Abstract
Allele-specific DNA methylation, histone acetylation and histone methylation are recognized as epigenetic characteristics of imprinted genes and imprinting centers (ICs). These epigenetic modifications are also used to regulate tissue-specific gene expression. Epigenetic differences between alleles can be significant either in the function of the IC or in the cis-acting effect of the IC on ‘target’ genes responding to it. We have now examined the epigenetic characteristics of NDN, a target gene of the chromosome 15q11–q13 Prader–Willi Syndrome IC, using sodium bisulfite sequencing to analyze DNA methylation and chromatin immunoprecipitation to analyze histone modifications. We observed a bias towards maternal allele-specific DNA hypermethylation of the promoter CpG island of NDN, independent of tissue-specific transcriptional activity. We also found that NDN lies in a domain of paternal allele-specific histone hyperacetylation that correlates with transcriptional state, and a domain of differential histone H3 lysine 4 di- and tri-methylation that persists independent of transcription. These results suggest that DNA methylation and histone H3 lysine 4 methylation are persistent markers of imprinted gene regulation while histone acetylation participates in tissue-specific activity and silencing in somatic cells.
INTRODUCTION
Epigenetic processes such as DNA methylation and histone modifications are associated with transcriptional regulation (1–3), X-inactivation (4–6) and genomic imprinting (7–10). Mechanistic links between DNA methylation and histone modification have been proposed whereby histone H3 K9 methylation can direct DNA methylation (11,12), which can in turn recruit histone deacetylases, thereby creating a closed chromatin conformation that inhibits transcription (1,13,14). DNA hypermethylation is generally associated with decreased gene activity, although the nature of the mechanistic link is controversial in endogenous autosomal genes (14–17). Histone hyperacetylation is associated with actively transcribed genes and genes poised for transcription (18). The effect of methylation of histones depends on the residue modified, as predicted by the histone code hypothesis (19). Histone H3 can be methylated on lysine 4 and lysine 9 residues to mark either active or inactive chromatin (3). Whether the same mechanisms operate in tissue-specific control and allele-specific control is less well understood.
The SNRPN gene, located in the Prader–Willi Syndrome (PWS) region on 15q11–q13, and the H19/IGF2 gene pair are among the most intensively studied imprinted genes (20). These genes contain imprinting control elements that control germline imprint resetting of genes located in cis, even over large distances (21–23). Allelic epigenetic differences found at these imprinted loci with closely associated imprinting centers (ICs) can therefore either be associated with the IC itself or be the result of a response to the IC (24–26). Current imprinting models do not address mechanisms for the extension of the epigenetic mark to target genes at a distance from their IC, nor the mechanisms for coordinate allele- and tissue-specific expression (27,28).
NDN is a small intronless gene located in the PWS region on human chromosome 15q11–q13, and is one of a set of PWS candidate genes that are expressed exclusively from the paternal allele (29–32). PWS genes are target genes of the PWS IC, located at the 5′ end of the SNRPN gene (21,32,33). We previously demonstrated developmentally dynamic patterns of maternal hypermethylation and paternal hypomethylation of the promoter CpG island in mouse Ndn, by sodium bisulfite sequencing (28). We have now characterized the allele-specific methylation of a similar promoter CpG island and a second downstream CpG island in human NDN by sodium bisulfite sequencing, and have finely mapped regions of histone acetylation and histone methylation surrounding NDN using antibody specificities previously shown to be differentially modified in imprinted regions. In contrast to SNRPN, NDN has a tissue-specific expression pattern, expressed in brain and fibroblasts among other tissues but silent in blood lymphocytes and derivative lymphoblastoid cell lines, thus allowing comparisons of DNA methylation and histone modification between tissues in which NDN is and is not expressed. We have evaluated the relative contribution of epigenetic changes associated with tissue-specific gene expression versus those associated with genomic imprinting. Our results suggest that DNA methylation and histone H3 K4 di- and trimethylation epigenetically differentiate alleles in this target gene, while histone acetylation acts in tissue-specific gene regulation.
MATERIALS AND METHODS
Tissues and cultured cell lines
Control fibroblasts from the NIGMS Human Genetic Cell repository (GM00650), PWS fibroblasts (University of Miami Brain and Tissue Bank for Developmental Disorders #1889), and AS fibroblasts (15q11–q13 deletion cell line, from Dr A. Beaudet, Baylor College of Medicine) were grown in DMEM supplemented with 10% fetal bovine serum (FBS). Control lymphoblastoid cell lines (LCLs) derived from primary blood lymphocytes (our laboratory number LCL10), PWS LCLs (GM09024B, GM09133) and AS LCLs (GM11515) were grown in RPMI supplemented with 15% FBS. Brain (cerebral cortex) samples were obtained from Dr Marc Lalande (University of Connecticut, AS sample) and the University of Miami Brain and Tissue Bank for Developmental Disorders (PWS sample). Blood was collected from PWS and AS patients with fluorescence in situ-hybridization-verified deletions and from control individuals, with informed consent.
DNA extraction
Genomic DNA was extracted from tissue culture cells by proteinase K/SDS digestion, phenol/chloroform extraction and ethanol precipitation (34). Blood was collected in sodium EDTA tubes and erythrocytes lysed in two successive washes with 4 vol of lysis buffer (1 mM EDTA, 10 mM KHCO3, 155mM NH4Cl, pH 7.4). After centrifugation, the cell pellet was washed with phosphate buffered saline (35). Genomic DNA was extracted as above for tissue culture cells.
Sodium bisulfite sequencing
Sodium bisulfite treatment of DNA samples was performed as previously described (32). For DNA methylation analysis, PCR and sequencing, 4 μl of the sodium-bisulfite-treated DNA was used in each 20 μl PCR reaction with primers designed to amplify bisulfite-converted DNA (see Supplementary Table). For the promoter CpG island, the first round PCR primers were NEC63F and NEC65R. Of the first round PCR, 1μl was used in the seminested amplification, with PCR primers NEC63F and NEC64R. Some of the reactions were done as a nested PCR reaction with the first round PCR primers NEC92F and NEC65R and second round primers NEC95F and NEC64R. For the 3′ CpG island, seminested PCR was performed as above, with first round primers NEC126F and NEC128R. The second round PCR primers were NEC127F and NEC128R.
PCR products were electrophoresed on 2% agarose gels, gel-purified using the QIAquick Gel Extraction Kit (Qiagen Inc.) and cloned into the pGEM–T vector (Promega Corp.). Plasmid DNA was prepared by alkaline lysis (36). At least 15 recombinant clones from at least 3 separate PCR reactions were sequenced using an Amersham kit with fluorescently labeled M13 forward and reverse primers, and analyzed on a LiCor automated sequencer.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitations were performed with reagents from Upstate Biotechnology [acetyl-histone H3 ChIP assay kit: #17-245, acetyl-histone H4 ChIP assay kit: #17-229, anti-dimethyl-histone H3 (Lys4): #07-030, anti-dimethy-histone H3 (Lys9): #07-212, anti-dimethyl-histone H3 (K79): #07-366, ChIP assay kit: #17-295] and Abcam [anti-histone H3 (trimethyl K4): ab8580, anti-histone H3 (trimethyl K9): ab8898]. The H3me3K9 antibody show significant cross-reactivity with H3me3K27 (37), therefore interpretations must include these modifications. The manufacturer's protocol was used with minor modifications. Crude lymphocyte preparations were made with Ficoll-Paque PLUS (Amersham Pharmacia Biotech) from 15 ml blood samples following the manufacturer's recommendations and expanded with phytohemagglutinin before being fixed as for other samples. LCL cultures and fibroblasts were fixed by the addition of formaldehyde to a final concentration of 1% for 10 min at 37°C and cells collected. Fixed chromatin was sonicated with three 10 s pulses at one quarter maximum power with a 2 mm tip on a Fisher Scientific Sonic Dismembrator 60. Samples were pre-cleared with protein A agarose beads prior to antibody addition. Mock control runs with no antibody were done and routinely gave no products from any primers used in this study. After reversal of the cross-links, DNA was extracted using commercially available binding columns (QIAquick PCR Purification Kit, Qiagen Inc.). The size of the resulting DNA fragments was determined by agarose gel electrophoresis. PCR was performed with 1 μl of template in each 20 μl reaction. SNRPN exon 1/IC primers were pair ‘A’ as previously published (24). PCRx (Invitrogen) was used as a PCR enhancer with a subset of primer pairs (see Supplementary Table 1). Quantification of ChIP experiments was done by densitometric analysis of 32P-end-labeled oligonucleotide probe hybridizations of slot-blotted PCR products detected on a Molecular Dynamics Typhoon and analyzed with ImageQuant 5.2 quantitation software. Band intensities were corrected to background then normalized to GAPDH or chromosome 16 centromeric sequence (CEN16) bands before calculating a paternal versus maternal allele ratio. ChIP experiments were done in quadruplicate for fibroblasts, while limited availability of patient blood allowed only duplicate analysis. PCR amplifications and detection were performed multiple times for each experiment. Primer and oligo probe sequences are provided as Supplementary Table 1.
RESULTS
CpG islands in the human NDN gene
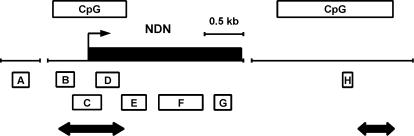
An imprinting center in the upstream region of the SNRPN gene (21) controls the imprinting of a large cluster of paternally expressed genes (38). One of these genes, NDN encoding necdin, is located ∼1.5 Mb proximal to SNRPN and is composed of a single 2.2 kb exon (29–31). To identify possible locations of cytosine methylation in NDN, we searched surrounding genomic DNA sequence (Genbank #AC006596) for CpG rich regions. One CpG island of 880 bp, containing 73 CpG sites, is located in the promoter region of NDN, extending from 250 bp upstream of the transcription start site into the open reading frame (Figure 1). A second CpG island is located ∼4.4 kb downstream of the NDN transcription start; no equivalent downstream CpG island was found in the mouse sequence for 30 kb downstream of Ndn (Genbank #AC027298). While the downstream NDN CpG island is contained within 1.2 kb, it contains an intervening sequence of 320 bp that is CpG poor.
Figure 1.
NDN and surrounding regions. Black box indicates the single exon of the NDN gene, the arrow indicates transcription start site and CpG islands are as indicated. Regions analyzed by bisulfite sequencing are indicated by double-headed arrows. Regions analyzed by ChIP are indicated by amplicons A through H. The two gaps each represent approximately 3 kb of DNA.
Patterns of DNA methylation in NDN
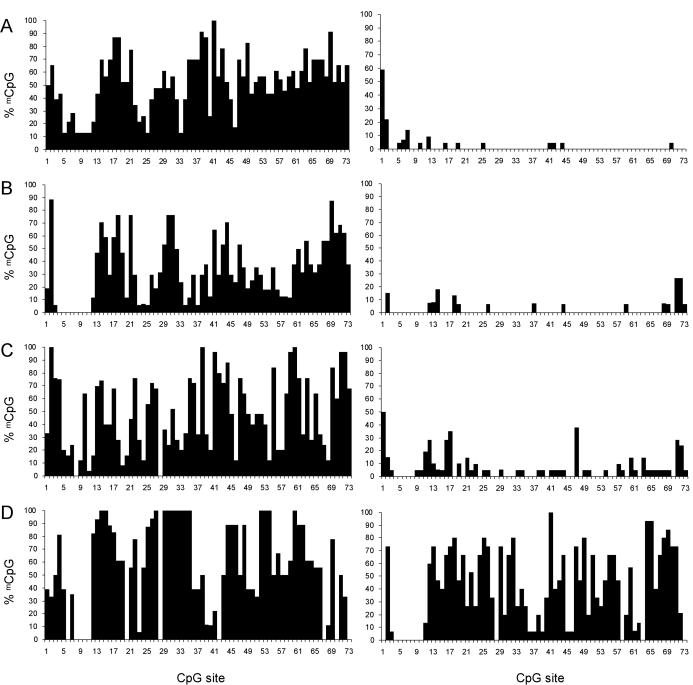
In a previous analysis of NDN promoter DNA methylation in lymphocytes from a control individual, a mixture of methylated and unmethylated clones were found that had an undetermined parent of origin (39). In order to analyze the maternal and paternal alleles in isolation, we used cell lines and tissues lacking either the maternal allele and derived from Angelman Syndrome (AS) individuals carrying maternal deletion of 15q11–q13, or lacking the paternal allele and derived from Prader–Willi Syndrome individuals carrying a paternal deletion of 15q11–q13 (32). We designed PCR primers appropriate for sodium bisulfite sequencing to analyze methylation of 73 CpG sites from the promoter CpG island and 19 CpG sites from a 350 bp segment in the 3′ end of the downstream CpG island. DNA was extracted from tissues or cells and treated with sodium bisulfite (28). Individual DNA strands produced by nested PCR of sodium bisulfite treated DNA were cloned and sequenced. Complete conversion was assessed by >99% of non-CpG sites converted to T.
To understand the correlation of DNA methylation with imprinted gene expression, we analyzed the same region in patient fibroblast cell lines and brain (cerebral cortex) samples. In these tissues in which NDN is expressed, the inactive maternal allele is hypermethylated in the promoter CpG island compared to the paternal allele (Figure 2A and B). The maternal allele in fibroblasts is less methylated than the maternal allele in brain, but in both samples the active paternal allele is almost completely unmethylated. Considerable heterogeneity in the methylation pattern among different clones was noted, and no CpG site was methylated in all maternal clones or unmethylated in all paternal clones. The heterogeneity among clones in brain may be due to a mixture of cell types in which NDN is or is not expressed.
Figure 2.
Methylation profile of 73 CpG sites in the promoter CpG island. The percentage of clones methylated at each CpG dinucleotide position was calculated and plotted against the CpG position (1 to 73). Profiles are of (A) brain, (B) fibroblasts, (C) lymphocytes and (D) LCLs. Data are from the maternal allele (left) and paternal allele (right) derived from PWS and AS deletion cells or tissues.
To verify that we could compare the epigenetic state of both alleles in both tissues in which NDN is active and those in which it is not, we first reconfirmed that NDN is not expressed in our LCLs or peripheral blood lymphocytes (29), and is expressed in fibroblasts (data not shown) by reverse transcription PCR of RNA from these cells. We then examined the DNA methylation of the NDN promoter CpG island in PWS and AS lymphocytes by sodium bisulfite sequencing. Overall, DNA hypermethylation of the maternal allele compared to the paternal allele was observed (Figure 2C), with the maternal allele methylation levels similar to that seen in fibroblasts. In PWS and AS LCLs, the levels of methylation on the maternal and paternal alleles are distinguishable, but the levels of methylation on both alleles are higher than in lymphocytes (Figure 2D). Notably, the paternal allele shows a considerable amount of methylation compared to that found in other cell types. A transition from maternal methylation to biparental methylation during culturing of T lymphocytes has also been shown for intron 7 of SNRPN (40). The observed methylation differences in NDN between cultured LCLs and lymphocytes may be a reflection of the same phenomenon of alteration of epigenetic characteristics during culture.
Histone modification of NDN
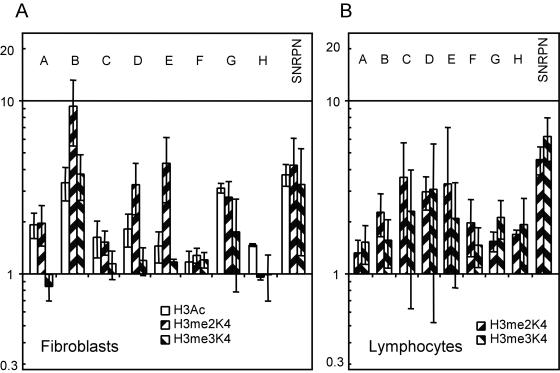
We next performed chromatin immunoprecipitation to investigate histone modifications across the NDN gene, using primer sets designed to give detailed coverage (Figure 1). Regions B to G cover NDN's transcription unit and 5′ CpG island, while regions A and H are several kilobases upstream and downstream respectively. We analyzed fibroblasts derived from PWS and AS patients. Consistent with previously identified patterns of histone H3 acetylation (H3Ac), we observed paternal bias in NDN in all regions assayed inside and outside of NDN's transcription unit (Figure 3A). A similar paternal bias in H4 acetylation was also present (data not shown). While differences in acetylation were present across NDN, consistent allelic differences were largest in region B, colocalizing with the promoter where there were greater than 4-fold differences between alleles. We then performed similar ChIP analysis with antibodies specific for di- and tri-methylated forms of lysine 4 and lysine 9 of the histone H3 tail (H3me2K4, H3me3K4, H3me2K9 and H3me3K9) and di-methylated lysine 79 of the histone H3 globular domain (H3me2K79). Consistent paternal bias in H3me2K4 was observed over regions B to E (Figure 3A). The most striking H3me2K4 difference was seen in region B with an average of greater than 7-fold paternal bias. Using trimethyl specific antibodies, a more restricted pattern of paternal bias in H3me3K4 was seen consistently over region B only. H3me3K4 showed approximately 3-fold paternal enrichment with very weak or inconsistent biases elsewhere in the gene. We did not observe any reproducible allelic biases in H3me2K9 although the H3me3K9 antibody did detect a variable and weak trend towards maternal bias (data not shown). Unambiguous analysis of this modification awaits commercial availability of more specific antibodies. No allelic differences were seen in methylation state of the H3me2K79 residue (data not shown).
Figure 3.
Histone modifications of active transcription. ChIP data from experiments using antibodies against acetylated histone H3 (H3Ac), di- and tri-methylated histone H3 at lysine 4 (H3me2K4 and H3me3K4, respectively). Paternal to maternal ratio of a representative trial plotted on logarithmic scale where 1 indicates no bias. Letters correspond to amplicons assayed as described in Figure 1. Error bars indicate variation of multiple rounds of detection. (A) PWS and AS patient fibroblasts data. (B) PWS and AS patient lymphocyte data.
We next performed similar experiments in patient blood lymphocytes to assay whether or not the H3me2K4 and H3me3K4 paternal biases were correlated with tissue type and NDN expression. It was previously reported that a region within region F is not associated with allelic histone acetylation in LCLs (41) and the paternal allele is associated with histone H3 dimethylation at K4 (H3me2K4) in region C in blood lymphocytes (10). We confirmed this lack of allelic histone acetylation in PWS and AS LCLs and lymphocytes in region F as well as paternal H3me2K4 of region C in patient blood lymphocytes. No other regions spanning regions A to G in NDN had any allelic histone acetylation in lymphocytes. H3me2K4 allelic differences were distributed over a wider region than previously reported, covering most regions analyzed, although with a weaker bias than seen in fibroblasts (Figure 3B). A trend towards paternal enrichment for H3me3K4 was also found in lymphocytes, although the degree and distribution of this bias was much more variable. Overall, these results define a domain of paternal H3me3K4 lying within a domain of paternal H3me2K4 which itself is contained within a large domain of paternal H3Ac in fibroblasts, while lymphocytes show a more general allelic bias in H3me2K4 and H3me3K4 without allelic H3Ac.
SNRPN allelic histone H3 lysine modification
To make comparisons between NDN and its imprinting center, we studied SNRPN/IC, which is expressed in fibroblasts and lymphocytes. We examined histone modification in exon 1 of SNRPN, previously described to be paternally enriched for histone H3me2K4 and maternally enriched for histone H3me2K9 in lymphocytes (10). In fibroblasts, a H3me2K4 paternal bias was also seen (Figure 3A), while we observed maternal bias in H3me2K9 in only some of our trials (data not shown). We next determined if this bias extended to the trimethylated forms, H3me3K4 and H3me3K9. H3me3K4 was found to be paternally enriched at SNRPN exon 1 at a level comparable to the enrichment seen with H3me2K4 (Figure 3A). Using antibodies specific to H3me3K9 however, did not show significant differences between alleles. In blood lymphocytes, we confirmed the paternal bias previously seen in H3me2K4, and discovered an H3me3K4 bias, as is seen in fibroblasts (Figure 3B). These observations are consistent with the fact that SNRPN/IC is expressed from the paternal allele in both fibroblasts and lymphocytes.
DISCUSSION
Our studies of histone acetylation are consistent with findings that developmentally regulated genes, such as NDN, are usually associated with domains of hyperacetylation (42,43) while changes in gene activity in response to stimuli are more frequently associated with localized changes in acetylation (44). In fibroblasts, in which NDN is actively transcribed, we identified allelic acetylation differences in a region of at least 10 kb surrounding NDN. Intriguingly, the paternal allele is hypoacetylated in the absence of DNA methylation in lymphocytes, suggesting that at this locus histone deacetylases are recruited by factors that are not dependent on DNA methylation, or that DNA methylation is lost after establishment of the hypoacetylated state. Limited studies of the murine Ndn promoter, in a region equivalent to human region D (Figure 1), indicate that neither allele is acetylated in liver, where necdin is inactive, whereas at least one allele is acetylated in brain, where necdin is expressed (43). Thus in both human and mouse, the acetylation state of NDN may act transiently in transcriptionally competent and transcriptionally active cells, and does not appear to remain as a longer lasting epigenetic imprinting mark.
In Saccharomyces cerevisiae, H3me2K4 has been associated with active regions of the genome whereas a H3me3K4 state is only seen in actively transcribed genes (2,3). In light of this association for H3me3K4, it is of great surprise that lymphocytes, not actively transcribing NDN, would carry any paternal bias in this modification. This paternal allele shows a striking resemblance to the b-globin cluster in that even in a tissue not expressing NDN, it carries more H3me2K4 and H3me3K4 as does inactive β-globin genes, in contrast to other developmentally regulated genes (45), which show little H3me2K4 and H3me3K4 modification. As those authors suggest, one possible explanation may be related to the long range function of the β-globin LCR, and at this locus, the PWS/AS imprinting center may share similar mechanisms of action. It is possible that the maintenance of the paternal state within the PWS/AS cluster requires all genes on that allele carry certain epigenetic marks regardless of tissue-specific transcriptional status.
Of greater interest are the wide region of paternal H3me2K4 and the nested region of H3me3K4 in fibroblasts. These modifications have been found to be markers of active regions and transcribed genes respectively (2,3). Unlike histone acetylation, histone methylation status has been implicated as an early event in chromatin control (46), with histone H3 methylation on residues lysine 4 and lysine 9 reciprocally marking active chromatin and heterochromatin respectively (19). As allelic H3me2K4 and H3me3K4 is also present in lymphocytes (10), we propose a model whereby histone H3 methylation at lysine 4 acts to mark allelic differential chromatin states at the NDN locus in response to the IC, and that this histone modification represents a persistent somatic mark of the active allele that allows histone acetylation to regulate expression of NDN in a tissue-specific manner (Figure 4). Interestingly, it has recently been shown that a promoter restricted distribution of H3me2K4 is a marker of monoallelic genes (47). While these authors were not able to distinguish parental alleles, we show here that a similar bias is present in NDN on the paternal allele regardless of expression. Restriction of the H3me2K4 modification to near the promoter of this single exon gene is also consistent with their observations. It remains to be seen whether H3me3K4 also display distribution patterns characteristic of imprinted or other monoallelic genes versus biallelic genes. The multiple levels of histone modification in expressing and non-expressing tissues and persistent allelic identity of this imprinted locus may indicate involvement of remodeling complexes implicated in cellular memory. For example, the human trithorax group ALL-1 complex contains HMT activity towards H3K4, histone acetyltransferase activity, as well as chromatin remodeling activity (48). It will be of great interest to study association of this or other regulatory complexes to maintenance of imprinting at the PWS/AS cluster.
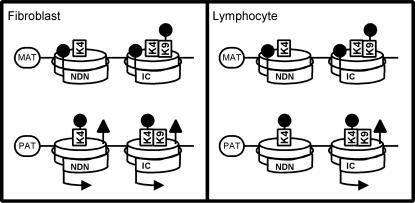
Figure 4.
Summary of epigenetic characteristics of the imprinted NDN and SNRPN/IC loci. MAT and PAT refer to alleles of maternal and paternal origin. Arrows indicate transcriptional status of NDN or SNRPN/IC (IC) in fibroblasts or lymphocytes. Lollipops on histone (cylinders) residues and spiral DNA strand indicate allelic biases in histone lysine methylation and in DNA CpG methylation respectively. Triangles indicate allelic biases in histone H3 acetylation.
DNA methylation in the imprinted target gene NDN appears to cooperate with other epigenetic alterations to regulate the final allele- and tissue-specific expression of NDN. The lack of DNA methylation on the paternal allele may signify that a different epigenetic mark, such as chromatin conformation or a specific protein–DNA interaction, serves as an imprint carried from the male germ line. For example, SNRPN and H19 have stable regions of nuclease hypersensitivity on the unmethylated paternal allele in the differentially methylated region (25,49). We observed consistent DNA hypermethylation on the maternal allele compared to the paternal allele in human lymphocytes, fibroblasts and brain. Less striking maternally biased hypermethylation was previously noted in mouse tissues, while both alleles are relatively hypermethylated in the 3′ end of the murine CpG island (28). Maternal DNA methylation may be important in maintaining allele-specific expression in tissues that express NDN/Ndn. Alternatively, the final methylation state may merely be a reflection of the competing tissue-dependent epigenetic modifications that may exclude DNA methylation. In support of the latter hypothesis, the murine Ndn promoter is relatively hypomethylated on both parental alleles in liver, in which Ndn is not expressed (28), but human NDN is differentially methylated in lymphocytes, in which NDN is inactive, and in fibroblasts in which NDN is active.
The developmental origins of somatic maternal epigenetic marks are not clear. In mouse and human oocytes, the necdin promoter is variably methylated, and at least in mouse, differences between the parental alleles are no longer present in blastocysts (28,39). Histone H3 K4 methylation on the active allele could serve as a candidate initial epigenetic mark of imprinted target genes, or could translate an initial DNA methylation imprint into a long-term mark that differentiates the two alleles. It remains to be tested whether differential histone H3 methylation exists during early embryogenesis and throughout development and if it acts at the top of a chromatin control hierarchy above allelic DNA methylation and histone acetylation or is simply correlated with these other epigenetic differences. While the mechanism of chromatin changes at target genes by the imprinting center is unknown, our data suggests that allele- and tissue-specific epigenetic changes are coordinated for proper gene expression.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge Aliyah Kanji for technical assistance, and the University of Miami Brain and Tissue Bank for Developmental Disorders (NICHD contract #N01-HD-8-3284), Dr Arthur Beaudet and Dr Marc Lalande for tissue samples. We thank the Edmonton area PWS and AS families for blood donations and the Canadian Angelman Syndrome Society for its support.
J.C.Y.L. is supported by Studentships from the AHFMR and the Natural Sciences and Engineering Research Council of Canada. M.L.H. is supported by a Studentship from the Alberta Heritage Foundation for Medical Research (AHFMR). R.W. is a Senior Scholar of the Alberta Heritage Foundation for Medical Research (AHFMR). This work was supported by operating grant MOP-14110 from the Canadian Institutes of Health Research (CIHR) to R.W.
REFERENCES
- 1.El-Osta A. and Wolffe,A.P. (2000) DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Expr., 9, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litt M.D., Simpson,M., Gaszner,M., Allis,C.D. and Felsenfeld,G. (2001) Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science, 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- 3.Noma K., Allis,C.D. and Grewal,S.I. (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science, 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 4.Beard C., Li,E. and Jaenisch,R. (1995) Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev., 9, 2325–2334. [DOI] [PubMed] [Google Scholar]
- 5.Keohane A.M., Lavender,J.S., O'Neill,L.P. and Turner,B.M. (1998) Histone acetylation and X inactivation. Dev. Genet., 22, 65–73. [DOI] [PubMed] [Google Scholar]
- 6.Boggs B.A., Cheung,P., Heard,E., Spector,D.L., Chinault,A.C. and Allis,C.D. (2001) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nature Genet., 30, 73–76. [DOI] [PubMed] [Google Scholar]
- 7.Mann J.R., Szabo,P.E., Reed,M.R. and Singer-Sam,J. (2000) Methylated DNA sequences in genomic imprinting. Crit. Rev. Eukaryot. Gene. Expr., 10, 241–257. [DOI] [PubMed] [Google Scholar]
- 8.Brannan C.I. and Bartolomei,M.S. (1999) Mechanisms of genomic imprinting. Curr. Opin. Genet. Dev., 9, 164–170. [DOI] [PubMed] [Google Scholar]
- 9.Grandjean V., O'Neill,L., Sado,T., Turner,B. and Ferguson-Smith,A. (2001) Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett., 488, 165–169. [DOI] [PubMed] [Google Scholar]
- 10.Xin Z., Allis,C.D. and Wagstaff,J. (2001) Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader–Willi syndrome imprinting center. Am. J. Hum. Genet., 69, 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamaru H. and Selker,E.U. (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature, 414, 277–283. [DOI] [PubMed] [Google Scholar]
- 12.Rice J.C. and Allis,C.D. (2001) Code of silence. Nature, 414, 258–261. [DOI] [PubMed] [Google Scholar]
- 13.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 14.Jones P.A. and Takai,D. (2001) The role of DNA methylation in mammalian epigenetics. Science, 293, 1068–1070. [DOI] [PubMed] [Google Scholar]
- 15.Reik W., Dean,W. and Walter,J. (2001) Epigenetic reprogramming in mammalian development. Science, 293, 1089–1093. [DOI] [PubMed] [Google Scholar]
- 16.Costello J.F. and Vertino,P.M. (2002) Methylation matters: a new spin on maspin. Nature Genet., 31, 123–124. [DOI] [PubMed] [Google Scholar]
- 17.Futscher B.W., Oshiro,M.M., Wozniak,R.J., Holtan,N., Hanigan,C.L., Duan,H. and Domann,F.E. (2002) Role for DNA methylation in the control of cell type specific maspin expression. Nature Genet., 31, 175–179. [DOI] [PubMed] [Google Scholar]
- 18.Razin A. (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson-Smith A.C. and Surani,M.A. (2001) Imprinting and the epigenetic asymmetry between parental genomes. Science, 293, 1086–1089. [DOI] [PubMed] [Google Scholar]
- 21.Dittrich B., Buiting,K., Korn,B., Rickard,S., Buxton,J., Saitoh,S., Nicholls,R.D., Poustka,A., Winterpacht,A., Zabel,B. et al. (1996) Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nature Genet., 14, 163–170. [DOI] [PubMed] [Google Scholar]
- 22.Leighton P.A., Ingram,R.S., Eggenschwiler,J., Efstratiadis,A. and Tilghman,S.M. (1995) Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature, 375, 34–39. [DOI] [PubMed] [Google Scholar]
- 23.Horsthemke B. (1997) Structure and function of the human chromosome 15 imprinting center. J. Cell. Physiol., 173, 237–241. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh S. and Wada,T. (2000) Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader–Willi syndrome. Am. J. Hum. Genet., 66, 1958–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweizer J., Zynger,D. and Francke,U. (1999) In vivo nuclease hypersensitivity studies reveal multiple sites of parental origin-dependent differential chromatin conformation in the 150 kb SNRPN transcription unit. Hum. Mol. Genet., 8, 555–566. [DOI] [PubMed] [Google Scholar]
- 26.Pedone P.V., Pikaart,M.J., Cerrato,F., Vernucci,M., Ungaro,P., Bruni,C.B. and Riccio,A. (1999) Role of histone acetylation and DNA methylation in the maintenance of the imprinted expression of the H19 and Igf2 genes. FEBS Lett., 458, 45–50. [DOI] [PubMed] [Google Scholar]
- 27.Hu J.F., Oruganti,H., Vu,T.H. and Hoffman,A.R. (1998) The role of histone acetylation in the allelic expression of the imprinted human insulin-like growth factor II gene. Biochem. Biophys. Res. Commun., 251, 403–408. [DOI] [PubMed] [Google Scholar]
- 28.Hanel M.L. and Wevrick,R. (2001) Establishment and maintenance of DNA methylation patterns in mouse Ndn: implications for maintenance of imprinting in target genes of the imprinting center. Mol. Cell. Biol., 21, 2384–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald H.R. and Wevrick,R. (1997) The necdin gene is deleted in Prader–Willi syndrome and is imprinted in human and mouse. Hum. Mol. Genet., 6, 1873–1878. [DOI] [PubMed] [Google Scholar]
- 30.Jay P., Rougeulle,C., Massacrier,A., Moncla,A., Mattei,M.G., Malzac,P., Roeckel,N., Taviaux,S., Lefranc,J.L., Cau,P. et al. (1997) The human necdin gene, NDN, is maternally imprinted and located in the Prader–Willi syndrome chromosomal region. Nature Genet., 17, 357–361. [DOI] [PubMed] [Google Scholar]
- 31.Sutcliffe J.S., Han,M., Christian,S.L. and Ledbetter,D.H. (1997) Neuronally-expressed necdin gene: an imprinted candidate gene in Prader–Willi syndrome. Lancet, 350, 1520–1521. [DOI] [PubMed] [Google Scholar]
- 32.Hanel M.L. and Wevrick,R. (2001) The role of genomic imprinting in human developmental disorders: lessons from Prader–Willi syndrome. Clin. Genet., 59, 156–164. [DOI] [PubMed] [Google Scholar]
- 33.Shemer R., Hershko,A.Y., Perk,J., Mostoslavsky,R., Tsuberi,B., Cedar,H., Buiting,K. and Razin,A. (2000) The imprinting box of the Prader–Willi/Angelman syndrome domain. Nature Genet., 26, 440–443. [DOI] [PubMed] [Google Scholar]
- 34.Ausubel F.M., Brent,R., Kingston,R.E., Seidman,J.G., Smith,J.A. and Struhl,K. (1993) Current Protocols in Molecular Biology. John Wiley and Sons, New York, Vol. 1. [Google Scholar]
- 35.Wevrick R. and Francke,U. (1996) Diagnostic test for the Prader–Willi syndrome by SNRPN expression in blood. Lancet, 348, 1068–1069. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbour Laboratory Press. [Google Scholar]
- 37.Perez-Burgos L., Peters,A.H., Opravil,S., Kauer,M., Mechtler,K. and Jenuwein,T. (2004) Generation and characterization of methyl-lysine histone antibodies. Meth. Enzymol, 376, 234–254. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls R.D., Saitoh,S. and Horsthemke,B. (1998) Imprinting in Prader–Willi and Angelman syndromes. Trends Genet., 14, 194–200. [DOI] [PubMed] [Google Scholar]
- 39.El-Maarri O., Buiting,K., Peery,E.G., Kroisel,P.M., Balaban,B., Wagner,K., Urman,B., Heyd,J., Lich,C., Brannan,C.I. et al. (2001) Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nature Genet., 27, 341–344. [DOI] [PubMed] [Google Scholar]
- 40.Balmer D. and LaSalle,J.M. (2001) Clonal maintenance of imprinted expression of SNRPN and IPW in normal lymphocytes: correlation with allele-specific methylation of SNRPN intron 1 but not intron 7. Hum. Genet., 108, 116–122. [DOI] [PubMed] [Google Scholar]
- 41.Fulmer-Smentek S.B. and Francke,U. (2001) Association of acetylated histones with paternally expressed genes in the Prader–Willi deletion region. Hum. Mol. Genet., 10, 645–652. [DOI] [PubMed] [Google Scholar]
- 42.Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forsberg E.C., Downs,K.M., Christensen,H.M., Im,H., Nuzzi,P.A. and Bresnick,E.H. (2000) Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA, 97, 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parekh B.S. and Maniatis,T. (1999) Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell, 3, 125–129. [DOI] [PubMed] [Google Scholar]
- 45.Schneider R., Bannister,A.J., Myers,F.A., Thorne,A.W., Crane-Robinson,C. and Kouzarides,T. (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell. Biol., 6, 73–77. [DOI] [PubMed] [Google Scholar]
- 46.Rice J.C. and Allis,C.D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol., 13, 263–273. [DOI] [PubMed] [Google Scholar]
- 47.Rougeulle C., Navarro,P. and Avner,P. (2003) Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum. Mol. Genet., 12, 3343–3348. [DOI] [PubMed] [Google Scholar]
- 48.Nakamura T., Mori,T., Tada,S., Krajewski,W., Rozovskaia,T., Wassell,R., Dubois,G., Mazo,A., Croce,C.M. and Canaani,E. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell, 10, 1119–1128. [DOI] [PubMed] [Google Scholar]
- 49.Khosla S., Aitchison,A., Gregory,R., Allen,N.D. and Feil,R. (1999) Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19 gene. Mol. Cell. Biol., 19, 2556–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.