Abstract
The high rates of recurrence and low median survival in many B-cell cancers highlight a need for new targeted therapeutic modalities. In dividing cells, eukaryotic translation initiation factor 5A (eIF5A) is hypusinated and involved in regulation of protein synthesis and proliferation, whereas the non-hypusinated form of eIF5A is a potent inducer of cell death in malignant cells. Here, we demonstrate the potential of modulating eIF5A expression as a novel approach to treating B-cell cancers. SNS01-T is a nonviral polyethylenimine-based nanoparticle, designed to induce apoptosis selectively in B-cell cancers by small interfering RNA–mediated suppression of hypusinated eIF5A and plasmid-based overexpression of a non-hypusinable eIF5A mutant. In this study, we show that SNS01-T is preferentially taken up by malignant B cells, inhibits tumor growth in multiple animal models of B-cell cancers without damaging normal tissues, and synergizes with the current therapies bortezomib and lenalidomide to inhibit tumor progression. The results collectively demonstrate the potential of SNS01-T as a novel therapeutic for treatment of a diverse range of B-cell malignancies.
Introduction
B-cell malignancies represent a significant proportion of lymphoid neoplasias diagnosed every year in North America. Neoplasms such as multiple myeloma (MM) and mantle cell lymphoma (MCL) are aggressive, incurable, and frequently relapse, contributing to short median survival.1,2 Even in diffuse large B-cell lymphoma (DLBCL), where the majority of patients respond to conventional treatments, a significant proportion of patients relapse, requiring stem cell transplants or secondary treatments to which some remain refractory.3 The current poor overall survival and the difficulty in achieving long-lasting remissions with conventional approaches highlight the urgency to develop novel therapeutic treatments to target B-cell cancers.
Initially identified as a translation initiation factor, eukaryotic translational initiation factor 5A (eIF5A) is now known to be involved in many cellular functions, including messenger RNA (mRNA) shuttling, stress granule formation, proliferation, and apoptosis.1,4,5,6,7,8,9,10,11,12,13,14,15 EIF5A is the only known protein to be modified by conversion of a lysine residue to the atypical, naturally occurring amino acid, hypusine. In dividing cells, most eIF5A is hypusinated and involved in protein synthesis and proliferation.4,5,16 Overexpression of the hypusinated form of eIF5A and the enzyme required for hypusine formation have been identified as markers of neoplastic growth.17,18 Conversely, overexpression of eIF5A mutants that cannot be hypusinated, including eIF5AK50A and eIF5AK50R, induces mitochondrial-dependent apoptosis9 in a number of cancer cell lines through activation of mitogen-activated protein kinase signaling pathways19 and p53.19,20 Numerous studies have demonstrated that the non-hypusinated form of eIF5A can induce apoptotic cell death in malignant cells, including MM cells.9,10,11,12,20,21 Small interfering RNAs (siRNAs) targeting eIF5A are potent anti-inflammatory agents,13,22 and siRNA-mediated suppression of eIF5A has been shown to reduce activation of nuclear factor-κB, an important regulator of survival in MM, and enhance apoptosis induced by eIF5AK50R overexpression in MM cells.11 As a regulator of proliferation and apoptosis, eIF5A stands out as an attractive molecular target for cancer therapies, as inhibiting expression of the hypusine-modified form might allow for induction of cell death by non-hypusinable forms of the protein.
SNS01-T, a nonviral polyethylenimine (PEI)-based nanoparticle comprised of both an RNAi-resistant DNA plasmid expressing non-hypusinable eIF5AK50R under the control of a B-cell–specific promoter/enhancer (pExp5A) and an eIF5A siRNA to reduce expression of endogenous hypusinated eIF5A, was designed to test the potential of targeting eIF5A in the treatment of B-cell malignancies. Here, we demonstrate that SNS01-T efficiently transfects, and is active, in a wide variety of B-cell tumor cells. As well, SNS01-T has a low level of toxicity at efficacious doses in healthy animals, and it is effective in inhibiting cancer growth in xenograft models of MM, MCL, and DLBCL, both as monotherapy and in combination with standard-of-care drugs such as bortezomib and lenalidomide. Together, these data demonstrate the relevance of eIF5A as a therapeutic target and the efficacy of SNS01-T as a novel approach to the treatment of B-cell cancers.
Results
Physical characterization of SNS01-T
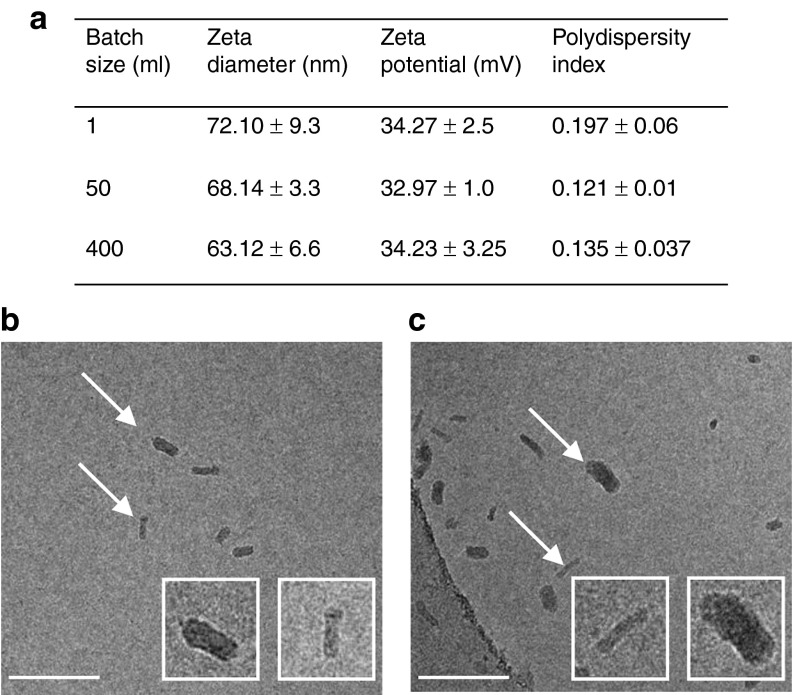
SNS01-T contains two active components: the pExp5A plasmid driven by the B-cell–specific B29 promoter and expressing eIF5AK50R, a mutant of eIF5A that is unable to undergo posttranslational modification of lysine 50 to hypusine and a siRNA that targets the untranslated region of the human eIF5A mRNA.11 SNS01-T contains 0.075 mg of nucleic acid/ml, is buffered in a 5 mmol/l Tris-HCl pH 7.4, 5% glucose solution, and has a polymer nitrogen/nucleic acid phosphorus (N/P) ratio of 6. SNS01-T nanoparticles are small, predominantly rod-shaped (Figure 1b,c), and relatively uniform in size (zeta diameter: 72 nm; polydispersity index: 0.197) (Figure 1), as determined by dynamic light scattering analysis. This is critical because surface charge and nanoparticle size and shape can have a significant impact on cellular internalization and particle stability.23,24 SNS01-T nanoparticles retain these characteristics when formulated in large batches of up to 400 ml (Figure 1).
Figure 1.
Chemical and physical properties of SNS01-T nanoparticles. (a) Comparison of different batch sizes of SNS01-T by dynamic light scattering. Values indicate the mean ± SD. (b,c) Two batches of SNS01-T were examined by electron microscopy. Representative images from (b) batch 1 and (c) batch 2 are shown at a magnification of ×52,000 (Bar = 200 nm). Insets show individual particles (white arrows) at a larger scale.
SNS01-T uptake and biological activity in B-cell tumor cells
To examine cellular uptake of SNS01-T, fluorescently-labeled nanoparticles (SNS01-T-fluor) having similar physical characteristics as that of SNS01-T (Supplementary Table S1) were transfected into MM, MCL, and DLBCL cell lines (Figure 2a). A large proportion of MM cells were transfected by nanoparticles containing both plasmid and siRNA, whereas MCL and DLBCL cell lines had increased uptake of nanoparticles containing only plasmid. Confocal analysis of Roswell Park Memorial Institute (RPMI) 8226 MM cells further confirmed that individual cells can take up the plasmid and siRNA simultaneously (Figure 2b). Conversely, as seen in Figure 2c, SNS01-T transfected naive B cells (cord blood CD19+ B cells) much less efficiently than RPMI 8226 MM cells (11 versus 53%, adjusted to PEI-negative controls), and cell death in response to SNS01-T was only observed in the MM cells (Figure 2d), likely due to differences in metabolism and proliferative capacity between naive B cells and cancer cells.25
Figure 2.
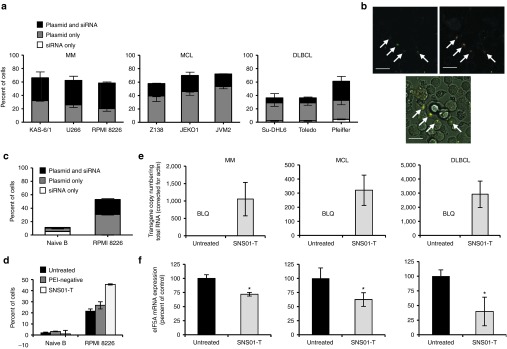
SNS01-T uptake into B-cell tumor cells. (a) Uptake of fluorescently-labeled SNS01-T into multiple myeloma (MM; n = 3), mantle cell lymphoma (MCL; n = 2), and diffuse large B-cell lymphoma (DLBCL; n = 3) cells was measured by flow cytometry. Data represent the means ± SD (b) Confocal images of nanoparticles in RPMI 8226 cells that contain both the plasmid (green) and small interfering RNA (siRNA; orange) (white arrows; Bar = 20 μm). (c) Mean uptake of fluorescently-labeled SNS01-T into naive B cells and RPMI 8226 cells ± SD (n = 2) is shown and is representative of two trials. (d) Mean cell death of naive B cells and RPMI 8226 cells that were treated with controls or SNS01-T ± SD (n = 2) is shown. (e,f) Biological activity of (e) transgene and (f) siRNA components of SNS01-T in MM (KAS-6/1), MCL (JVM2), and DLBCL (SuDHL-6) cells was assessed by quantitative real time PCR. The means ± SEM are plotted; asterisks indicate P values < 0.05 (Student's t-test; n = 3). BLQ, below limit of quantification; RPMI, Roswell Park Memorial Institute.
We further confirmed that the nucleic acid components of SNS01-T were biologically active using reverse transcriptase quantitative polymerase chain reaction (qPCR) to quantify expression of eIF5AK50R mRNA from the pExp5A plasmid and knockdown of endogenous eIF5A mRNA resulting from siRNA activity (Figure 2e,f). Transgene expression and knockdown of endogenous eIF5A were detected in all MM, MCL, and DLBCL lines tested. The effects of SNS01-T and control nanoparticles containing a nonexpressing control plasmid and a nontargeting control siRNA were compared to ensure that PEI does not affect expression (Supplementary Figure S1a). Transfection of hepatocellular carcinoma cells26 with SNS01-T demonstrated that the B-cell–specific plasmid was expressed at extremely low levels compared with a nanoparticle containing a plasmid expressing the transgene under the control of the human EF1 promoter (Supplementary Figure S1b). Finally, SNS01-T was cytotoxic to a variety of MM, MCL, and DLBCL cell lines, having an inhibitory concentration (IC50)range of 3.12–12.57 ng/μl for all cell lines tested (Supplementary Table S2). Taken together, these results indicate that SNS01-T transfects malignant, but not naive, B cells causing expression of the eIF5AK50R transgene and suppression of endogenous eIF5A expression, which results in death of the B-cell tumor cells.
SNS01-T inhibits tumor progression in xenograft models of MM, MCL, and DLBCL
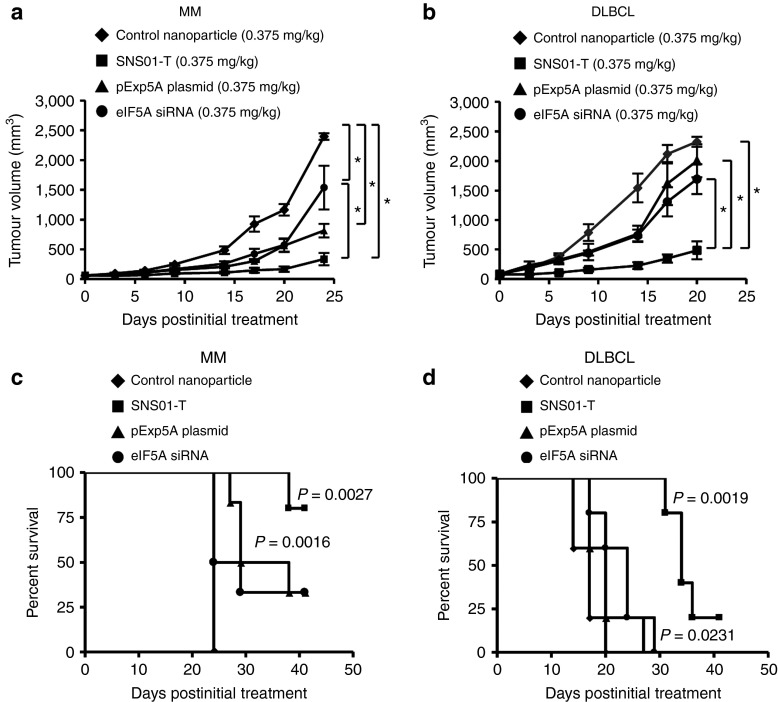
Because SNS01-T exhibited potent cell-killing effects in cell culture experiments, we evaluated its antitumoral activity in animal models. Subcutaneous xenograft models were used to assess the effectiveness of SNS01-T as an anticancer therapeutic in vivo. The combination of an eIF5AK50R-expressing plasmid and an eIF5A siRNA has been shown to effectively inhibit tumor growth in a KAS-6/1 MM xenograft model.11 In this study, twice-weekly intravenous administration of either the pExp5A plasmid or the eIF5A siRNA alone slowed the growth of RPMI 8226 MM tumors compared with control nanoparticles (one-way analysis of variance, Bonferroni posttest, P ≤ 0.05), whereas SNS01-T inhibited tumor growth more effectively than the siRNA alone (one-way analysis of variance, Bonferroni posttest, P ≤ 0.05) (Figure 3a). Administration of the single agents or SNS01-T also increased duration of survival (Figure 3c). Similar results were obtained in the Su-DHL6 model of DLBCL (Figure 3b,d), indicating that both the plasmid and siRNA components contribute to the activity of SNS01-T in vivo. SNS01-T administration inhibited tumor growth in MCL (JVM2) and DLBCL (Su-DHL6) tumor models in a dose-dependent manner (Figure 4a,b). Of note, treatment with SNS01-T at doses ≥ 0.18 mg/kg significantly extended the life span of mice compared with the control-treated mice (Figure 4c,d). Importantly, SNS01-T treatment resulted in decreased levels of hypusinated eIF5A and total eIF5A in tumor tissues, consistent with drug activity (Figure 4e). Collectively, these data demonstrate that SNS01-T strongly inhibits tumor progression in a number of B-cell malignancies.
Figure 3.
The plasmid and small interfering RNA (siRNA) components of SNS01-T both contribute to its antitumoral activity in multiple myeloma (MM) and diffuse large B-cell lymphoma (DLBCL) xenografts. SCID mice were injected subcutaneously with (a) RPMI 8226 or (b) Su-DHL6 cells, and treatments were initiated once tumors became palpable. (a,b) Tumor growth for mice receiving intravenous injections of polyethylenimine nanoparticles twice per week at a dose of 0.375 mg/kg. Nanoparticles contained either: a nonexpressing control plasmid and nontargeting control siRNA (control nanoparticle); the pExp5A plasmid and control siRNA (pExp5A plasmid); control vector and eukaryotic translation initiation factor 5A (eIF5A) siRNA (eIF5A siRNA); or pExp5A and eIF5A siRNA (SNS01-T). Error bars represent the SEM; asterisks indicate statistically significant differences (one-way analysis of variance, Bonferroni posttest; n = 5 per group, P value ≤ 0.05). (c,d) Survival data for the mice treated in a and b. Significant P values, calculated by nonparametric log-rank test comparing control nanoparticle and treatment groups, are indicated. (d) SNS01-T treatment also enhanced survival relative to pExp5A or eIF5A siRNA alone (log-rank test; n = 5 per group, P = 0.0019, 0.0021, respectively) RPMI, Roswell Park Memorial Institute.
Figure 4.
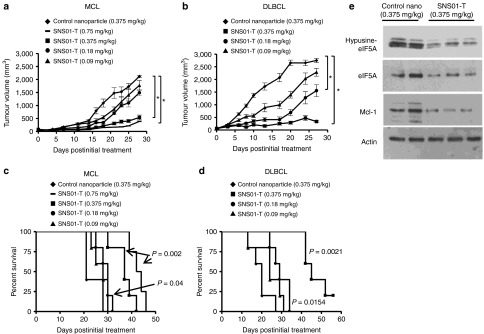
SNS01-T inhibits tumor growth in a dose-dependent manner in B-cell xenograft models. (a,b) Tumor response to different doses of SNS01-T in JVM2 (mantle cell lymphoma (MCL)) and Su-DHL6 (diffuse large B-cell lymphoma (DLBCL)) tumors, respectively. In a and b, error bars represent the SEM; asterisks indicate P values ≤ 0.05 (one-way analysis of variance, Dunnett's posttest; n = 5). (c,d) Kaplan–Meier survival curves corresponding to the experiments outlined in a and b. Significant P values, calculated by nonparametric log-rank test comparing control nanoparticle and treatment groups, are indicated (n = 5). (e) Western blot analysis of individual Su-DHL6 tumor samples 48 hours after control nanoparticle or SNS01-T treatment (0.375 mg/kg).
SNS01-T synergizes with standard-of-care drugs to induce apoptosis and inhibit tumor progression
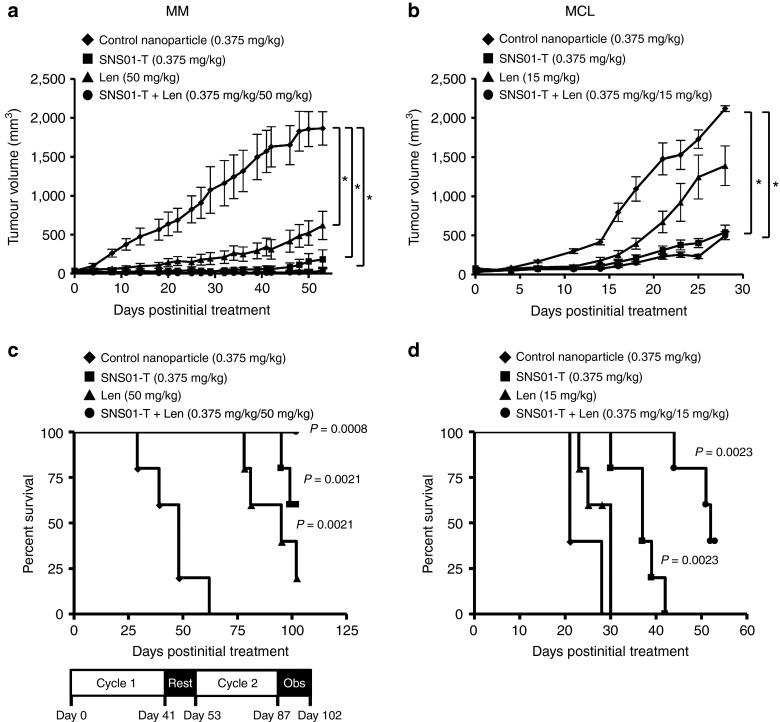
SNS01-T was also effective in combination with standard-of-care drugs for MM and MCL. SNS01-T enhanced efficacy of the immunomodulatory drug, lenalidomide, in xenograft models of MM and MCL (Figure 5). The combination treatment with SNS01-T and lenalidomide did not reduce tumor growth beyond that of single drug treatments (Figure 5a,b); however, it did significantly enhance survival compared with either drug alone (Figure 5c,d). This drug combination was particularly effective in the MM model. Responses after one round of treatment were so robust that after an 11-day rest period, another round of dosing was administered (Figure 5a,c). One hundred percent of the mice treated with SNS01-T and lenalidomide survived the 102-day study and 67% remained tumor free.
Figure 5.
Combinatorial treatment with SNS01-T and lenalidomide (Len) is more effective than either drug alone. (a) RPMI 8226 or (b) JVM2 xenografts were treated with control nanoparticles, SNS01-T, Len, or a combination of SNS01-T and Len at the indicated dosages. Means ± SEM are shown; asterisks indicate statistically significant differences (one-way analysis of variance, Bonferroni posttest; n = 5 per group, P value < 0.05). (c,d) Kaplan–Meier survival curves for the data obtained in a and b, respectively. Significant P values, calculated by nonparametric log-rank test comparing control nanoparticle and treatment groups, are indicated (n = 5 per group). (c) Survival was obtained for mice given two cycles of treatment with an 11-day rest period between cycles and a 2-week observation window (Obs) after the second cycle was complete. Here, the combination of SNS01-T and Len enhanced survival relative to Len treatment alone (log-rank test; n = 5 per group, P = 0.0068). (d) The SNS01-T/Len combination treatment enhanced survival relative to either Len or SNS01-T alone (log-rank test; n = 5 per group, P = 0.0066 and P = 0.0018, respectively) RPMI, Roswell Park Memorial Institute.
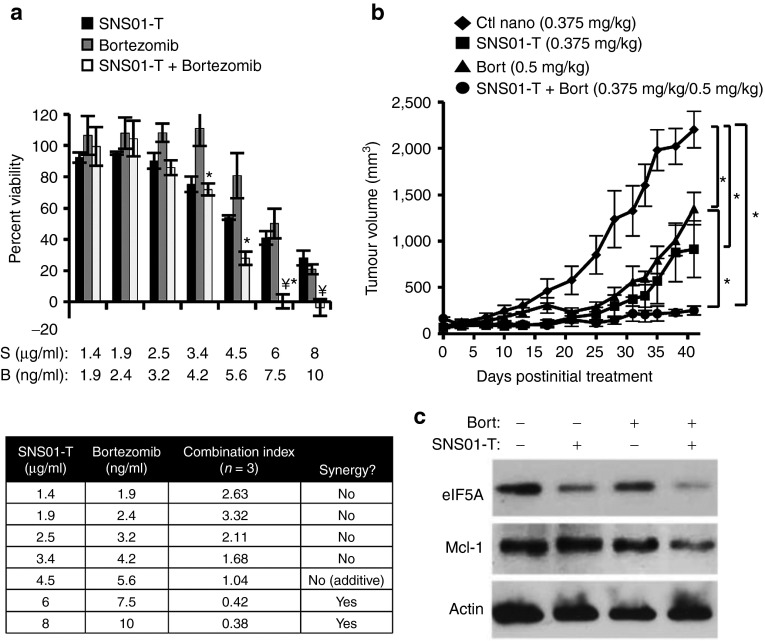
SNS01-T and the proteasomal inhibitor, bortezomib, synergistically induced cytotoxicity in RPMI 8226 and KAS-6/1 MM cells (Figure 6a and Supplementary Figure S2a). Even more striking was the response of the two drugs together in an RPMI 8226 xenograft model. While treatment with either SNS01-T or 0.5 mg/kg bortezomib alone slowed tumor growth relative to the control nanoparticle-treated mice (59 and 39% respectively), a combination of SNS01-T (0.375 mg/kg) and bortezomib (0.5 mg/kg) delayed tumor growth by 89%, a statistically significant response compared with bortezomib alone (Figure 6b) (one-way ANOVA, Bonferroni posttest, P < 0.05). Western blot analysis of RMPI 8226 showed that the combination of SNS01-T and bortezomib led to decreased expression of MCL-1 (Figure 6c), a prosurvival member of the BCL2 family that is associated with disease progression and drug resistance in MM and MCL.27,28,29 Similar results were seen in KAS-6/1 cells (Supplementary Figure S2b). In that case, bortezomib treatment stimulated MCL-1 expression and concurrent use of SNS01-T prevented the overexpression of the prosurvival MCL-1. In accordance, there was a significant increase in apoptosis when KAS-6/1 cells were treated with both drugs compared with either drug alone (Supplementary Figure S2c and Supplementary Materials and Methods). Together, these data provide a rationale for clinical investigations of SNS01-T in combination with current therapies against B-cell cancers.
Figure 6.
SNS01-T synergizes with bortezomib (Bort) to induce cytotoxicity and inhibit multiple myeloma (MM) tumor growth. (a) RPMI 8226 cells were treated with increasing doses of SNS01-T and Bort at a constant ratio of 1:1.25 for 48 hours. Cytotoxicity was measured by XTT, and synergy was determined using CompuSyn software. Mean ± SEM is shown (n = 3); an asterisk indicates P values ≤ 0.05 (two-way analysis of variance (ANOVA), Bonferroni posttest) compared with Bort, and a yen symbol indicates P values ≤ 0.05 compared with SNS01-T. S, SNS01-T; B, Bort. (b) Effect of SNS01-T, Bort, or both on the growth of RPMI 8226 xenografts. Error bars represent the SEM; asterisks indicate P values < 0.05 (one-way ANOVA, Bonferroni posttest; n = 5). Ctl nano, control nanoparticle. (c) Western blot analysis of RPMI 8226 cells after single or combination treatments of SNS01-T and Bort for 48 hours. RPMI, Roswell Park Memorial Institute.
SNS01-T is well tolerated in healthy animals
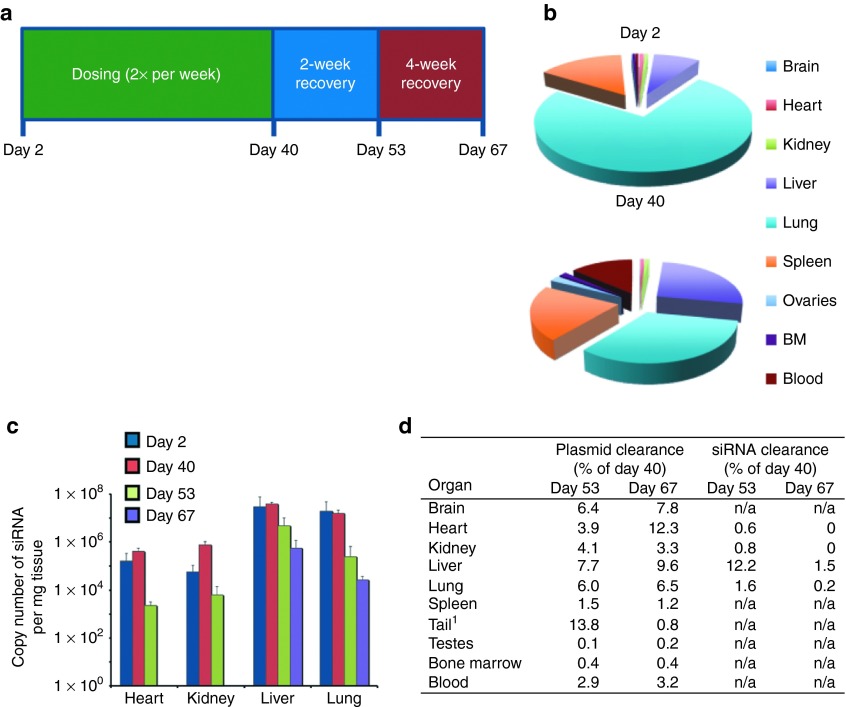
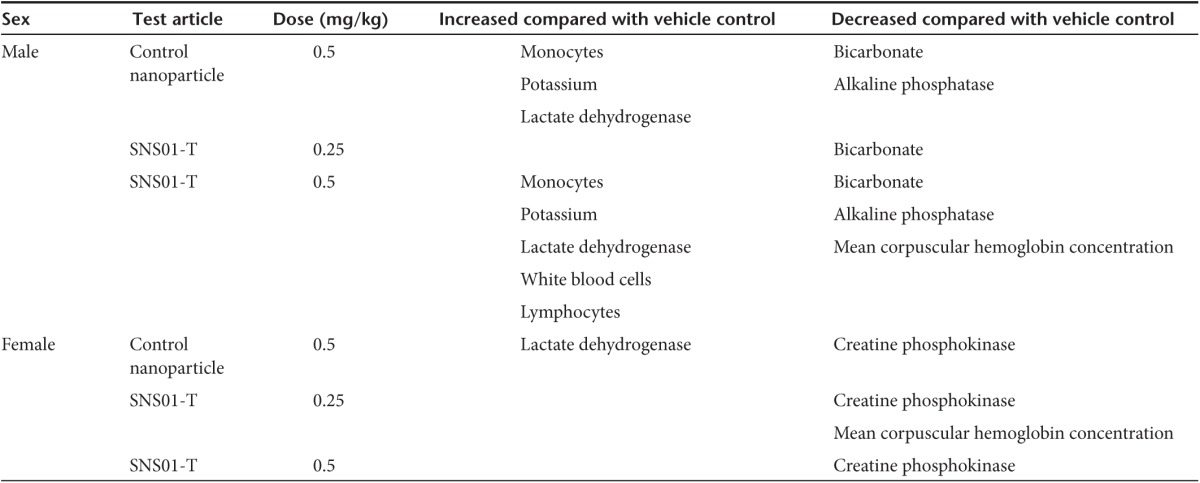
SNS01-T was well tolerated at the doses and schedules examined in the xenograft studies as assessed by no significant changes in body weight and behavior (data not shown). We also examined the effects of SNS01-T in healthy animals. The distribution and persistence of the siRNA and plasmid components of SNS01-T were assessed by qPCR following either single or repeat-dose intravenous administration of 0.5 mg/kg of SNS01-T to healthy mice followed by a 4-week recovery period (Figure 7a). Analysis of various organs revealed that the plasmid DNA was present in all tissues tested, including the bone marrow, after a single dose and multiple doses of SNS01-T (Figure 7b and Supplementary Figure S3a). The siRNA component of SNS01-T was present in liver, lung, heart, and kidney on days 2 and 40 of the study (Figure 7c and Supplementary Figure S3b). There were no obvious signs of accumulation of SNS01-T within tissues, and both the plasmid DNA and siRNA were rapidly and effectively cleared from tissues after treatment was ended (Figure 7d and Supplementary Figure S3c). SNS01-T also had few toxic effects in healthy mice. No treatment-related changes in body weight or mortality were observed (data not shown). There were few significant changes in hematology and clinical chemistry after 6 weeks of treatment, and most of the changes were present in the PEI control group and were dose related (Table 1). None of the changes were consistent between the sexes, and no histopathological abnormalities were found that were consistent with the observed changes in clinical chemistry. Together, these data demonstrate that SNS01-T is well tolerated in mice even at doses that exceeded the therapeutic levels used in the xenograft studies.
Figure 7.
SNS01-T is well tolerated in healthy animals. (a) Timeline of toxicological and pharmacological testing. (b) Plasmid biodistribution in male mice after a single dose (day 2) or 12 doses (day 40) of SNS01-T (n = 5). BM, bone marrow. (c) Biodistribution of small interfering RNA (siRNA) in tissues from male mice after 1 and 12 doses (days 2 and 40), and 2 and 4 weeks recovery posttreatment (days 53 and 67). Data indicate the mean ± SEM (n = 5). (d) Plasmid and siRNA clearance as a percentage of the quantity on day 40, based on the data in panels b and c. 1Injection site; n/a, not available.
Table 1. Changes in hematology and clinical chemistry after 40 days of SNS01-T treatment.
Discussion
This study examines the efficacy of a novel antitumoral agent for B-cell cancers. The SNS01-T nanoparticles were effectively taken up and induced potent cytotoxicity in cell lines from a diversity of B-cell malignancies, yet the same nanoparticles were taken up less effectively by, and not cytotoxic to, their normal B-cell counterparts. In animal studies, this translated into a broad biodistribution with a favorable toxicology profile in healthy mice, yet resulted in inhibition of tumor growth in xenograft models of MM, MCL, and DLBCL. Because the nucleic acid components of SNS01-T selectively target eIF5A, our results highlight eIF5A as an important molecular target in MM, MCL, and DLBCL and SNS01-T as a therapeutic candidate to treat B-cell cancers.
An important aspect of this work is the differential uptake between B-cell tumor cells and normal B cells. The uptake efficiency of cationic polymers like SNS01-T depends on the charge differential between the relatively positive nanoparticle and negative charges on the cell surface.30 Cancer cells can display an increase in net negative surface charge due to changes in phospholipid composition, fatty acid content, and glyocoslyation,31,32,33,34,35,36 which could result in differential interactions at the cell surface and consequent uptake of the SNS01-T nanoparticle. Whatever the mechanism, the enhanced tumor cell uptake correlates well with our biodistribution and toxicology data, suggesting that SNS01-T preferentially targets malignant cells for cell death.
Analysis by flow cytometry indicated that lymphoma, but not myeloma, cell lines preferentially took up nanoparticles in which only plasmid DNA was detectable rather than both plasmid and siRNA when transfected with SNS01-T. However, endogenous eIF5A was knocked down comparably in MM, MCL, and DLBCL cell lines, suggesting that siRNA uptake was adequate to affect biological activity. Furthermore, treatment of both RPMI 8226 MM and SuDHL6 DLBCL xenograft models with nanoparticles containing an inactive plasmid and an eIF5A siRNA demonstrated an antitumoral effect, suggesting sufficient uptake of the siRNA component of SNS01-T in lymphoma cells in vivo. Overall, our data indicate that both the siRNA and plasmid components add to the effectiveness of SNS01-T in all three B-cell malignancies examined.
Our work appears to contradict a report by Scuoppo et al.37, which implicated hypusinated eIF5A as a lymphoma-specific tumor suppressor. However, these contradictory results may be explained by differences in experimental approach. We used SNS01-T to knock down endogenous eIF5A while expressing a non-hypusinable form of the protein, inhibiting tumor growth in MM, MCL, and DLBCL xenograft models. Scuoppo et al. focused on the loss of eIF5A in an Eμ-Myc overexpression mouse model of lymphogenesis, which most closely resembles Burkitt lymphoma. In that specific context, loss of eIF5A enhanced c-Myc–driven tumorigenesis. This is surprising given that hypusinated eIF5A can enhance proliferation,34 is overexpressed in several tumor types,17,38 and blocking hypusination of eIF5A can inhibit proliferation in leukemia cell lines.39 Our findings are in line with Nilsson et al.40 who showed that inhibiting upstream components of the eIF5A pathway in the Eμ-Myc model protects against lymphomagenesis. There may be a unique property of Eμ-myc–driven mouse tumors that allows for such seemingly contradictory results, as there are other examples of known oncogenes having tumor-suppressive properties in this background.41 It cannot be extrapolated from the results of Scuoppo et al. that depletion of eIF5A induces lymphoma in the absence of C-Myc overexpression. We have demonstrated that siRNA-induced knockdown of eIF5A inhibits tumor growth, and this tumor-suppressive effect is enhanced when combined with expression of the non-hypusinable pExp5A transgene.11 Thus, within the context of our experiments, loss of endogenous eIF5A does not promote tumor growth. Future work designed to elucidate factors that influence eIF5A function will provide a clearer understanding of the contexts where eIF5A modulation can be most effective and allow identification of patients most likely to benefit from SNS01-T treatment.
Our work provides evidence that targeting eIF5A with SNS01-T could become a viable treatment strategy for MM, MCL, and DLBCL. SNS01-T is currently being investigated in a multisite, open-label phase 1b/2a dose-escalation study in patients with relapsed or refractory MM, MCL, or DLBCL (http://www.clinicaltrials.gov; Identifier: NCT01435720). The synergy we demonstrate between SNS01-T and current standard-of-care drugs further emphasizes the potential therapeutic utility of SNS01-T. Finally, because the promoter of the pExp5A plasmid can be exchanged to selectively induce expression in specific cell types, there may be utility for SNS01-T–like therapeutics in a range of human malignancies.
Materials and Methods
siRNAs and plasmids. Construction of the pExp5A plasmid and sequences of the eIF5A siRNA used in this study were described previously.10,11 The control siRNA used in this study is a validated nontargeting siRNA (Dharmacon; Thermo Fisher Scientific, Waltham, MA).
SNS01-T nanoparticle formation and characterization. To prepare 1 ml of SNS01-T nanoparticles, 50 μg of plasmid DNA and 25 μg of siRNA were diluted in 5 mmol/l tris(hydroxymethyl)aminomethane (Tris) (pH 7.4) and 5% glucose to a total volume of 500 μl. Nine microliters of in vivo-jetPEI (Polyplus Transfection, Illkirch, France) was diluted in 5 mmol/l Tris (pH 7.4) and 5% glucose to a total volume of 500 μl in a separate tube. The diluted nucleic acids and diluted PEI were then mixed to create nanoparticles with an N/P ratio of 6. Larger volumes (50 and 400 ml) of SNS01-T were prepared in the same manner by slow addition of diluted nucleic acids to a solution of diluted in vivo-jetPEI stirring in disposable spinner flasks (Corning, Corning, NY). Particle size and zeta potential were determined as previously described.11
Cell culture. The KAS-6/1 human MM cell line was obtained from John Lust (Mayo Clinic, Rochester, MN). KAS-6/1 cells were maintained in S10 culture media (RPMI-1640 + 10% fetal bovine serum (FBS) + 4 ng/ml recombinant human interleukin 6). All other B-cell lines were obtained from American Type Culture Collection (Manassas, VA) and maintained in RPMI-1640 media + 10% FBS. Cord blood CD19+ B cells were obtained from Lonza (Allendale, NJ) and maintained in lymphocyte growth media 3. Normal B cells were not passaged prior to treatments. Hep3B-βhCG-C4 cells were maintained in Dulbecco's modified Eagle medium + 10% FBS and 0.3 µg/ml puromycin.26 Cell lines were maintained in a humidified chamber at 37 °C and 5% CO2.
Cryo–transmission electron microscopy. Cryo–transmission electron microscopy was performed by NanoImaging Services (La Jolla, CA). SNS01-T samples were preserved in vitrified ice supported by carbon-coated holey carbon films on 400-mesh copper grids. Samples were prepared by applying a 3 μl drop of undiluted sample suspension to a cleaned grid, blotting excess with filter paper, and immediately vitrifying in liquid ethane. Electron microscopy was performed using a cryostage maintained below −170 °C, using an FEI Tecnai T12 electron microscope (FEI, Hillsboro, OR), operating at 120 keV equipped with an FEI Eagle 4K × 4K charge-coupled device camera.
Potency assay/reverse transcriptase qPCR. KAS-6/1, JVM2, and Su-DHL6 cells were seeded in media containing 1% FBS at 1 × 106 cells per ml on six-well plates and incubated with 33 µl (Su-DHL6) or 50 µl (KAS-6/1, JVM2) of SNS01-T or controls for 4 hours. For Hep3B-βhCG-C4 cells, 1 × 105 cells per ml in Dulbecco's modified Eagle Medium + 5% FBS were seeded on 24-well plates and transfected with 7.5 µl of SNS01-T or controls for 4 hours. The cells were incubated in media with 10% FBS for a further 24 hours. Total RNA was isolated (GenElute Mammalian Total RNA Miniprep Kit, Sigma, St Louis, MO) and used as a template for complementary DNA synthesis (Omniscript RT Kit, Qiagen, Hilden, Germany). Reverse transcriptase qPCR was performed as previously described.11
XTT assay. XTT assays were performed using the Cell Proliferation Kit II(11465015001, Roche, Mannheim, Germany). Cells were treated with a dilution series of SNS01-T or the appropriate controls for 48 hours prior to XTT treatment. Drug combination studies were performed using the Chou-Talalay method at a 1:1.25 constant ratio.42,43 Absorbances were measured on a Multiskan Spectrum UV/Vis microplate spectrophotometer (Thermo Scientific) at 450 nm and a reference wavelength of 650 nm. IC50 values were calculated using GraphPad Prism 4.0 software (Prism GraphPad Software, La Jolla, CA), and synergy was determined by calculating the combination index using CompuSyn software (ComboSyn, Paramus, NJ).
Western blot analysis. RPMI 8226 cells were treated with SNS01-T (2µg/ml), bortezomib (2.5 ng/ml), or a combination of SNS01-T and bortezomib at a ratio of 1:1.25 for 48 hours. Cell lysate was prepared and quantified as previously described.11 Forty-eight hours after treatment with 0.375 mg/kg of control nanoparticle or SNS01-T, Su-DHL6 tumor tissue was homogenized in radio immunoprecipitation assay buffer (Sigma) using a dounce homogenizer. Lysates were sonicated, centrifuged at 4 °C for 15 minutes, and the supernatant was quantified using a BCA kit (Sigma). Proteins were detected using the following antibodies: eIF5A (611977, BD Transduction Laboratories, BD Biosciences, Mississauga, Canada), hypusine-eIF5A (Mirmira lab, Indiana University, Indianapolis, IN),44 β-actin (612657, BD Transduction Laboratories), and MCL-1 (4572, Cell Signaling, Danvers, MA).
Flow cytometry. For flow cytometric analysis of nanoparticle transfection, pExp5A plasmid was labeled with fluorescein isothiocyanate (FITC) following the protocol provided (Label IT Tracker Fluorescein Kit, Mirus Bio, Madison, WI), MIR7025) to a final plasmid (pExp5A-FITC) concentration of 2 μg/μl. EIF5A siRNA conjugated with DY547 was synthesized by Thermo Scientific and diluted to a concentration of 1 μg/μl. SNS01-T nanoparticles were prepared using either labeled plasmid (with unlabeled siRNA), labeled siRNA (with unlabeled plasmid), or both (SNS01-T-Fluor), and 10 μl was added to cell suspensions diluted to 1 × 106 cells per ml in media and seeded at 0.3 ml per well in a 24-well plate. Four hours later, the cells were harvested, washed, and resuspended with 500 μl fluorescence-activated cell sorting (FACS) buffer. The percentage of SNS01-T uptake in cells was assessed on a FACSVantage SE or FACSCalibur flow cytometer (BD Scientific, Franklin Lakes, NJ). Cells transfected with siRNA and plasmid without PEI were used as a control, and the data shown represent the values with this PEI-negative control subtracted.
To examine viability, normal B cells and RPMI 8226 cells were left untreated or transfected with pExp5A and siRNA without PEI (PEI-negative) or transfected with SNS01-T for 24 hours. The unfixed cells were then stained with propidium iodide, and propidium iodide–positive cells were quantified on a FACSCalibur flow cytometer.
For the SNS01-T/bortezomib apoptosis analysis, 0.4 ml of cells was seeded at 0.5 × 106 cells/ml in a 24-well plate. SNS01-T (1.25 µg/ml) and bortezomib (1.56 ng/ml) were given either as single agents or at a ratio of 1:1.25. Forty-eight hours later, the cells were harvested and stained with propidium iodide and annexin as per the manufacturer's instructions (BD Biosciences). The percentage propidium iodide- and annexin-positive cells was analyzed on a FACSCalibur flow cytometer (BD Scientific).
Confocal microscopy. For microscopic analysis of SNS01-T uptake, RPMI 8226 cells were grown in RPMI-1640 media without phenol red supplemented with 10% FBS. RPMI 8226 cells were seeded at 300,000 cells per well in four-chamber culture dishes and incubated at 37 °C overnight. Fluorescently-labeled SNS01-T (SNS01-T-fluor) consisted of FITC-conjugated pExp5A prepared as outlined above and eIF5A siRNA conjugated with DY647 (Thermo Scientific). Each well was treated with 10 μl of SNS01-T-fluor or controls. After 4 hours, treated cells were transferred to sterile microcentrifuge tubes, centrifuged for 5 minutes at 1,000g and resuspended in 100 μl of phosphate-buffered saline. The cell suspensions were transferred to microscope slides and examined using a Zeiss LSM S10 confocal microscope (Carl Zeiss, Oberkochen, Germany) with a Blue 488 nm argon–ion laser with 505LP and 530/30 band pass for FITC and using an FL-4 filter/channel with 650/670 band pass for DY647.
Toxicology and biodistribution studies. Pharmacokinetic and toxicology studies were performed by BioReliance Corporation (Rockville, MD) under Good Laboratory Practices. ICR mice were randomized into groups by body weight. Animals received either a single intravenous injection of vehicle, control nanoparticle (0.5 mg/kg), or SNS01-T (0.25 or 0.5 mg/kg) or were dosed by intravenous injection twice per week for 6 weeks. Animals were either sacrificed immediately following the end of dosing or were allowed to recover for 2 or 4 weeks. Body weight and clinical observations were monitored weekly or daily, respectively, throughout the study. Blood samples were collected for evaluation of whole blood hematology, serum chemistry parameters, and clinical pathology on days 2, 40, 53, and 67. For biodistribution samples, the PCR necropsy procedure involved a systematic dissection of the animal viscera and carcass to avoid cross contamination of tissues. Blood was collected from the retro-orbital sinus into sterile tubes and snap frozen in liquid nitrogen immediately after collection. For DNA analysis, a portion of each tissue was snap frozen, stored at −60 °C and used for detection of pExp5A plasmid DNA by TaqMan (Integrated DNA Technologies, Coralville, IA). Tissues collected for siRNA analysis were stored in RNALater, and micro RNA/siRNA was isolated using miRNeasy Mini kit (Qiagen) and used to generate complementary DNA using a stem-loop RT primer followed by qPCR to quantify siRNA copies. Clinical chemistry and hematology data were analyzed by Dunnett's t-test.
Tumor models. All studies involving animals were conducted in accordance with the guidelines set out by the University of Waterloo Animal Care Committee (Waterloo, Ontario, Canada) as established by the Canadian Council on Animal Care and the Province of Ontario Animals for Research Act. The mice were housed in a pathogen-free animal facility at the University of Waterloo. To generate the RPMI 8226, JVM2, and Su-DHL6 tumor models, female CB17-SCID mice (Charles River Laboratories, Saint-Constant, Canada) at 4–6 weeks of age were implanted subcutaneously with 1.2 × 107, 2.5 × 106, and 1.2 × 107 cells, respectively. Animals were randomized into groups based on tumor size, and treatment was initiated when tumor volumes approximated 50 mm3. The mice were treated by intravenous tail vein injection twice weekly for 6 weeks. Animals were dosed with 0.09 mg (nucleic acid)/kg to 0.75 mg/kg of PEI nanoparticles. Nanoparticles contained either: a nonexpressing control plasmid and nontargeting control siRNA (control nanoparticle); the pExp5A plasmid and control siRNA (pExp5A plasmid); control vector and eIF5A siRNA (eIF5A siRNA); or pExp5A and eIF5A siRNA (SNS01-T). In the drug combination studies, 0.5 mg/kg of bortezomib were injected intraperitoneally twice weekly for 6 weeks, while one round of lenalidomide dosing consisted of intraperitoneal injections five times per week at 15 or 50 mg/kg for 6 weeks.45,46,47 Tumors were measured twice weekly with digital calipers. Tumor volume was calculated using the equation: tumor volume (mm3) = length * width2 * 0.5. Mice were euthanized by CO2 inhalation at the termination of each study or upon reaching end point.
SUPPLEMENTARY MATERIAL Figure S1. SNS01-T plasmid activity is specific to B-cells. Figure S2. SNS01-T and bortezomib synergize in KAS-6/1 cells. Figure S3. Biodistribution of SNS01-T components in healthy female mice. Table S1. Comparison of SNS01-T and SNS01-T-fluor. Table S2. Cytotoxicity of SNS01-T in B cell cancer cell lines. Materials and Methods
Acknowledgments
The authors thank many colleagues for advice and discussions during preparation of the manuscript. The authors are grateful to John Lust of the Mayo Clinic in Rochester, MN for providing the KAS-6/1 cell line; to Raghu Mirmira and Bernhard Maier for providing the anti-hypusine antibody; to Mishi Savulescu and the Quadrilatero lab at the University of Waterloo for assistance with flow cytometry; and to Daryl Enstone and Simon Cheung for assistance with microscopy.
S.M.F., C.A.T., T.T., R.D., and J.E.T. designed the experiments; S.M.F., T.T. Z.L., C.A.T., and Q.Z. performed the research; and S.M.F., C.A.T., and J.E.T. wrote and edited the manuscript.
This work was supported by a research contract from Senesco Technologies. J.E.T. is Chief Scientific Officer for Senesco Technologies and holds Senesco shares. R.D. is Vice President, Research and Development for Senesco. Senesco Technologies holds patents pertaining to the use of SNS01-T as an anticancer agent. All other authors declare no potential conflict of interest.
Supplementary Material
References
- Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol. 2012;9:135–143. doi: 10.1038/nrclinonc.2012.15. [DOI] [PubMed] [Google Scholar]
- Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–762. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- Friedberg JW. Relapsed/refractory diffuse large B-cell lymphoma. Hematology Am Soc Hematol Educ Program. 2011;2011:498–505. doi: 10.1182/asheducation-2011.1.498. [DOI] [PubMed] [Google Scholar]
- Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Henderson A, Hershey JW. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:6415–6419. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One. 2010;5:e9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K. Independent roles of eIF5A and polyamines in cell proliferation. Biochem J. 2005;385 Pt 3:779–785. doi: 10.1042/BJ20041477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Cheng Z, Taylor CA, McConkey BJ, Thompson JE. Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J Cell Physiol. 2010;223:798–809. doi: 10.1002/jcp.22100. [DOI] [PubMed] [Google Scholar]
- Taylor CA, Sun Z, Cliche DO, Ming H, Eshaque B, Jin S, et al. Eukaryotic translation initiation factor 5A induces apoptosis in colon cancer cells and associates with the nucleus in response to tumour necrosis factor alpha signalling. Exp Cell Res. 2007;313:437–449. doi: 10.1016/j.yexcr.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Taylor CA, Liu Z, Tang TC, Zheng Q, Francis S, Wang TW, et al. Modulation of eIF5A expression using SNS01 nanoparticles inhibits NF-κB activity and tumor growth in murine models of multiple myeloma. Mol Ther. 2012;20:1305–1314. doi: 10.1038/mt.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CA, Senchyna M, Flanagan J, Joyce EM, Cliche DO, Boone AN, et al. Role of eIF5A in TNF-alpha-mediated apoptosis of lamina cribrosa cells. Invest Ophthalmol Vis Sci. 2004;45:3568–3576. doi: 10.1167/iovs.03-1367. [DOI] [PubMed] [Google Scholar]
- Maier B, Ogihara T, Trace AP, Tersey SA, Robbins RD, Chakrabarti SK, et al. The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J Clin Invest. 2010;120:2156–2170. doi: 10.1172/JCI38924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosorius O, Reichart B, Krätzer F, Heger P, Dabauvalle MC, Hauber J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci. 1999;112 (Pt 14):2369–2380. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, et al. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A, Hershey JW. The role of eIF5A in protein synthesis. Cell Cycle. 2011;10:3617–3618. doi: 10.4161/cc.10.21.17850. [DOI] [PubMed] [Google Scholar]
- Cracchiolo BM, Heller DS, Clement PM, Wolff EC, Park MH, Hanauske-Abel HM. Eukaryotic initiation factor 5A-1 (eIF5A-1) as a diagnostic marker for aberrant proliferation in intraepithelial neoplasia of the vulva. Gynecol Oncol. 2004;94:217–222. doi: 10.1016/j.ygyno.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Taylor CA, Zheng Q, Liu Z, Thompson JE. Role of p38 and JNK MAPK signaling pathways and tumor suppressor p53 on induction of apoptosis in response to Ad-eIF5A1 in A549 lung cancer cells. Mol Cancer. 2013;12:35. doi: 10.1186/1476-4598-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AL, Li HY, Jin BF, Ye QN, Zhou T, Yu XD, et al. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J Biol Chem. 2004;279:49251–49258. doi: 10.1074/jbc.M407165200. [DOI] [PubMed] [Google Scholar]
- Caraglia M, Park MH, Wolff EC, Marra M, Abbruzzese A. eIF5A isoforms and cancer: two brothers for two functions. Amino Acids. 2013;44:103–109. doi: 10.1007/s00726-011-1182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CC, Martin EN, Lee G, Taylor C, Dondero R, Reznikov LL, et al. Eukaryotic translation initiation factor 5A small interference RNA-liposome complexes reduce inflammation and increase survival in murine models of severe sepsis and acute lung injury. J Infect Dis. 2008;198:1407–1414. doi: 10.1086/592222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- Moghimi SM, Hunter AC, Andresen TL. Factors controlling nanoparticle pharmacokinetics: an integrated analysis and perspective. Annu Rev Pharmacol Toxicol. 2012;52:481–503. doi: 10.1146/annurev-pharmtox-010611-134623. [DOI] [PubMed] [Google Scholar]
- Mortimer I, Tam P, MacLachlan I, Graham RW, Saravolac EG, Joshi PB. Cationic lipid-mediated transfection of cells in culture requires mitotic activity. Gene Ther. 1999;6:403–411. doi: 10.1038/sj.gt.3300837. [DOI] [PubMed] [Google Scholar]
- Tang TC, Man S, Lee CR, Xu P, Kerbel RS. Impact of metronomic UFT/cyclophosphamide chemotherapy and antiangiogenic drug assessed in a new preclinical model of locally advanced orthotopic hepatocellular carcinoma. Neoplasia. 2010;12:264–274. doi: 10.1593/neo.91872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho-Vega JH, Rassidakis GZ, Admirand JH, Oyarzo M, Ramalingam P, Paraguya A, et al. MCL-1 expression in B-cell non-Hodgkin's lymphomas. Hum Pathol. 2004;35:1095–1100. doi: 10.1016/j.humpath.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Khoury JD, Medeiros LJ, Rassidakis GZ, McDonnell TJ, Abruzzo LV, Lai R. Expression of Mcl-1 in mantle cell lymphoma is associated with high-grade morphology, a high proliferative state, and p53 overexpression. J Pathol. 2003;199:90–97. doi: 10.1002/path.1254. [DOI] [PubMed] [Google Scholar]
- Wuillème-Toumi S, Robillard N, Gomez P, Moreau P, Le Gouill S, Avet-Loiseau H, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19:1248–1252. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- Hillaireau H, Couvreur P. Nanocarriers' entry into the cell: relevance to drug delivery. Cell Mol Life Sci. 2009;66:2873–2896. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamakosmanović A, Loewenstein WR. Cellular uncoupling in cancerous thyroid epithelium. Nature. 1968;218:775. doi: 10.1038/218775a0. [DOI] [PubMed] [Google Scholar]
- Cone CD., Jr The role of the surface electrical transmembrane potential in normal and malignant mitogenesis. Ann N Y Acad Sci. 1974;238:420–435. doi: 10.1111/j.1749-6632.1974.tb26808.x. [DOI] [PubMed] [Google Scholar]
- Johnstone BM. Micro-electrode penetration of ascites tumour cells. Nature. 1959;183:411. doi: 10.1038/183411a0. [DOI] [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abercrombie M, Ambrose EJ. The surface properties of cancer cells: a review. Cancer Res. 1962;22:525–548. [PubMed] [Google Scholar]
- Dobrzyńska I, Szachowicz-Petelska B, Sulkowski S, Figaszewski Z. Changes in electric charge and phospholipids composition in human colorectal cancer cells. Mol Cell Biochem. 2005;276:113–119. doi: 10.1007/s11010-005-3557-3. [DOI] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gharib TG, Thomas DG, Huang CC, Misek DE, Kuick RD, et al. Proteomic analysis of eIF-5A in lung adenocarcinomas. Proteomics. 2003;3:496–504. doi: 10.1002/pmic.200390063. [DOI] [PubMed] [Google Scholar]
- Balabanov S, Gontarewicz A, Ziegler P, Hartmann U, Kammer W, Copland M, et al. Hypusination of eukaryotic initiation factor 5A (eIF5A): a novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood. 2007;109:1701–1711. doi: 10.1182/blood-2005-03-037648. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- Nishiki Y, Farb TB, Friedrich J, Bokvist K, Mirmira RG, Maier B. Characterization of a novel polyclonal anti-hypusine antibody. Springerplus. 2013;2:421. doi: 10.1186/2193-1801-2-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocio EM, Vilanova D, Atadja P, Maiso P, Crusoe E, Fernández-Lázaro D, et al. In vitro and in vivo rationale for the triple combination of panobinostat (LBH589) and dexamethasone with either bortezomib or lenalidomide in multiple myeloma. Haematologica. 2010;95:794–803. doi: 10.3324/haematol.2009.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Zhang L, Cai Z, Sun L, Wang H, Yi Q, et al. Lenalidomide synergizes with dexamethasone to induce growth arrest and apoptosis of mantle cell lymphoma cells in vitro and in vivo. Leuk Res. 2011;35:380–386. doi: 10.1016/j.leukres.2010.09.027. [DOI] [PubMed] [Google Scholar]
- Zhang L, Qian Z, Cai Z, Sun L, Wang H, Bartlett JB, et al. Synergistic antitumor effects of lenalidomide and rituximab on mantle cell lymphoma in vitro and in vivo. Am J Hematol. 2009;84:553–559. doi: 10.1002/ajh.21468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.