Abstract
We report a method for conformal nanopatterning of extracellular matrix proteins onto engineered surfaces independent of underlying microtopography. This enables fibronectin, laminin, and other proteins to be applied to biomaterial surfaces in complex geometries inaccessible using traditional soft lithography techniques. Engineering combinatorial surfaces that integrate topographical and biochemical micropatterns enhances control of the biotic-abiotic interface, used here to understand cardiomyocyte response to competing physical and chemical cues in the microenvironment.
The nano- and microscale patterning of biomaterial surfaces has enabled the development of advanced systems to control cell structure and function. Specifically, engineering topographical, chemical and/or mechanical cues in defined geometries has demonstrated the ability to directly regulate cell adhesion, morphology, cytoskeletal organization, and cell-cell interactions. The technology to do this is based primarily on photolithographic techniques used to create nano- or micropatterned masters (typically silicon wafers) that are replica molded to create topographically patterned surfaces in other materials such as hydrogels and elastomers. These are used directly for cell culture or are formed into stamps and microfluidic systems to pattern ECM proteins, growth factors, and other bioactive molecules onto surfaces1. Researchers have shown that these nanometer and micrometer scale patterns of topography and biochemistry can each align cells, organize anisotropic tissue sheets, and modulate gene expression profiles2, 3. There is also evidence of the synergistic effect of combining these patterned cues into an integrated surface, such as for the enhanced alignment of neurons4 and endothelial cells5. However, to date, the ability to independently engineer microtopography and patterned chemistry into hierarchically structured surfaces has been limited due to the technical challenge of chemical patterning onto rough surfaces.
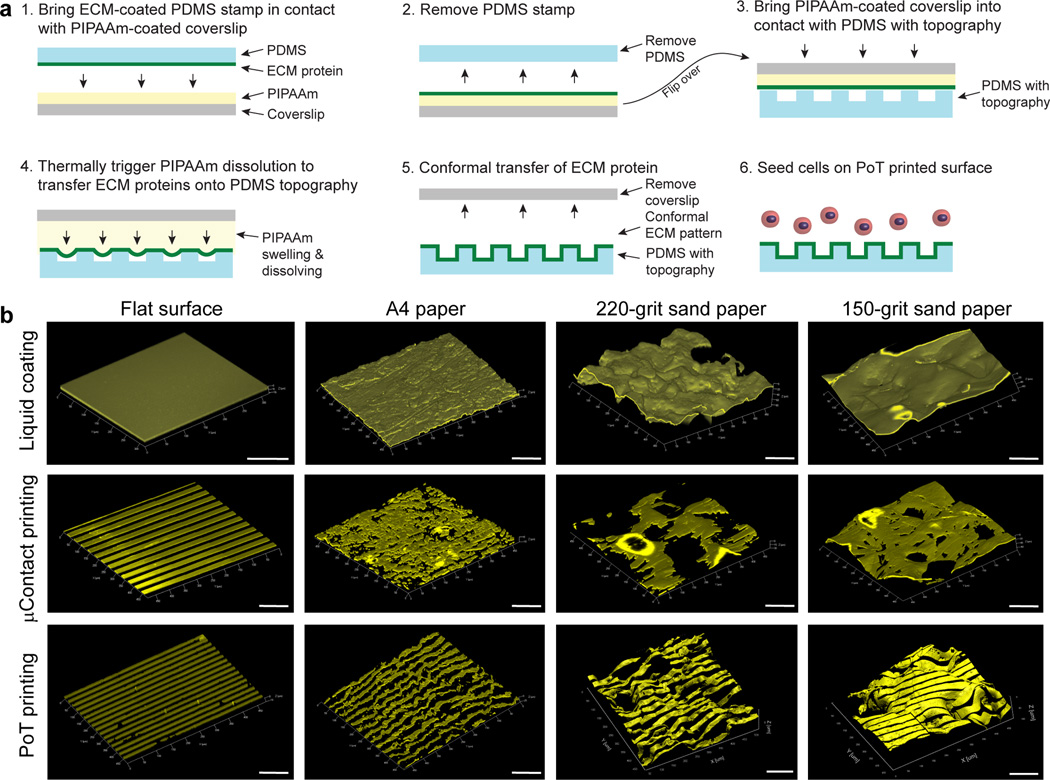
Here we report development of the Patterning on Topography (PoT) printing technique, which is able to directly transfer ECM proteins in defined geometries from a smooth release surface onto a microtopographically complex surface while substantially maintaining pattern fidelity (Fig. 1a and Online Methods). Briefly, thermally-sensitive poly(N-isopropylacrylamide) (PIPAAm) is spincoated onto glass coverslips (Fig. 1a step 1 and Supplementary Fig. 1) and then an ECM protein is patterned onto the PIPAAm using microcontact printing (µCP) with a polydimethylsiloxane (PDMS) stamp (Fig. 1a step 2). Next, a topographically patterned surface is brought into contact with the ECM patterned PIPAAm-coated coverslip (Fig. 1a step 3), submerged in distilled water at 40°C and then slowly cooled to room temperature. As the PIPAAm transitions through its lower critical solution temperature at ~35°C, the PIPAAm swells and pushes the patterned ECM protein as an ~5 nm thick layer6, 7 onto the adjacent, topographically patterned surface where it adheres due to hydrophobic interactions (Fig. 1a step 4). As the PIPAAm continues to swell it eventually dissolves (Fig. 1a step 5) and the PoT printed surface can be used for cell seeding and culture (Fig. 1a step 6).
Figure 1.
The Patterning on Topography (PoT) printing technique can transfer nano- and micropatterns of ECM proteins onto microtopographically patterned surfaces. (a) A schematic of the PoT process shows that (1) microcontact printing with a PDMS stamp is used to transfer ECM proteins onto a thin layer of PIPAAm spincoated onto a coverslip and then (2) the PDMS stamp is removed. (3) Then a PDMS substrate with surface microtopography is placed in conformal contact. (4) Distilled water at 40°C is used to hydrate the PIPAAm and then thermally-controlled dissolution of the PIPAAm causes it to swell and push the ECM pattern onto the microtopography. (5) Once release has occurred, the patterned ECM adheres to the microtopography (via non-specific hydrophobic binding) and (6) is used as a substrate for cell culture. (b) Representative 3D confocal images of flat PDMS controls and PDMS negatives of A4 paper, 220-grit sandpaper and 150-grit sandpaper coated with FN adsorbed from solution, microcontact printed with 20 µm wide, 20 µm spaced FN lines or PoT printed with 20 µm wide, 20 µm spaced FN lines. Only PoT printing can transfer the pattern with good fidelity on all surfaces and conformally follow the surface topography. Scale bars are 100 µm.
The unique capabilities of PoT printing to pattern ECM proteins on topographically patterned surfaces are clearly demonstrated when compared to standard µCP and protein coatings adsorbed from solution. To show this, we used PDMS either spin coated on glass coverslips as a flat control surface or cast against A4 paper, 150-grit sandpaper or 220-grit sandpaper. These surfaces were chosen because the heterogeneous distribution of feature width, depth and morphology enabled us to simultaneously evaluate the ability to pattern a wide range of microscale feature dimensions. We examined the full range of test surfaces and used confocal imaging and 3D rendering to evaluate PoT printing fidelity (Fig. 1b). As expected, the spincoated PDMS surface could be patterned with PoT or µCP, with no discernible difference. In comparison, even the A4 paper was rough enough to present challenges to µCP with a collapse of the line pattern and gaps in pattern transfer, causing a loss of fidelity. Results were worse on the rougher 220- and 150-grit sandpaper surfaces, with FN transferred in patches and large gaps on the order of 100’s of micrometers. In contrast, the PoT printed surfaces had well-transferred and conformal FN lines that maintained pattern fidelity and followed surface contours, even on the sandpaper surfaces (Fig. 1b and Supplementary Fig. 2).
Next, we used PoT to pattern ECM protein lines onto micro-ridges with defined geometries in order to determine the limits of the technique. Test surfaces with 20 µm wide, 20 µm spaced micro-ridges demonstrated that we could PoT print up to 37 µm deep, a depth-to-width aspect ratio of 1.85 (Supplementary Fig. 3). The FN lines were conformal to the ridge tops, bottom of the trenches and the vertical sidewalls, even on undercuts. Further, we could PoT print on deeper, 48 µm, micro-ridges with a 2.4 aspect ratio corresponding to nearly 500% strain of the FN, however, there were gaps in the FN on the sidewalls (see Supplementary Note for further discussion on PoT limitations). Varying feature dimensions demonstrated PoT printing on micro-ridges 2 µm wide, 2 µm spaced and 2.5 µm deep up to 200 µm wide, 200 µm spaced and 36 µm deep (Supplementary Fig. 4) and we expect that smaller (sub-micron) and larger features can also be patterned based on the sandpaper results (Fig. 1b and Supplementary Fig. 2). We also validated that other ECM proteins could be PoT printed including laminin and collagen type IV and that multiple proteins could be combined, such as FN and laminin lines orthogonal to each other and 45° to the micro-ridges (Supplementary Fig. 5). While we refer to PoT as a nanopatterning technique because the transferred ECM proteins are ~5 nm thick6, 7, we verified that PoT can also print conformal FN lines 100–500 nm wide and 5 µm spaced on 15 µm deep micro-ridges (Supplementary Fig. 6). Finally, control experiments confirmed (i) that µCP the FN lines using a PDMS stamp with a low Young’s modulus (E ~50 kPa) to better conform to the micro-ridges failed to achieve results comparable to PoT (Supplementary Fig. 7) and (ii) that FN lines could also be PoT printed on stiffer photoresist micro-ridges (Supplementary Fig. 8, see Supplementary Note for further discussion on µCP techniques compared to PoT).
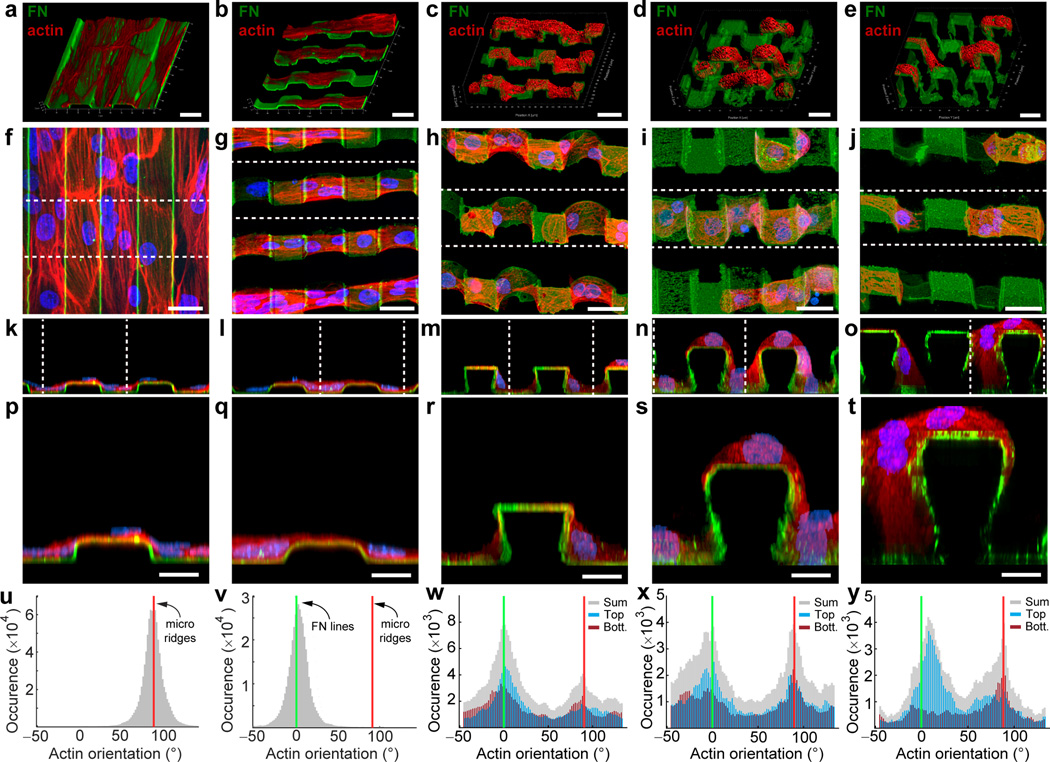
Next, we used PoT printing to investigate cell response to surfaces presenting orthogonal topographic and chemical guidance cues. This is important for applications such as cardiac tissue engineering, where cardiomyocytes (CMs) can be engineered into anisotropic 2D tissues using surfaces with either micro-ridges8 or micropatterned9, 10 FN lines, but what happens when CMs are presented with both alignment cues? Control 20 µm wide, 20 µm spaced micro-ridges uniformly coated with FN and seeded with chick CMs resulted in cell alignment parallel to the microtopography (Fig. 2a). In contrast, CMs seeded on micro-ridges with orthogonal PoT printed FN lines followed the FN lines despite having to traverse relatively large topographic features (Fig. 2b–e). Notably, the CMs were able to spread and deform around the micro-ridges (Fig. 2f–t) while displaying a characteristic sarcomeric structure (Supplementary Fig. 9). To quantify this difference in CM response, we analyzed actin alignment relative to the micro-ridges and FN lines (Fig. 2f–j). On the uniformly FN coated surface, CMs aligned in parallel with the underlying micro-ridges (Fig. 2u). However, on the PoT printed surface with 5 µm micro-ridges CMs aligned with the underlying FN lines (Fig. 2v). Interestingly, as the depth increased CM alignment became bimodal, with cells on the ridge tops continuing to align to the FN lines and cells in the trenches shifting alignment 90° to the microtopography (Fig. 2w–y). This demonstrated that CM response was depth dependent and cross-sectional views suggested that adhesion to the ridge sidewalls influenced this process (Fig. 2r–t). When vertical sidewalls were not present and changes in height were more gradual, such as for FN lines PoT printed on a PDMS replica of 220-grit sandpaper, CM alignment followed the protein pattern (Supplementary Fig. 10). Control experiments confirmed that stability in culture and cell response to PoT printed FN lines were similar to standard µCP FN lines (Supplementary Fig. 11).
Figure 2.
PoT printing provides independent control of micropatterned chemistry and microtopography to direct cell alignment on surfaces. (a) Example of 20 µm wide, 20 µm spaced, and 5 µm tall micro-ridges in PDMS uniformly coated with FN (green). The actin cytoskeleton (red) of CMs cultured on this surface aligns with the topography. 20 µm wide FN lines were also PoT printed orthogonal to micro-ridges that were (c) 15.5 µm, (d) 26 µm, and (e) 33 µm tall. Note that CMs follow the FN lines that conform to the top, bottom and sides of the ridges. (f–j) Top view of the surfaces in (a–e) with CMs additionally stained for nuclei (blue). (k–o) Cross-sections of the areas highlighted by dashed lines in (f–j). (p–t) Single ridges from the cross-sections in (k–o) show that the CMs are able to conform around the topography, even for 33 um tall micro-ridges. (u–y) Histograms of actin orientation angles show CMs are aligned along the topography on (u) FN adsorbed micro-ridge surfaces but follow the orthogonal PoT printed FN lines (v) on 5 um tall micro-ridges. (w) At the 15.5 µm height actin orientation starts to become bimodal, with CMs in the trenches showing some alignment to the micro-ridges. At the taller (x) 26 µm and (y) 33 µm heights CMs show increased actin alignment to the micro-ridges. Images are representative of n=3 samples for each depth. Histograms (u–y) are based on the actin alignment of the images in (f–j).
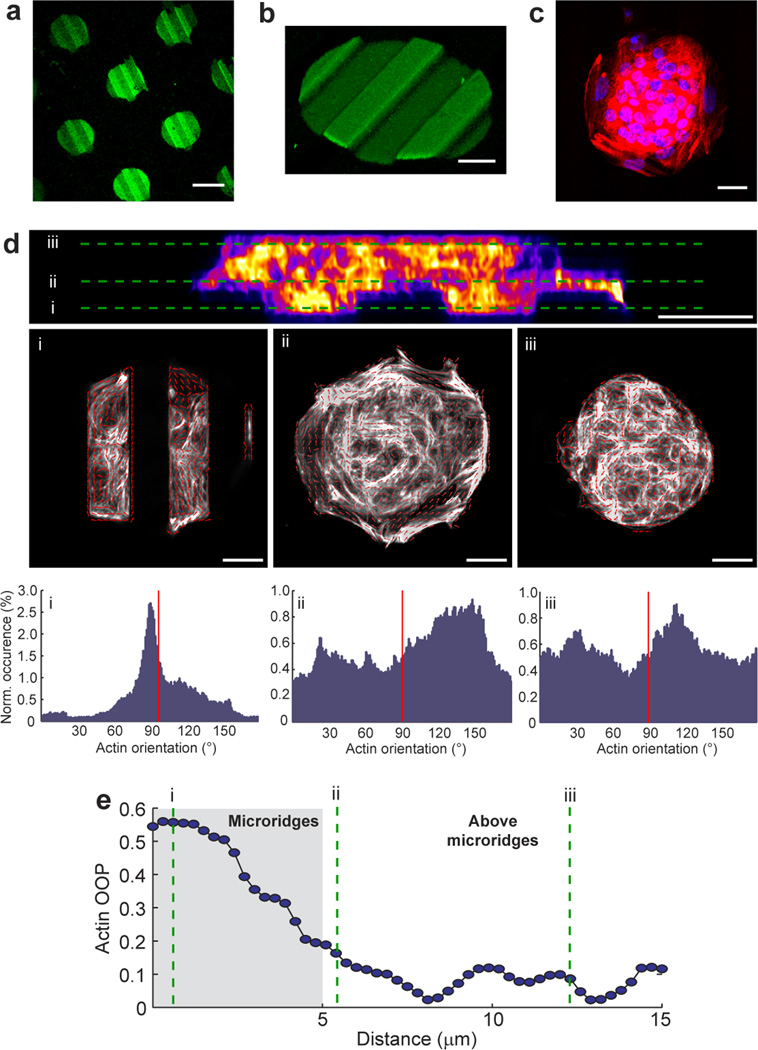
Finally, we used PoT printed FN circles to produce spatially variable guidance cues where the circle edge changed from being parallel to orthogonal to the underlying micro-ridges at a π/2 interval. To do this, we patterned an array of 100 µm diameter FN circles on 20 µm wide, 20 µm spaced and 5 µm tall micro-ridges (Fig. 3a). As expected, the FN circles conformally coated the ridge tops, sidewalls and trench bottoms (Fig. 3b) and the seeded CMs formed a cardiac microtissue that covered the entire surface (Fig. 3c). The microtissue was ~15 µm thick with distinct differences in CM alignment based on vertical position (Fig. 3d). Analysis of actin alignment (Fig. 3e) showed that CMs in the grooves were aligned to the micro-ridges, above the micro-ridges were aligned to the edge of the FN circle and above that appeared completely isotropic with no apparent guidance from the aligned CMs below. Increasing the diameter of the FN circle from 100 µm to 250 or 500 µm (Supplementary Figs. 12 and 13) showed that CM alignment to the edge of the FN circle extended ~100 µm from the periphery. While this is a basic example of combining spatially variable microtopographical and chemical cues, it clearly demonstrates that (i) in our system patterned FN overrides topography in terms of CM alignment and (ii) that CM alignment to either cue depends on direct physical contact with limited ability to propagate the alignment to neighboring CMs
Figure 3.
Circular areas of fibronectin can be patterned onto micro-ridges to investigate the interplay between micropatterned topography and chemistry. (a) An array of 100 µm diameter FN circles patterned on top of 20 µm wide, 20 µm spaced, 5 µm tall micro-ridges in PDMS. (b) 3D rendering of a 100 µm diameter FN circle on the micro-ridges. (c) Fluorescent image of CMs on the 100 µm diameter FN circle stained for nuclei (blue) and actin (red). (d) Analysis of actin alignment throughout the thickness of a cardiac micro-tissue showing a cross-section through the tissue (false colored, actin staining) with threes region selected; (i) in the micro-ridge area, (ii) just above the micro-ridge area and (iii) well above the micro-ridge area. Each region has a gray scale image with an overlay of actin orientation vectors (red arrows) and a histogram of actin orientation throughout the image (red line indicates micro-ridge orientation). (e) Quantification of the actin orientational order parameter (OOP) of the cardiac micro-tissue in (d) demonstrates that the topography is the dominant guidance cue when cells are in direct contact, but as soon as cell are above the topography the FN pattern becomes the dominant guidance cue. Images are representative of n=10 samples for each condition. Scale bars are 100 µm in (a) and (e) and 20 µm in all other panels.
In summary, PoT printing enables the nano- to microscale patterning of ECM proteins onto microtopographically complex surfaces. Unique is the independent control of physical and chemical surface properties to modulate cell response. This provides a methodology to probe how cells respond to competing topographical and chemical cues and to engineer surfaces that maximize a given behavior, such as organization of CM into an aligned cardiac tissue. Looking forward, PoT printing enables improved control over the cellular microenvironment, which will be useful for understanding basic cell-surface interactions as well as engineering more advanced biomaterial surfaces where topography and chemistry can be independently optimized.
ONLINE METHODS
Fabrication of microtopographically modified surfaces
Polydimethylsiloxane (PDMS, Sylgard 184, Dow-Corning) was mixed in 10:1 base resin to curing agent ratio, degassed and then used to create the different microtopographically modified surfaces in this study. Smooth control surfaces were fabricated by spin coating the PDMS on 25 mm diameter glass coverslips at 4,000 RPM and cured at 65°C for 4 h to create ~15 µm thick films11. Surfaces with random microtopographies were fabricated by creating PDMS replicas (negatives) of A4 printer paper and 220-grit and 150-grit sandpaper. To do this a 4x4 cm square of the paper was placed in the bottom of a 10 cm petri dish and PDMS was poured on top to an approximate thickness of 0.5 cm. The PDMS was cured at 65°C for 4 h, peeled off and then cut into 1x1 cm squares. Surfaces with micro-ridge topographies were fabricated by using photolithography according to previously published methods7, 11. The materials used for photolithography varied depending on the photoresist layer thickness. For photoresist layers with a thickness between 2 and 10 µm, SPR 220.3 photoresist (Microchem) was spincoated onto glass wafers. Thicker photoresist layers between 10 and 40 µm were prepared by spin coating SU-8 2015 photoresist (Microchem) onto silicon wafers. SU-8 2050 photoresist (Microchem) was spincoated onto silicon wafers to generate photoresist layers thicker than 40 µm. All photoresist layers were exposed to UV light through a transparency photomask and then developed using SU8 developer or MF-26A developer (SU8 and SPR respectively, Microchem) to create SU-8 or SPR micro-ridges on top of silicon or glass wafers. Patterned wafers were then coated with PDMS to a thickness of 0.5 cm, cured at 65°C for 4 h, peeled off and then cut into 1x1 cm squares.
Uniform coating and microcontact printing (µCP) of ECM proteins
For uniform protein coatings adsorbed from solution, the PDMS substrates were sonicated in 50% ethanol for 30 min and dried by nitrogen. The ECM protein fibronectin (FN) (Corning, catalog number 356008) was diluted to a concentration of 50 µg/mL in distilled water; 40% of the FN was conjugated to Alexa Fluor 488 fluorescent dye (Life Technologies). A 300 µL droplet of FN solution was incubated on the PDMS substrates for 15 minutes, rinsed 2 times with distilled water and then dried under nitrogen. For µCP, PDMS stamps with micro-ridges 20 µm wide, 20 µm spaced and 2 µm tall were fabricated using photolithography as described above for the microtopographically modified surfaces. PDMS stamps were sonicated in 50% ethanol for 30 min and dried by nitrogen, incubated with FN solution for 1 h, rinsed 2x with distilled water and then dried under nitrogen. The µCP of laminin (LAM, Invitrogen) and collagen type IV (COL IV, Sigma) were performed similarly except both were diluted to working concentrations of 100 µg/mL in distilled water. The PDMS substrates to be patterned were UV/Ozone treated for 15 minutes and then brought into conformal contact with the PDMS stamps for 5 min. Gentle pressure by light tapping the back of the PDMS stamps with tweezers was used to ensure contact.
Patterning on topography (PoT) printing of ECM proteins
The PoT printing process is a modification of surface-initiated assembly 7 where the release of the ECM protein occurs directly onto a microtopographically structured surface. To do this, we first spincoated 25 mm diameter glass coverslips with a 10% poly (N-isopropylacrylamide) (PIPAAm, Polysciences, Inc.) in 1-butanol (w/v) solution at 6,000 RPM. To PoT print features with a depth-to-width aspect ratio higher than 0.5, we increased the thickness of the PIPAAm layer by spin coating glass coverslips with a 40% PIPAAm in 1-butanol (w/v) solution at 2,000 RPM. PIPAAm layer thickness was determined by first spin coating PIPAAm at different concentrations and then making a small scratch through the PIPAAm layer with a razor blade. Next, we used atomic force microscopy (AFM, MFP3D, Asylum Research) in air using AC mode with AC160TS-R3 cantilevers (Olympus Corporation) to scan over the scratch. The thickness of the PIPAAm layer was then quantified using the IGOR Pro software environment (WaveMetrics, Inc.). Prior to PoT printing, the PIPAAm coated coverslips were sterilized using high-intensity UV light for 15 min. Fluorescently labeled FN, LAM and COLIV were then patterned onto the PIPAAm coated coverslips by µCP with various geometries including, (i) 20 µm wide, 20 µm spaced lines; (ii) 100 µm diameter circles; (iii) 250 µm diameter circles; and 500 µm diameter circles. Next, PDMS substrates with microtopography on the surface were sterilized using high-intensity UV light for 15 min and placed in conformal contact with the protein patterned, PIPAAm coverslips for 10 min. Making sure to keep the PDMS stamp and PIPAAm surface in contact, the samples were placed within the well of a 6-well plate, 40°C sterile distilled water was poured onto the stamp and PIPAAm, and then the plate was placed in 37°C incubator for at least 3 hours to allow time for water to enter the channels or spaces and diffuse into the PIPAAm. The plate was then removed from the incubator and placed in the biosafety cabinet at room temperature for 1 hour in order to dissolve the PIPAAm layer and release the ECM patterns onto the PDMS substrates. Once the PIPAAm dissolved, the PDMS and coverslip were no longer attached and the PoT printed PDMS substrate could then be removed and used for cell culture or analysis. We also tested the ability to PoT print onto very stiff substrates by placing an SPR 220.3 master mold containing 20 µm wide, 20 µm spaced, and 3 µm tall micro-ridges into conformal contact with a PIPAAm coated coverslip patterned with 20 µm wide and 20 µm spaced FN lines. For all experiments, PoT printed substrates that had defects due to poor contact with the PIPAAm-coated coverslip or to improper water penetration within the topography were discarded. At least three samples per condition were used for imaging and cell seeding.
Cardiomyocyte isolation and culture
Chick cardiomyocytes (CMs) were isolated from day 7–8 (HH stages 30–32) White Leghorn embryos and are not considered live vertebrate animals under PHS policy. For each isolation, 7–15 hearts were dissected, cut in small pieces and digested in 3 mL of 0.05% Trypsin-EDTA (Sigma-Aldrich) for 7 minutes. The supernatant was carefully collected, added to seeding media (M199 media, 10% heat-inactivated fetal bovine serum, 1% Penicillin/streptomycin) and filtered through a 40 µm cell strainer to isolate individual cells. The 7 minute incubation was repeated 3 times so that the hearts were completely digested while minimizing cytotoxicity by long exposure to trypsin. The cell solution was then centrifuged at 50 g for 10 minutes and resuspended in 30 mL of seeding media. Fibroblasts were removed by selective adhesion by pre-plating successively for 45 minute incubations in T75 cell culture flasks. The supernatant, enriched in CMs, was centrifuged at 50 g for 10 min then resuspended in seeding media at 7.5x105 cells/mL. PDMS substrates were placed in a 6-well plate, 200 µL of the cell suspension was pipetted on top and incubated for 2 hours to let the cells attach and then the wells were filled with additional seeding media. The next day the media was replaced with a maintenance media with a reduced level of 2% heat-inactivated fetal bovine serum to minimize fibroblast proliferation.
Fluorescent staining and imaging
The CMs and ECM proteins were fluorescently labeled in order to analyze the surface structure and cell response. The FN (human, Corning, catalog number 356008) was fluorescently labeled prior to use with fluorescent dyes, either Alexa Fluor 546 maleimide or Alexa Fluor 488 carboxylic acid, succinimidyl ester (Life Technologies) according to published methods12. The fluorescently labeled FN was combined with unlabeled FN at volume concentration of 40%. PDMS surfaces PoT printed with LAM or COL IV were fluorescently stained using monoclonal primary antibodies (Sigma, catalog number L9393 for LAM, C1926 for COL IV) followed by staining using Alexa Fluor 488 conjugated goat anti-mouse secondary antibodies (Life Technologies, catalog number A-11001). The CMs were cultured for 2 days, removed from the incubator, briefly washed with 37°C PBS and then fixed for 15 minutes in 4% paraformaldehyde with 0.05% TritonX-100. Next, samples were blocked for 1 hour with 5% goat serum and then incubated with 1:200 DAPI, 1:100 primary monoclonal antibody against sarcomeric alpha-actinin (Sigma, catalog number A7811) and 1:60 Phalloidin conjugated to Alexa Fluor 633 (Life Technologies) for 2 hours at 37°C. Samples were subsequently washed 3 times for 10 min in PBS and then incubated with goat anti-mouse secondary antibody conjugated to Alexa Fluor 488 for 2 hours at 37 °C. Finally, samples were washed 3 times for 10 min in PBS and mounted against No. 1.5 glass coverslips with Prolong Gold (Life Technologies). Substrates were imaged using a Zeiss LSM 700 laser scanning confocal microscope with a 40x (NA = 1.3) and 63x (NA=1.4) oil immersion objectives or a Leica SP5 multiphoton microscope with a 25x (NA=0.95) water immersion objective. 3D image stacks were deconvolved using AutoQuant X3 (Media Cybernetics, Inc.). Image processing and 3D rendering were performed using Zen 2010 (Zeiss), ImageJ image processing software (NIH) and Imaris (Bitplane, Inc.).
Image analysis of actin alignment
The alignment of actin filaments was analyzed using a modified ridge detection algorithm according to previously published methods13. Briefly, Phalloidin labeled images of actin filaments were captured as Z-stacks for CMs on the PoT printed surfaces. The 8-bit gray scale images were normalized and an orientation vector map was generated from the pixel intensity gradients. These orientation vectors were found parallel to the actin filaments and used to generate a histogram where cytoskeletal anisotropy was indicated be a distinct peak at a specific angle. For deeper micro-ridges, sub Z-stacks for the top and bottom halves of the samples were created and analyzed. The 2D orientational order parameter (OOP) was used as a metric of alignment9, which scales from 0 to 1, with 0 indicating complete isotropic orientation and 1 indicating perfect anisotropic orientation.
PoT printing of nanoscale ECM patterns
To fabricate FN nanofibers with a submicron width, we used a Photonic Professional GT 3D printer (Nanoscribe GmbH). Briefly, a master mold containing 250 µm long, 5 µm wide, 2 µm high, and 500 nm spaced features was first designed in AutoCAD (Autodesk Inventor) and exported into DeScribe 2.0 software. The master mold was then 3D printed using IP-L photoresist. Unexposed regions were then removed by immersion in a PGMEA developer solution (Microchem). PDMS stamps containing the negative of the master mold (yielding 500 nm wide features) were prepared by casting the PDMS pre-polymer solution over the master mold as previously mentioned. To accurately assess their nanoscale dimensions, the FN was µCP onto PIPAAm-coated glass coverslips and scanned using AFM in air using AC mode with AC160TS-R3 cantilevers (Olympus Corporation). High-resolution AFM images were obtained using a scan size of 1024×1024 lines over a scan area of 8 µm×8 µm. The nanofiber width was quantified using the IGOR Pro software environment (WaveMetrics, Inc.).
Stability of PoT printed ECM patterns in culture
PDMS coated coverslips were either µCP or PoT printed with 20 µm wide and 20 µm spaced FN lines that were fluorescently labeled as previously described. All substrates were treated with 1% Pluronics F-127 (Sigma-Aldrich) for 15 min, washed three times with PBS, and then placed in separate wells of a 6-well plate. For each condition, three coverslips were seeded with 50,000 C2C12 mouse myoblasts (ATCC, CRL-1772) per well in growth medium (DMEM, 10% fetal bovine serum, 1% Penicillin/streptomycin, 1% L-glutamine) while three coverslips were kept in growth medium without cells. After three days in culture, all samples were fixed and cell samples were stained for nuclei and F-actin and imaged as described earlier. C2C12 cells were authenticated and certified mycoplasma-free by the manufacturer.
Fabrication of low Young’s modulus PDMS stamps for µCP
To determine the depth-to-width aspect ratio at which µCP is unable to pattern topographical surfaces, we fabricated low Young’s modulus PDMS stamps as described previously11. Briefly, stamps with an elastic modulus of ~50 kPa were prepared by mixing Sylgard 184 and Sylgard 527 in a 10:1 ratio (w:w). Sylgard 184 was prepared as described previously while Sylgard 527 was prepared by mixing equal parts of A and B components per manufacturer’s instructions. The 10:1 mixture of Sylgard 184 and Sylgard 527 was then cast over a SPR 220.3 master mold and cured at 65 °C for 4 hours. Prior to casting, the SPR 220.3 master mold was silanized (PlusOne Repel-Silane ES, GE Healthcare) in a vacuum desiccator for at least 4 hours. After curing, the stamps were peeled off of the master mold, coated with FN as previously described and then µCP orthogonally onto PDMS micro-ridges.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge financial support from China Scholarship Council (CSC) for State Scholarship Fund No. [2008]3027 to Y.S., the Carnegie Mellon University Dowd-ICES Fellowship to Q.J., the NIH Biomechanics in Regenerative Medicine T32 training grant (2T32EB003392) and the Bertucci Graduate Fellowship to J.M.S. and the NIH Director’s New Innovator Award (1DP2HL117750) to A.W.F.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
A.W.F. conceived and supervised the project. A.W.F. and Y.S. designed the experiments. Y.S., Q.J. and J.M.S. performed the experiments. A.W.F, Q.J., J.M.S. and Y.S. analyzed the data and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
A.W.F. is a co-inventor on a filed patent application covering the surface-initiated assembly process that is part of the PoT technique described in the present study.
REFERENCES
- 1.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 2.Bettinger CJ, Langer R, Borenstein JT. Angewandte Chemie International Edition. 2009;48:5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falconnet D, Csucs G, Michelle Grandin H, Textor M. Biomaterials. 2006;27:3044. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Greene AC, Washburn CM, Bachand GD, James CD. Biomaterials. 2011;32:8860–8869. doi: 10.1016/j.biomaterials.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AW, et al. Journal of Biomedical Materials Research Part A. 2007;86A:522–534. doi: 10.1002/jbm.a.31626. [DOI] [PubMed] [Google Scholar]
- 6.Szymanski JM, Jallerat Q, Feinberg AW. Journal of Visualized Experiments. 2014:e51176. doi: 10.3791/51176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feinberg AW, Parker KK. Nano Letters. 2010;10:2184–2191. doi: 10.1021/nl100998p. [DOI] [PubMed] [Google Scholar]
- 8.Chung C-y, et al. The FASEB Journal. 2011;25:851–862. doi: 10.1096/fj.10-168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg AW, et al. Biomaterials. 2012;33:5732–5741. doi: 10.1016/j.biomaterials.2012.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg AW, et al. Science. 2007;317:1366–1370. doi: 10.1126/science.1146885. [DOI] [PubMed] [Google Scholar]
- 11.Palchesko RN, Zhang L, Sun Y, Feinberg AW. PLoS ONE. 2012;7:1–13. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klotzsch E, et al. Proceedings of the National Academy of Sciences. 2009;106:18267–18272. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray M-AP, et al. Biomaterials. 2010;31:5143–5150. doi: 10.1016/j.biomaterials.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.