Abstract
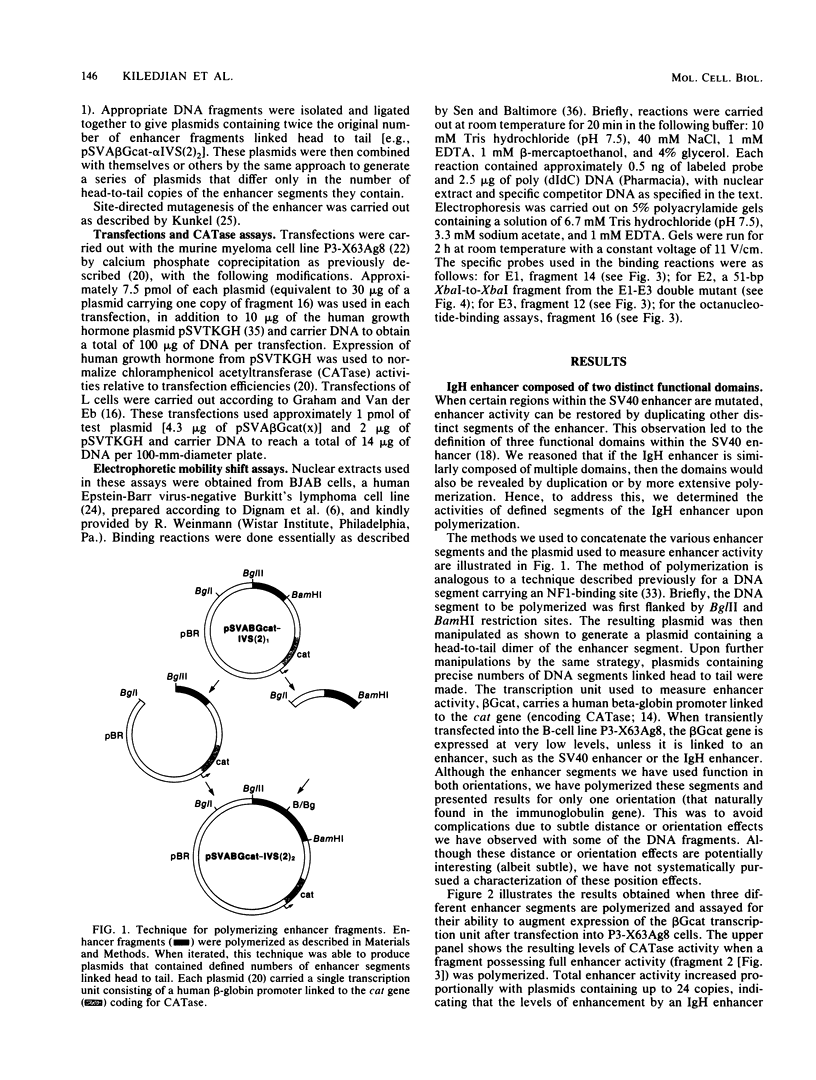
We have investigated the effect of polymerizing defined segments of the immunoglobulin heavy-chain enhancer on the activity of a single, linked transcription unit. Transient assays in lymphoid cells have led to the following observations. First, polymerizing the entire enhancer led to an increase in overall transcription. Second, polymerizing defined DNA segments revealed two distinct functional domains within the enhancer. Although each domain alone possessed only partial enhancer activity, greater than wild-type levels of activity could be obtained upon polymerization. One of these domains contains three regions thought to be involved in protein binding in vivo and in vitro (E motifs E1, E2, and E3). The other domain contains the fourth E motif (E4) and the conserved octanucleotide, ATTTGCAT. We have tested the functional importance of these motifs by determining the effect of mutating these elements singly or in combination in the context of the isolated domains. Although E2, E3, E4, and the octanucleotide are clearly important for enhancer function, mutation of the E1 motif did not appear to have an effect on enhancer activity in our assay. Transient assays in mouse L cells indicate that nonlymphoid cells are able to use a distinct subset of these motifs.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Augereau P., Chambon P. The mouse immunoglobulin heavy-chain enhancer: effect on transcription in vitro and binding of proteins present in HeLa and lymphoid B cell extracts. EMBO J. 1986 Aug;5(8):1791–1797. doi: 10.1002/j.1460-2075.1986.tb04428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Borrelli E., Hen R., Wasylyk C., Wasylyk B., Chambon P. The immunoglobulin heavy chain enhancer is stimulated by the adenovirus type 2 E1A products in mouse fibroblasts. Proc Natl Acad Sci U S A. 1986 May;83(9):2846–2849. doi: 10.1073/pnas.83.9.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I., Fromental C., Augereau P., Wildeman A., Zenke M., Chambon P. Cell-type specific protein binding to the enhancer of simian virus 40 in nuclear extracts. Nature. 1986 Oct 9;323(6088):544–548. doi: 10.1038/323544a0. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Falkner F. G., Mocikat R., Zachau H. G. Sequences closely related to an immunoglobulin gene promoter/enhancer element occur also upstream of other eukaryotic and of prokaryotic genes. Nucleic Acids Res. 1986 Nov 25;14(22):8819–8827. doi: 10.1093/nar/14.22.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T., Matthias P., Thali M., Jiricny J., Schaffner W. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 1987 May;6(5):1323–1330. doi: 10.1002/j.1460-2075.1987.tb02371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Rigby P. W., Lane D. P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985 Sep;42(2):519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Imler J. L., Lemaire C., Wasylyk C., Wasylyk B. Negative regulation contributes to tissue specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1987 Jul;7(7):2558–2567. doi: 10.1128/mcb.7.7.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch T., Zervos P., Ruezinsky D. Functional analysis of the murine IgH enhancer: evidence for negative control of cell-type specificity. Nucleic Acids Res. 1986 Oct 24;14(20):8209–8221. doi: 10.1093/nar/14.20.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Haslinger A., Holtgreve H., Richards R. I., Krauter P., Westphal H. M., Beato M. Characterization of DNA sequences through which cadmium and glucocorticoid hormones induce human metallothionein-IIA gene. Nature. 1984 Apr 5;308(5959):513–519. doi: 10.1038/308513a0. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Klein G., Giovanella B., Westman A., Stehlin J. S., Mumford D. An EBV-genome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5(6):319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Lennon G. G., Perry R. P. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5'-nontranslatable exon. Nature. 1985 Dec 5;318(6045):475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir U., Walker M. D., Rutter W. J. Regulation of rat insulin 1 gene expression: evidence for negative regulation in nonpancreatic cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3180–3184. doi: 10.1073/pnas.83.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Orth K., Calame K. L. Binding in vitro of multiple cellular proteins to immunoglobulin heavy-chain enhancer DNA. Mol Cell Biol. 1986 Dec;6(12):4168–4178. doi: 10.1128/mcb.6.12.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Selden R. F., Howie K. B., Rowe M. E., Goodman H. M., Moore D. D. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol Cell Biol. 1986 Sep;6(9):3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C., Wasylyk B. The immunoglobulin heavy-chain B-lymphocyte enhancer efficiently stimulates transcription in non-lymphoid cells. EMBO J. 1986 Mar;5(3):553–560. doi: 10.1002/j.1460-2075.1986.tb04246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger J., Baltimore D., Sharp P. A. Distinct factors bind to apparently homologous sequences in the immunoglobulin heavy-chain enhancer. 1986 Aug 28-Sep 3Nature. 322(6082):846–848. doi: 10.1038/322846a0. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]





