Abstract
Asperger disorder (ASP) is one of the autism spectrum disorders (ASD) and is differentiated from autism largely on the absence of clinically significant cognitive and language delays. Analysis of a homogenous subset of families with ASP may help to address the corresponding effect of genetic heterogeneity on identifying ASD genetic risk factors. To examine the hypothesis that common variation is important in ASD, we performed a genome-wide association study (GWAS) in 124 ASP families in a discovery data set and 110 ASP families in a validation data set. We prioritized the top 100 association results from both cohorts by employing a ranking strategy. Novel regions on 5q21.1 (P = 9.7 × 10−7) and 15q22.1–q22.2 (P = 7.3 × 10−6) were our most significant findings in the combined data set. Three chromosomal regions showing association, 3p14.2 (P = 3.6 × 10−6), 3q25–26 (P = 6.0 × 10−5) and 3p23 (P = 3.3 × 10−4) overlapped linkage regions reported in Finnish ASP families, and eight association regions overlapped ASD linkage areas. Our findings suggest that ASP shares both ASD-related genetic risk factors, as well as has genetic risk factors unique to the ASP phenotype.
Keywords: asperger, susceptibility, genetics
Introduction
Asperger disorder (ASP) is one of the autism spectrum disorders (ASD). Neurodevelopmental in origin, ASP is characterized by deficits in social interaction and the presence of repetitive behaviors or circumscribed interests. Unlike in autism, the prototypical ASD, language acquisition and cognition are typically spared [American Psychiatric Association, 2000; World Health Organization, 1992]. Despite formal recognition as a diagnostic category, there has been continued debate as to whether ASP represents a separate diagnostic entity or is simply a milder version of autism [Bennett et al., 2008;Woodbury-Smith & Volkmar, 2009]. Differentiation of ASP from other ASD relies on behavioral methods as consistent biological markers unique to ASP (e.g., serum markers or neuroimaging) have not been found. Clearly, individuals with ASP show relatively superior language and cognitive traits [Bennett et al., 2008; Witwer & Lecavalier, 2008]. However, it is unclear if these traits reflect underlying biological differences. There are few reports that have tied biological variation to specific symptoms or behavioral traits in ASP. Neuroimaging of metabolic activity in regions implicated in ASD was able to distinguish ASD cases with and without language impairment [Gabis et al., 2008]. Recently, Kleinhans et al. [2009] showed positive correlation of IQ and neurometabolites in the amygdala region of high-functioning ASD patients. As this work is preliminary, there are presently no laboratory tests specific to the ASP diagnosis or that differentiate clinical subgroups.
Given the lack of consensual definitions of the ASP diagnosis until recently, it is not surprising that the prevalence of the condition is unclear. Recent studies of English and Finnish cohorts estimated ASP prevalence as 2.5–2.9 per 1000 [Mattila et al., 2007; Williams, Thomas, Sidebotham, & Emond, 2008]. Consistent with other ASD, males are reported to have higher risk for ASP than females. However, the reported estimated male:female ratios vary between 1.7:1 and 22:1 in different samples [Mattila et al., 2007; Williams et al., 2008]. Initially described in 1944, ASP was defined as a highly heritable disease [Asperger, 1944]; yet there remains no consensus on its underlying genetic architecture.
More broadly, it is well known that ASD traits are highly heritable [Ronald et al., 2006; Ronald, Simonoff, Kuntsi, Asherson, & Plomin, 2008]. However, gene mapping has been hindered by significant clinical and genetic heterogeneity. One solution has been to identify clinically homogeneous subgroups for genetic mapping. Several investigators have looked to ASD subphenotypes with promising results. For instance, Liu, Paterson, Szatmari [2008] found significant linkage on chromosome 15q13.3–q14 in a subgroup of individuals with ASD with IQ≥70 (LOD = 4.01) and on chromosome 11p15.4–p15.3 in a subgroup with delayed onset of phrase speech (LOD = 3.4). In contrast, none of the LOD scores were >3 in their overall ASD data set. Buxbaum et al. [2004] showed linkage on chromosome 1q24.2 in a subgroup of autism-affected relative pairs with more severe obsessive–compulsive behavior. Another strategy has been to examine clinical subphenotypes such as ASP. The genetic risk for the ASP subphenotype may be due to fewer or a different set of genetic loci compared to the broader ASD diagnostic group. A genome-wide linkage study of ASP in 13 Finnish families [Ylisaukko-oja et al., 2004] found Zmax >1.5 on nine chromosomal regions, 1q21–22, 3p14–24, 3q25–27, 4p14, 4q32, 6p25, 6q16, 13q31–33 and 18p11. In the fine mapping stage, the highest two-point LOD scores were observed on chromosomes 1q21–22, 3p14–24 and 13q31–33 [Ylisaukko-oja et al., 2004]. Their linkage finding on 3p21–24 was validated in an additional sample of 12 Finnish families [Rehnström et al., 2006]. As with linkage studies in the broader ASD phenotype, no consistent regions have emerged as containing ASD risk genes. However, two recent genome-wide association studies (GWAS) revealed a common allele on chromosome 5p14.1 associated with ASD risk [Ma et al., 2009; Wang et al., 2009]. The application of GWAS to complex disorders, such as ASD, results in increased resolution and power in mapping risk loci, especially in the ability to detect smaller effect sizes.
The goal of our study was to identify susceptibility loci for ASP. To achieve this goal, we performed a GWAS in nuclear families ascertained for ASP and validated results from this screening in an independent sample.
Material and Methods
Discovery Data Set
The discovery sample consisted of families ascertained through a proband with ASP by the University of South Carolina and the John P. Hussman Institute for Human Genomics (HIHG). In total, 392 individuals from 124 families were ascertained, including 107 complete trios and 7 families with two or more affected individuals. Ten unaffected sibs were included in the study. Ascertainment protocols were approved by the appropriate Institutional Review Boards. Parents provided written informed consent for all minors and individuals who were not able to give informed consent. Assent was obtained whenever possible. Core inclusion criteria were as follows: (1) chronological age between 3 and 21 years; (2) presumptive diagnosis of ASP (3) expert clinical determination of ASP diagnosis using DSM-IV criteria supported by the Autism Diagnostic Interview (ADI-R) [Rutter, LeCouteur, & Lord, 2003]. All cases were reviewed by the HIHG clinical team consisting of two clinical psychologists (J. L. R. and M. L. C.) and a pediatric medical geneticist (S. S.) to assure that they met criteria for inclusion as an ASP case. In those instances where an ADI-R was not available (n = 27), a best-estimate diagnosis was assigned using all available clinical information, including clinician summaries, caregiver report on other measures, and medical records; (4) IQ equivalent >70; and (5) acquisition of first words before 24 months of age. We excluded participants with severe sensory problems (e.g., visual impairment or hearing loss), significant motor impairments (e.g., failure to sit by 12 months or walk by 24 months), or identified metabolic, genetic, or progressive neurological disorders. These research diagnostic criteria capture the ASP clinical phenotype, which is defined by the presence of ASD social and behavioral impairments in the presence of intact language acquisition.
Validation Data Set
The validation sample consisted of 110 families (468 individuals) from the Autism Genetic Resource Exchange (AGRE) [AGRE, accessed 2008]. This sample was composed of the following: 37 trios; 27 trios with unaffected sibs; 22 families with multiple affecteds; 6 families with 1 parent and siblings available; 8 parent–child pairs; 8 probands with 2 discordant sibs and 2 families including 2 generations. The ASP research diagnostic classification was assigned using the same criteria as above as derived from all available clinical data including the ADI-R. The ascertainment scheme in the AGRE data set was slightly different from the discovery data set, since it was ascertained through patients with ASD (i.e., not exclusively ASP). Thus, from the AGRE validation data set we classified as ASP only those affected individuals with demonstrated social and behavioural impairments, normal acquisition of first words, and IQ equivalent >70. Genotypes on 550K Illumina Human Beadchip were available for the validation data set [AGRE, 2008].
Genotyping of the Discovery Data set
Genomic DNA was purified from whole blood using Puregene chemistry on the Qiagen Autopure LS according to standard automated Qiagen protocols (Valencia, CA). Samples were genotyped using Illumina’s Human 1M Beadchip, containing 1,072,820 SNPs. The samples were processed according to Illumina Procedures for processing of the Infinium II® assay. Samples with call rates below 98% were excluded from analysis and a GenCall cutoff score of 0.15 was used for all Infinium II® products.
Sample Quality Control
After genotyping, samples underwent quality control (QC) tests. We used the same protocol for both the discovery and the validation data sets. Reported and genetic sex were examined using X-chromosome linked SNPs. Relatedness between samples, sample contaminations, swaps and duplications were tested using genome-wide IBD estimation. As a next step we tested for Mendelian inconsistencies on all SNPs and samples. Mendelian errors (ME) can emerge from sample swaps, DNA contamination, copy-number variation (CNV), genotype calling errors and other reasons. We excluded individuals and families with ME >2% from the analysis. This threshold would still allow for small deletions and duplications that are common in the human genome.
SNP QC
Additional QCs were applied to SNP results. First, we removed from analysis SNPs with minor allele frequencies (MAF) below 10%. Since association testing was conditioned on heterozygous parents, we would expect at most 40 informative parent–child pairs in our data sets for markers with MAF = 0.1. We observed a negative correlation between the proportion of MEs per SNP and P-values for HWE. To minimize genotyping errors, we excluded SNPs with P-value <10−6 for HWE and ME>4%. Remaining genotypes with MEs were set as missing. PLINK software was used for all the steps described above [Purcell et al., 2007].
Genotype Imputation
Since the validation data set was genotyped on a different platform with a smaller number of SNPs (558,183), we imputed missing SNPs in the validation data set using the program IMPUTE [Marchini, Howie, Myers, McVean, & Donnelly, 2007] and the phased CEU HapMap data set as a reference [International HapMap Consortium, 2007; accessed February 2009]. Individual genotypes with certainty less than 0.90 were set as missing. To avoid possible bias, we used only genotypes with a call rate of 0.98 for association testing. All individuals were treated independently while doing imputation.
Whole-Genome Association Analysis
Association analysis was performed using the CAPL algorithm for autosomal markers [Chung, Hauser, & Martin, 2006; Chung, Schmidt, Morris, & Martin, 2010] and X-APL algorithm for markers on the X-chromosome [Chung, Morris, Zhang, Li, & Martin, 2007] in the discovery and the validation data sets separately and in the combined data set. These methods provide valid and robust tests for allelic association across both trio and multiplex families. In order to address possible differences in genetic background and ascertainment schemes, we also combined P-values using Fisher’s method [Fisher, 1925]. In order to prioritize the results from the GWAS for followup, we ranked the results in both the discovery and the validation data sets. We explored the top 100 SNPs with the smallest rank sum over both cohorts with the same over-transmitted risk allele. Genomic positions for SNPs and genes are described according to the NCBI Human Genome build 36.1, October 2008. Available information about expression in human brain tissues was used from open source databases GNF expression Atlas2 [accessed June 2009] and NCBI UniGene [accessed June 2009].
Linkage Disequilibrium
Linkage disequilibrium (LD) between SNPs was investigated using pair-wise LD measures (r2 and D’) that were calculated in using Haploview [Barrett, Fry, Maller, & Daly, 2005]. Solid spine LD method was used for LD block definition [Barrett et al., 2005].
Power Estimation
To estimate power in our study, we simulated pedigree data with the same family structure and sample sizes as the discovery and validation data sets. SIMLA software (version 3.2 build 24) [Schmidt, Hauser, Martin, & Schmidt, 2005] was used for 1,000 simulations with the following parameters: prevalence = 0.0025; risk allele frequency (RAF) = 0.3 and 0.6; recessive model (GRRAA = 2) and multiplicative model (GRRAA = 2); LD between marker and disease allele r2 = 0.8. We used CAPL software [Chung et al., 2006, 2010] to obtain P-values for each simulation.
Results
All 124 families (392 individuals) in the discovery data set met our QC criteria and were used in association analysis. Of the initial 467 samples in the validation data set, 11 were dropped; 6 individuals (two families) were excluded from association analysis because of Mendelian inconsistencies while 5 individuals were excluded because the genotypic sex disagreed with the clinical information. The median of ME per family in both investigated cohorts was below 0.005%. More than 99% of the discovery families and 98% of the validation families had ME below 0.02%.
Using our QC criteria on each data set independently, we removed 382,733 SNPs from the discovery data set and 133,235 SNPs from the validation data set. After all QC steps, genotype call rates for each individual were >99.8% in both data sets. We were able to impute 124,294 SNPs with MAF >0.1 in the validation data set. The average missing rate within imputed SNPs was 0.30. After stringent QC, only 14,334 imputed SNPs had a missing rate lower than 0.02. In total, 439,282 SNPs survived QC in both data sets and were available for statistical analysis. The quantile–quantile plot for the association P-values is shown in Supplementary Figure 1. The top-ranked P-values for association with ASP ranged between 0.00014 and 0.025 in the discovery data set, between 0.00003 and 0.022 in the validation data set, and between 9.7 × 10−7 and 0.0072 in the combined data set. Supplementary Table I summarizes results for the 100 top-ranked SNPs using our ranking strategy in discovery, validation and combined data sets. Meta-analysis P-values do not significantly differ from combined data set results. SNP names for which genotypes were imputed are presented in italic text format.
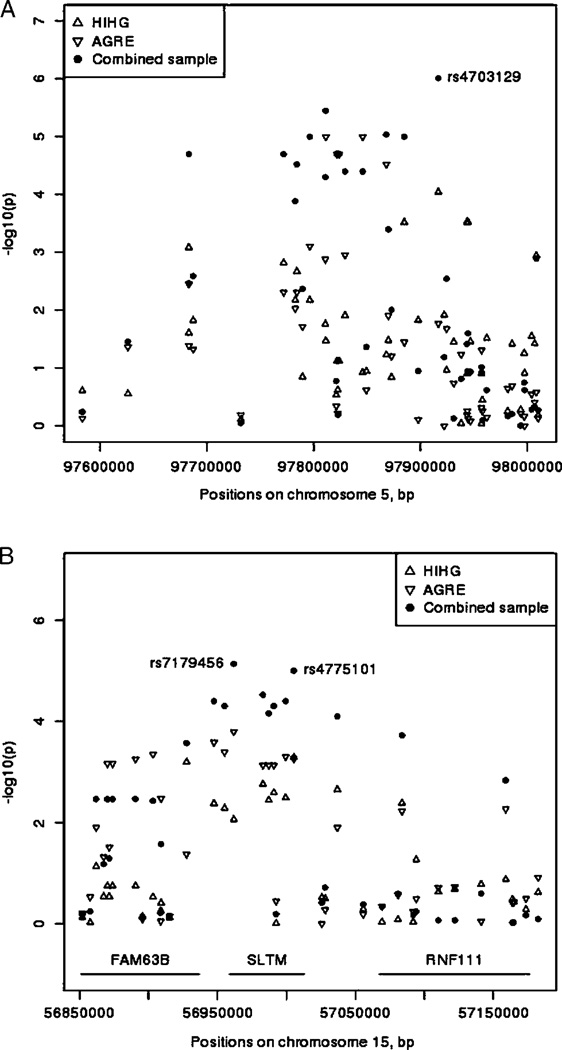
The smallest P-value in the discovery data set was 6.81 × 10−7 for rs2326253 and 2.05 × 10−7 for rs724655 in the validation sample. Neither SNP showed evidence for association in the second data set and did not appear in top 100 results. None of tested regions showed genome-wide significance using a P-value cutoff 7.2 × 10−8 [Dudbridge & Gusnanto, 2008]. The smallest rank sum in both cohorts showed rs4775101 on chromosome 15q22.1 (ranked in HIHG as 332th best and in AGRE as 258th) with the P-value 0.00055 in the discovery and 0.00052 in the validation cohort (Fig. 1B). In total, ten SNPs in this region could be validated (supplementary Table I). Nine of ten SNPs fell into one LD block spanning the SAFB-like transcription modulator (SLTM) gene. The smallest P-value in the combined data set was 9.7 × 10−7 for rs4703129 on chromosome 5q21.1 (Fig. 1). This region had eight SNPs within the top-ranked hundred association results representing two LD blocks (supplementary Table I). The majority of the top-ranked SNPs (59) are not within annotated coding genes [RefSeq, accessed June 2009], whereas 41 SNPs fell into the transcription units of 24 known genes (Table I). Eight of these genes were reported to be highly expressed in human brain [GNF expression atlas 2, UniGene]: kelch domain containing 10 (KLHDC10) on 7q32.2; regulating synaptic membrane exocytosis 2 (RIMS2) on 8q22.3; hypothetical protein (FAM171A1) on 10p13; glial cell line-derived neurotrophic factor family receptor alpha 1 (GFRA1) on 10q25.3; neurotrimin (NTM) on 11q25; transmembrane protein 132B (TMEM132B) on 12q24.32; junctophilin type 4 (JPH4) on 14q11.2 and modulator of estrogen induced transcription (SLTM) on 15q22.1. The complete list of the resulting 26 candidate genes is represented in Table I. Power estimation to detect association was highest for a recessive model when the risk allele was the major allele (β = 0.84) in the combined data set at P-value threshold of 0.05 (supplementary Table II). Power to detect association with genome-wide significance (7.2 × 10−8 [Dudbridge & Gusnanto, 2008]) in the discovery and validation samples separately was less than 0.1% for all tested models (data not shown). Estimated power at genome-wide significance level in the combined data set was 1.2% for recessive model with RAF = 0.6.
Figure 1.
Association results in two Asperger cohorts and combined sample. (A) in locus 5q21.1; (B) in locus 15q22.1–22.2.
Table I.
Candidate Genes for Asperger Prioritized by Ranking Strategy in Both Discovery and Validation
| Locus | SNP | Gene | Location | Expression in brain |
|---|---|---|---|---|
| 1q25.3 | rs1174657, rs1174658 | APOBEC4 | Exon, nonsyn. | Moderately |
| 1q25.3 | rs6424904 | RGL1 | Exon, nonsyn | Moderately |
| 3p14.2 | rs10510837 | FHIT | Intron | Moderately |
| 5p14.3 | rs6879343 | Next to CDH12 | 202679 bp 5′ | Yes |
| 5q14.1 | rs404375 | THBS4 | Intron | Yes |
| 5q21.1 | rs17165849, rs1378441, rs11241318, rs1455420, rs1563317, rs1037597, rs1195473, rs4703129 | Next to RGMB | 216263–449709 bp 5′ | Broadly |
| 6q24.3 | rs9390411, rs9322069, rs2226067 | 6orf103 | Introns | No |
| 7q32.2 | rs10251765, rs17133163 | KLHDC10 | Introns | Broadly |
| 8q22.3 | rs2080610 | RIMS2 | Intron | Yes |
| 8q24.3 | rs4358783 | TRAPPC2 | Intron | No |
| 9q21.2 | rs1929402 | CEP78 | Intron | Moderately |
| 10p13 | rs4750628, rs10796285, rs11599615 | FAM171A1 | Introns | Yes |
| 10q25.3 | rs11197571 | GRFA1 | Intron | Yes |
| 11q25 | rs1550976, rs1448363 | NTM | Introns | Yes |
| 12q24.32 | rs11058197 | TMEM132B | Intron | Yes |
| 14q11.2 | rs222682 | THTPA | Intron | Moderately |
| 14q11.2 | rs11844366 | JPH4 | Yes | |
| 15q15.1 | rs12903690 | GANC | Intron | Moderately |
| 15q22.1–q22.2 | rs7179456, rs12595482, rs17302045, rs10518992, rs3145, rs4775101, rs1446240 | SLTM | Introns, 3′ and 5′ | Yes |
| 17q32 | rs7218167 | SKAP1 | Intron | No |
| 17q23.1 | rs3902107, rs1034911 | BCAS3 | Introns | Moderately |
| 18q21.1 | rs9954152, rs2457965 | MYO5B | Intron | Broadly |
| 18q22.3 | rs2581644 | TSHZ1 | Intron | Yes |
| 19p13.3 | rs2392794, rs892055, rs4803455 | ODF3L2 | Intron | Moderately |
| 20q11.23 | rs6022419 | KIAA0406 | Intron | Yes |
| 21q22.3 | rs6517673 | TMPRSS2 | Intron | Yes |
Forty-one SNPs are located in the transcription units of 24 known genes. Imputed SNPs marked in Italic.
Discussion
Several genome-wide screens have been performed in families with ASD and ASP, but only a few loci have been replicated in independent studies [Freitag, 2007]. Stratification by clinical phenotypes has been used to create more homogenous groups and improve the likelihood of identifying true susceptibility loci [Alarcón, Yonan, Gilliam, Cantor, & Geschwind, 2005; Buxbaum et al., 2004; Rehnström et al., 2006; Ylisaukko-oja et al., 2004]. In the present study, genetic heterogeneity has been limited by selecting a subset of families with ASP.
It is not surprising that the smallest P-value in our discovery data set neither withstands multiple testing correction nor replicates in the validation data set [IMSGC, 2007]. In order to capture the most reproducible results from our two independent samples, we employed a ranking strategy in both data sets. Further, we did not focus on gene-only regions. Most of the validated loci in this study, found to be associated with ASP, fell outside of protein coding regions and have not been previously related to ASP or ASD. This was the case for the two new associated regions on chromosomes 5q21.1 and 15q22.1–q22.2. The association region on chromosome 5q21.1 does not contain any validated genes (Fig. 1). The SNP (rs4703129) showing the smallest P-value in the combined data set (9.7 × 10−7) is located 216.2 kb from the 5′ end of the next annotated gene, RGMB, which encodes for RGM domain family, member B isoform 1 precursor. RGMB is a glycosylphosphatidylinositol (GPI)-anchored member of the repulsive guidance molecule family and contributes to the patterning of the developing nervous system [Samad et al., 2005]. The observed association may reflect changes in long-range regulatory elements of RGMB, since such elements have been described for other genes [Kleinjan et al., 2006] or non-annotated transcripts, as described in the ENCODE pilot project in regions of the genome previously thought to be transcriptionally silent [Birney et al., 2007].
The second association region on chromosome 15q22.1–q22.2, with the best rank over both samples, spans the SLTM gene, coding for estrogen-induced transcription modulator gene, and part of RNF111 gene coding for ring finger protein 111. Since RNF111 is known to be involved in early stage embryonic development [RefSeq, 2009], it could contribute to ASP risk. Both 5q21.1 and 15q22.1–q22.2 are new candidate regions for ASP.
Our results provide independent confirmation of three susceptibility loci previously reported to be linked with ASP. The three genomic regions published by the Finnish group 3p14.2, 3q25–26 [Ylisaukko-oja et al., 2004], and 3p23 [Rehnström et al., 2006] also were associated with ASP in our two samples. Their reported marker D3S1766 on 3p14 being located 1307 kb from our top-ranked SNP rs10510837. This SNP is located in an intron of FHIT, a gene encoding a diadenosine triphosphate hydrolase and involved in purine metabolism. FHIT encompasses the common fragile site FRA3B on chromosome 3. There is one reported case of FHIT deletion in an autistic patient [Sebat et al., 2007]. The SNP rs7649494 in the region on 3q25.22 is within a non-transcribed region, 55.6 kb away from P2RY1 (Homo sapiens purinergic receptor P2Y, G-protein coupled, 1) gene. P2RY1 is a receptor for extracellular adenine nucleotides, such as ATP and ADP, which controls mobilization of intracellular calcium ions via activation of phospholipase C. While the Finnish linkage analysis suggests broad genomic candidate regions spanning several million base pairs, our results provide more accurate localization of candidate regions for ASP on chromosomes 3p14.2, 3q25–26 and 3p23.
Our data did not confirm the remaining Finnish ASP linkage results in chromosomal regions 1q21–22, 4p14, 4q32, 6p25, 6q16 and 13q31–33 [Ylisaukko-oja et al., 2004]. This lack of confirmation may be explained by insufficient power to detect an effect or the different ethnic composition of the American and Finnish samples. Further replication will help to distinguish false positives from true associations, but low prevalence and recent recognition of ASP in clinical classification make similar data sets scarce.
The ASP subgroup in our study and the high-functioning autism subgroup in Liu et al. [2008] are clinically similar with respect to IQ level (>70). We find it encouraging that our association of ASP with chromosome 15q15.1 (rs12903690) is within 10Mb of linkage region 15q13.3–q14 (rs1454985) reported in families with ASD individuals with IQ>70. The linkage peak on 15q13.3–14 was seen in this subgroup only, but not in the entire ASD data set [Liu et al., 2008]. This demonstrates that testing phenotypic subgroups is a potentially useful strategy for heterogeneity dissection.
In addition, the findings from our GWAS may be relevant to other published linkage signals. For instance, the linkage region confirmed in our study on chromosome 7q32 is known as the autism susceptibility locus (AUTS1) and was implicated by several research groups [IMGSAC, 2001a,b; Lamb et al., 2005; Vourc’h et al., 2003]. The UBE2H gene is located in the same 7q32 region, just 130 kb away from the SNP rs10251765 in the KLHDC10 gene associated with ASP in our study, although in a different LD block. The function of the KLHDC10 gene is unknown, but the protein has been shown to be expressed in fetal and adult brains [Su et al., 2002]. Locus 9q34 showed suggestive linkage in families with ASD [Lamb et al., 2005; Liu et al., 2008]. Their genotyped markers D9S185 and rs1544105 are respectively 6.8Mb and 1.7Mb away from our top-ranked SNP rs1228439. Reported association of the rs1544105 with ASD status did not replicate in our combined sample (P = 0.86). Five linkage regions 4q21–31, 5p15–q12, 7q33–36, 8q22–24 and 19p13–q13 published by Ylisaukko-oja et al. [2006] in combined Finnish and AGRE sample overlap with the top hundred association regions in our study. These overlapping linkage findings in ASD and results from our ASP association study support the hypothesis that ASP and ASD share common genetic features.
The majority of validated results from our study provide new candidate regions for ASP. Some of them are within well-characterized genes. Best-investigated among these are GFRA1 [Cacalano et al., 1998; Pozas & Ibáñez, 2005], NTM [Struyk et al., 1995], RIMS2 [Wang, Sugita, & Sudhof, 2000] and JPH4 [Moriguchi et al., 2006]. These genes are highly expressed in the human brain [Struyk et al., 1995, GNF expression atlas 2]. Their expression profiles partially overlap with brain regions showing abnormalities in postmortem and in vivo imaging studies of ASD patients [Gabis et al., 2008; Kleinhans et al., 2009; Saitoh, Karns, & Courchesne, 2001], as well as with findings of cellular studies [Bailey et al., 1998; Bauman & Kemper, 1985; Ritvo et al., 1986; Rodier, Ingram, Tisdale, Nelson, & Romano, 1996] this makes GFRA1, NTM, and JPH4 promising candidate genes for ASP.
In conclusion, dissection of homogenous subphenotypes may be advantageous in disorders such as ASD, which is often conceived as a spectrum of disorders composed of several dimensions [Liu et al., 2008]. While none of the investigated loci showed a large disease risk, supporting the hypothesis that many genes each of small effect are involved in ASP, several potentially interesting candidates emerged.
Supplementary Material
Acknowledgments
We thank the patients with Asperger disorder and their family members who participated in this study and the personnel at the John P. Hussman Institute for Human Genomics (HIHG), including the staff at the HIHG Biorepository and members of the HIHG. We also thank Michael Schmidt and Susan Slifer for their contributions to the data analysis and simulation. This research was supported by a gift from the Hussman Foundation. We also wish to gratefully acknowledge the resources provided by the AGRE consortium and the participating Autism Genetic Resource Exchange (AGRE) families. The AGRE resource is supported by Autism Speaks. A subset of the participants was ascertained while Dr. Pericak-Vance was a faculty member at Duke University.
Grant sponsor: National Institutes of Health (NIH); Grant numbers: NS26630; MH080647; Grant sponsor: Autism Speaks.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-IV), text revision. Washington, DC: American Psychiatric Press, Inc.; 2000. [Google Scholar]
- Alarcón M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and Ordered-Subsets Analysis of autism endophenotypes support language QTLs. Molecular Psychiatry. 2005;10:747–757. doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- Asperger H. Die “Autistischen Psychopatien” im Kinderalter. Archiv fur Psychiatrie und Nervenkrankheiten. 1944;117:73–136. [Google Scholar]
- Bailey A, Luthert P, Dean A, Harding B, Janota I, et al. A clinicopathological study of autism. Brain. 1998;121:889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bauman M, Kemper TL. Histoanatomic observations of the brain in early infantile autism. Neurology. 1985;35:866–874. doi: 10.1212/wnl.35.6.866. [DOI] [PubMed] [Google Scholar]
- Bennett T, Szatmari P, Bryson S, Volden J, Zwaigenbaum L, et al. Differentiating autism and Asperger syndrome on the basis of language delay or impairment. Journal of Autism and Developmental Disorders. 2008;38:616–625. doi: 10.1007/s10803-007-0428-7. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman J, Keddache M, Smith CJ, Hollander E, et al. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: Evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Molecular Psychiatry. 2004;9:144–150. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- Cacalano G, Fariñas I, Wang LC, Hagler K, Forgie A, et al. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998;21:53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung R-H, Hauser ER, Martin ER. The APL test: Extension to general nuclear families and haplotypes and the examination of its robustness. Human Heredity. 2006;61:189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Chung R-H, Morris RW, Zhang L, Li Y-J, Martin ER. X-APL: An Improved Family-Based Test of Association in the Presence of Linkage for the X Chromosome. The American Journal of Human Genetics. 2007;80:59–68. doi: 10.1086/510630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung R-H, Schmidt MA, Morris RW, Martin ER. CAPL: A novel association test using case-control and family data and accounting for population stratification. Genetic Epidemiology. 2010;34:747–755. doi: 10.1002/gepi.20539. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genetic Epidemiology. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. Statistical methods for research workers, 13e. London: Oliver and Loyd, Ltd.; 1925. pp. 99–101. [Google Scholar]
- Freitag CM. The genetics of autistic disorders and its clinical relevance: A review of the literature. Molecular Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- Gabis L, Huang W, Azizian A, DeVincent C, Tudorica A, et al. 1H-magnetic resonance spectroscopy markers of cognitive and language ability in clinical subtypes of autism spectrum disorders. Journal of Child Neurology. 2008;23:766–774. doi: 10.1177/0883073808315423. [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Human Molecular Genetics. 2001a;10:973–982. doi: 10.1093/hmg/10.9.973. [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) A genome-wide screen for autism: Strong evidence for linkage to chromosomes 2q, 7q, and 16p. American Journal of Human Genetics. 2001b;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds D, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium. Risk Alleles for Multiple Sclerosis Identified by a Genomewide Study. NEJM. 2007;357 doi: 10.1056/NEJMoa073493. www.nejm.org. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Mella S, Carr CB, Tyas DA, et al. Long-range downstream enhancers are essential for Pax6 expression. Development Biology. 2006;299:563–581. doi: 10.1016/j.ydbio.2006.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhans NM, Richards T, Weaver KE, Liang O, Dawson G, Aylward E. Brief report: Biochemical correlates of clinical impairment in high functioning autism and Asperger’s disorder. Journal of Autism and Development Disorders. 2009;39:1079–1086. doi: 10.1007/s10803-009-0707-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Barnby G, Bonora E, Sykes N, Bacchelli E, et al. International Molecular Genetic Study of Autism Consortium (IMGSAC) Analysis of IMGSAC autism susceptibility loci: Evidence for sex limited and parent of origin specific effects. Journal of Medical Genetics. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Paterson AD, Szatmari P. Autism Genome Project Consortium. Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biological Psychiatry. 2008;64:561–570. doi: 10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Annals of Human Genetics. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics. 2007;39:906–1013. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Mattila ML, Kielinen M, Jussila K, Linna SL, Bloigu R, et al. An epidemiological and diagnostic study of Asperger syndrome according to four sets of diagnostic criteria. Journal of the American Academy of Child and Adolescent Psychiatry. 2007;46:636–646. doi: 10.1097/chi.0b013e318033ff42. [DOI] [PubMed] [Google Scholar]
- Moriguchi S, Nishi M, Komazaki S, Sakagami H, Miyazaki T, et al. Functional uncoupling between Ca2+ release and afterhyperpolarization in mutant hippocampal neurons lacking junctophilins. PNAS. 2006;103:10811–10816. doi: 10.1073/pnas.0509863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozas E, Ibáñez CF. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic. Neurons. 2005;45:701–713. doi: 10.1016/j.neuron.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehnström K, Ylisaukko-oja T, Nieminen-von Wendt T, Sarenius S, Källman T, et al. Independent replication and initial fine mapping of 3p21-24 in Asperger syndrome. Journal of Medical Genetics. 2006;43:e6. doi: 10.1136/jmg.2005.033621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, et al. Lower Purkinje cell counts in the cerebella of four autistic subjects: Initial findings of the UCLA-NSAC Autopsy Research Report. American Journal of Psychiatry. 1986;143:862–866. doi: 10.1176/ajp.143.7.862. [DOI] [PubMed] [Google Scholar]
- Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: Developmental anomalies of the cranial nerve motor nuclei. The Journal of Comparitive Neurology. 1996;370:247–261. doi: 10.1002/(SICI)1096-9861(19960624)370:2<247::AID-CNE8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happé F, Bolton P, Butcher LM, Price TS, et al. Genetic heterogeneity between the three components of the autism spectrum: A twin study. Journal of the American Academy of Child and Adolescent Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviors in a community twin sample. Journal of Child Psychology Psychiatry. 2008;49:535–542. doi: 10.1111/j.1469-7610.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- Rutter M, LeCouteur A, Lord C. Autism diagnostic interview, revised (ADI-R) Los Angeles, CA: Western Psychological Services; 2003. [Google Scholar]
- Saitoh O, Karns CM, Courchesne E. Development of the hippocampal formation from 2 to 42 years: MRI evidence of smaller area dentata in autism. Brain. 2001;124:1317–1324. doi: 10.1093/brain/124.7.1317. [DOI] [PubMed] [Google Scholar]
- Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, et al. DRAGON, a bone morphogenetic protein co-receptor. The Journal of Biological Chemistry. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Hauser ER, Martin ER, Schmidt S. Extension of the SIMLA package for generating pedigrees with complex inheritance patterns: Environmental covariates, gene-gene and gene-environment interaction. Statistical Applications in Genetics and Molecular Biology. 2005;4 doi: 10.2202/1544-6115.1133. article 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyk AF, Canoll PD, Wolfgang MJ, Rosen CL, D’Eustachio P, Salzer JL. Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. Journal of Neuroscience. 1995;15:2141–2156. doi: 10.1523/JNEUROSCI.15-03-02141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, et al. Large-scale analysis of the human and mouse transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourc’h P, Martin I, Bonnet-Brilhault F, Marouillat S, Barthélémy C, et al. Mutation screening and association study of the UBE2H gene on chromosome 7q32 in autistic disorder. Psychiatric Genetics. 2003;13:221–225. doi: 10.1097/00041444-200312000-00005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sugita S, Sudhof TC. The RIM/NIM family of neuronal C2 domain proteins: Interactions with Rab3 and a new class of Src homology 3 domain proteins. The Journal of Biological Chemistry. 2000;275:20033–20044. doi: 10.1074/jbc.M909008199. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E, Thomas K, Sidebotham H, Emond A. Prevalence and characteristics of autistic spectrum disorders in the ALSPAC cohort. Developmental Medicine and Child Neurology. 2008;50:672–677. doi: 10.1111/j.1469-8749.2008.03042.x. [DOI] [PubMed] [Google Scholar]
- Witwer AN, Lecavalier L. Examining the validity of autism spectrum disorder subtypes. Journal of Autism and Developmental Disorders. 2008;38:1611–1624. doi: 10.1007/s10803-008-0541-2. [DOI] [PubMed] [Google Scholar]
- Woodbury-Smith MR, Volkmar FR. Asperger syndrome. European Child & Adolescent Psychiatry. 2009;18:2–11. doi: 10.1007/s00787-008-0701-0. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Geneva, Switzerland: World Health Organization; 1992. The ICD-10 classification of mental and behavioural disorders. [Google Scholar]
- Ylisaukko-oja T, Nieminen-von Wendt T, Kempas E, Sarenius S, Varilo T, et al. Genome-wide scan for loci of Asperger syndrome. Molecular Psychiatry. 2004;8:879–884. doi: 10.1038/sj.mp.4001385. [DOI] [PubMed] [Google Scholar]
- Ylisaukko-oja T, Alarcón M, Cantor RM, Auranen M, Vanhala R, et al. Search for autism loci by combined analysis of autism genetic resource exchange and Finnish families. Annals of Neurology. 2006;59:145–155. doi: 10.1002/ana.20722. [DOI] [PubMed] [Google Scholar]
Web site references
- 1. http://www.agre.org/; Autism Genetics Resource Exchange (AGRE).
- 2. http://expression.gnf.org; Gene Expression Atlas.
- 3. http://www.hapmap.org/; International Hap-Map Consortium.
- 4. http://www.ncbi.nlm.nih.gov/RefSeq/; RefSeq.
- 5. http://www.ncbi.nlm.nih.gov/UniGene; UniGene.
- 6. http://www.chg.duke.edu/research/simla3_3gui.html SIMLA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.