Rep. 2009;58:1–207, quiz CE1–CE4. [PubMed] [Google Scholar]
- 39.Morris A, Alexander T, Radhi S, Lucht L, Sciurba FC, Kolls JK, Srivastava R, Steele C, Norris KA. Airway obstruction is increased in Pneumocystis-colonized human immunodeficiency virus-infected outpatients. J Clin Microbiol. 2009;47:3773–3776. doi: 10.1128/JCM.01712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crothers K, Huang L, Goulet JL, Goetz MB, Brown ST, Rodriguez-Barradas MC, Oursler KK, Rimland D, Gibert CL, Butt AA, et al. HIV infection and risk for incident pulmonary diseases in the combination antiretroviral therapy era. Am J Respir Crit Care Med. 2011;183:388–395. doi: 10.1164/rccm.201006-0836OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris A, George MP, Crothers K, Huang L, Lucht L, Kessinger C, Kleerup EC Lung HIV Study. HIV and chronic obstructive pulmonary disease: is it worse and why? Proc Am Thorac Soc. 2011;8:320–325. doi: 10.1513/pats.201006-045WR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, Cooper J, Sin DD, Mohn WW, Hogg JC. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185:1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris A, Sciurba FC, Lebedeva IP, Githaiga A, Elliott WM, Hogg JC, Huang L, Norris KA. Association of chronic obstructive pulmonary disease severity and Pneumocystis colonization. Am J Respir Crit Care Med. 2004;170:408–413. doi: 10.1164/rccm.200401-094OC. [DOI] [PubMed] [Google Scholar]
- 45.Christensen PJ, Preston AM, Ling T, Du M, Fields WB, Curtis JL, Beck JM. Pneumocystis murina infection and cigarette smoke exposure interact to cause increased organism burden, development of airspace enlargement, and pulmonary inflammation in mice. Infect Immun. 2008;76:3481–3490. doi: 10.1128/IAI.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St Leger RJ, Screen SE, Shams-Pirzadeh B. Lack of host specialization in Aspergillus flavus. Appl Environ Microbiol. 2000;66:320–324. doi: 10.1128/aem.66.1.320-324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]