SUMMARY
The him-8 gene is essential for proper meiotic segregation of the X chromosomes in C. elegans. Here we show that loss of him-8 function causes profound X chromosome-specific defects in homolog pairing and synapsis. him-8 encodes a C2H2 zinc-finger protein that is expressed during meiosis and concentrates at a site on the X chromosome known as the meiotic pairing center (PC). A role for HIM-8 in PC function is supported by genetic interactions between PC lesions and him-8 mutations. HIM-8 bound chromosome sites associate with the nuclear envelope (NE) throughout meiotic prophase. Surprisingly, a point mutation in him-8 that retains both chromosome binding and NE localization fails to stabilize pairing or promote synapsis. These observations indicate that stabilization of homolog pairing is an active process in which the tethering of chromosome sites to the NE may be necessary but is not sufficient.
INTRODUCTION
Physical interactions between homologous chromosomes are essential for their proper segregation during meiosis. In most species, extensive pairing of homologous chromosomes is a meiosis-specific phenomenon that must be established de novo during meiotic prophase. Meiotic pairing is normally coordinated with the assembly of a protein scaffold called the synaptonemal complex (SC), which polymerizes between paired homologs and regulates their interactions. The mechanisms that bring homologs into physical contact, enable them to recognize each other as partners, and control the polymerization of the SC remain obscure.
In C. elegans, specific chromosome sites have been implicated in meiotic chromosome segregation. In an accompanying paper (MacQueen et al., 2005 [this issue of Cell]), we demonstrate that these sites, known as homolog recognition regions or pairing centers (PCs), play at least two distinct roles to promote homologous chromosome synapsis. Specifically, PCs stabilize homologous chromosome pairing locally and also promote SC formation (synapsis) between paired homologs.
To discover molecular components involved in PC function, we used a genetic strategy to identify loci for which a reduction in dose further compromises the partial function of a single, unpaired X chromosome PC. We found that mutations at the him-8 locus show strong synergistic interactions with lesions of the X chromosome PC. This finding catalyzed the experiments we describe here to investigate the role of him-8 in chromosome pairing and synapsis.
The him-8 locus was first identified in a general screen for meiotic mutations in C. elegans (Hodgkin et al., 1979). This screen was based on the Him (high incidence of males) phenotype that results from meiotic missegregation of the X chromosomes. Most mutations that cause this Him phenotype affect the segregation of all six chromosome pairs and consequently produce many dead (aneuploid) progeny. him-8 mutations are unusual in that they specifically impair segregation of the X chromosomes during hermaphrodite meiosis. Although a very high fraction of the self-progeny of him-8 hermaphrodites are X0 males (39%) or triplo-X hermaphrodites (6%) (Broverman and Meneely, 1994), these offspring usually receive the correct number of autosomes, resulting in very high viability rates (Table 1). For this reason, him-8 mutations are frequently introduced into worm strains as a way to generate males for genetic manipulation.
Table 1.
Fraction of Male Self-Progeny and Dead Eggs Produced by Wild-Type, him-8, and meDf2 Hermaphrodites
| Genotype | Percent Males (Total Number of Adults Scored) |
Percent Viable Embryos (Total Number of Embryos Scored) |
|---|---|---|
| Wild-type | 0.1 (1954) | 100.0 (372) |
| him-8(e1489) | 38.9 (1567) | 96.1 (1631) |
| him-8(me4) | 39.7 (1081) | 100.0 (1029) |
| him-8(mn253) | 36.2 (1315) | 100.0 (228) |
| him-8(tm611) | 37.3 (1636) | 100.0 (1560) |
| mnDp66;meDf2 | 33.9 (2078) | 100.0 (1944) |
|
mnDp66;him-8 (mn253);meDf2 |
38.2 (1408) | 100.0 (310) |
When other chromosome-specific meiotic segregation defects have been analyzed, they have usually been found to result from structural aberrations of the affected chromosome that perturb their ability to pair or synapse with their homologous partners (Baker and Carpenter, 1972; Villeneuve, 1994; Zetka and Rose, 1992). However, the him-8 locus has been mapped to the middle of chromosome IV, indicating that it cannot be a structural component of the X chromosome but instead presumably encodes a trans-acting factor specifically required for X chromosome segregation. Although X chromosome recombination is dramatically reduced in him-8 mutant hermaphrodites, there are modest levels of residual crossovers that are skewed relative to their normal distribution toward the PC end of the chromosome (Broverman and Meneely, 1994). While him-8 does not appear to provide an essential function in wild-type males, which have only a single X chromosome, mutations in him-8 do reduce crossing-over between X chromosome fragments and a normal X chromosome during male meiosis (Herman and Kari, 1989).
Here we have investigated the function of him-8 during meiosis. In addition to describing the loss-of-function phenotype of him-8 mutants, we show that him-8 encodes a C2H2 zinc-finger protein that concentrates specifically at the X chromosome PC during meiosis. HIM-8 immunostaining reveals that this locus is also associated with the nuclear envelope (NE) during meiotic prophase. Analysis of different him-8 alleles indicates that localization of this protein to the PC region and to the NE may be necessary but is not sufficient for pairing and synapsis of the X chromosomes.
RESULTS
him-8 Mutations Show Genetic Interactions with X Chromosome Pairing-Center Deficiencies
To identify components required for function of the C. elegans pairing centers, we tested genes with known or potential roles in meiosis for interactions with X chromosome PC lesions. Deletion of one copy of the X chromosome PC causes a partial loss of PC function and a modest segregation defect of 5%–7% male self-progeny (Villeneuve, 1994; MacQueen et al., 2005). A strong enhancement of X chromosome missegregation was seen when him-8 mutations were introduced into meDf2/+ heterozygotes. All known him-8 mutations behave recessively, meaning that him-8/+ heterozygous hermaphrodites do not produce elevated numbers of males. However, the loss of a single copy of him-8 increases the male production in meDf2/+ heterozygotes from 7% (n = 1649) to 18% (n = 1679) (see Experimental Procedures). This effect is not allele specific; we observed similar levels of X chromosome nondisjunction when we combined meDf2/+ with two previously characterized alleles (e1489 and mn253) and one new allele (me4) of him-8 (data not shown). This enhancement of the meDf2/+ phenotype suggested that him-8 might function in the same process as the meiotic PC.
him-8 Mutants Exhibit Defective Pairing and Synapsis of the X Chromosome
Previous work has shown that him-8 mutations lead to defects in X chromosome segregation similar to those resulting from deletion of the X chromosome PC. Specifically, mutations in him-8 produce a very high incidence of male self-progeny (Broverman and Meneely, 1994; Herman and Kari, 1989; Hodgkin et al., 1979; this work), a phenotype that is diagnostic for X chromosome nondisjunction during meiosis. Furthermore, both him-8 mutations and cis-acting PC lesions display a strong X chromosome-specific reduction in crossing-over with no accompanying autosomal segregation defects (Broverman and Meneely, 1994; Herman and Kari, 1989; Hodgkin et al., 1979). Nondisjunction of the X chromosome is slightly but consistently more severe in him-8 mutants than in animals lacking both X chromosome PCs (Broverman and Meneely, 1994; Villeneuve, 1994; Table 1).
We investigated whether him-8 mutations disrupt physical interactions between X chromosomes during meiotic prophase. At the onset of meiotic prophase in C. elegans, nuclei normally adopt a polarized morphology with the chromosomes concentrated asymmetrically toward one side. It is at this stage, known as the “transition zone,” that homologs pair and SC polymerization is initiated. Once all chromosomes have paired and fully synapsed with their homologs, nuclei exit from this stage and chromosomes redistribute around the nuclear periphery. In wild-type gonads stained with DAPI, there is a clear demarcation between the transition zone and the pachytene region. By contrast, in him-8 mutant hermaphrodites, entry into the transition zone occurs normally, but nuclei retain the polarized appearance typical of transition-zone nuclei well into pachytene despite the fact that most of their chromosomes appear to be synapsed (Figure 1A). A similar extended region of polarized nuclei has been described in a few other meiotic mutants that disrupt synapsis in C. elegans (Colaiácovo et al., 2003; Couteau et al., 2004; MacQueen et al., 2002). This apparent delay in chromosome reorganization at the onset of pachytene has been postulated to result from defects in chromosome synapsis. In the case of him-8, this delay is likely to be a consequence of asynapsis of a single chromosome pair.
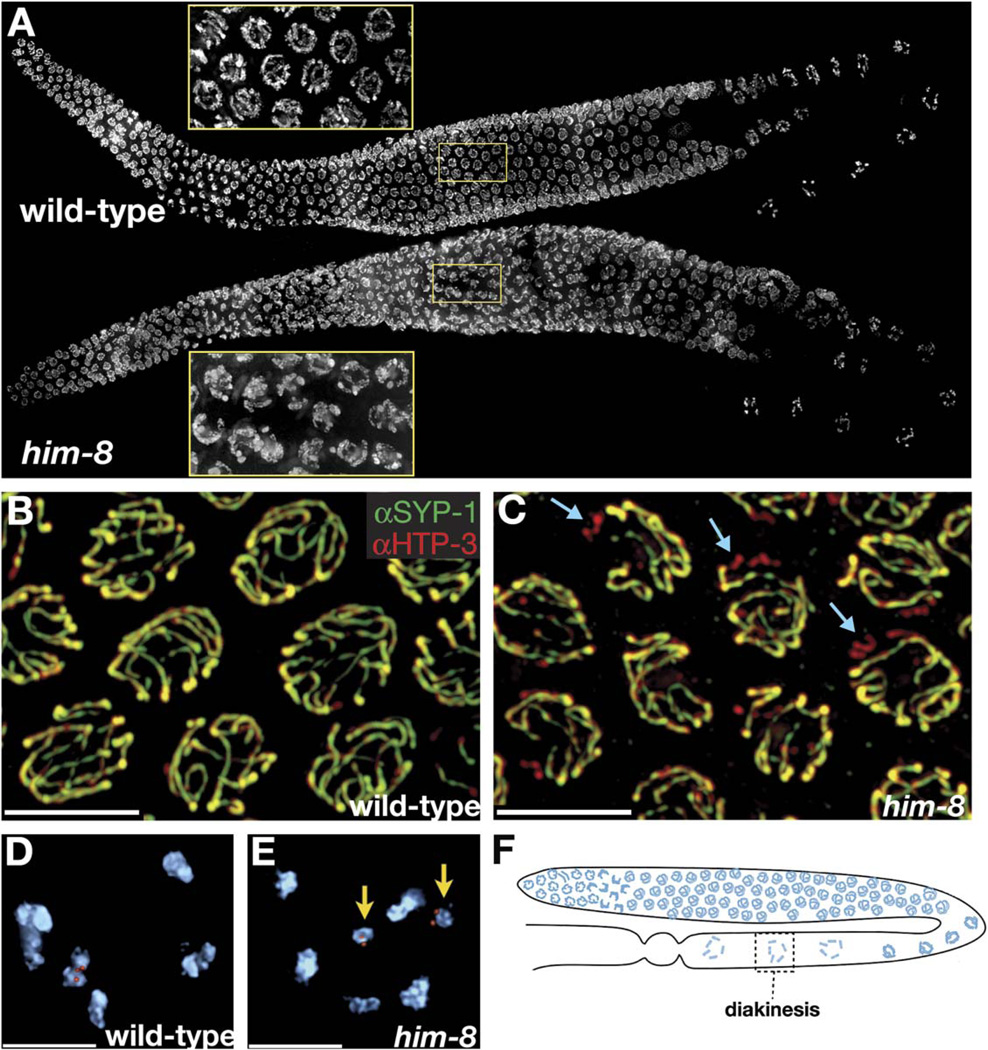
Figure 1. him-8 Mutants Display X Chromosome-Specific Defects in Synapsis and Chiasma Formation.
(A) Whole gonads from wild-type and him-8 hermaphrodites stained with DAPI. Insets magnify the pachytene region to show that him-8 nuclei retain a polarized appearance that is usually restricted to transition-zone nuclei.
(B and C) Pachytene nuclei stained with antibodies to SYP-1 (green) and HTP-3 (red).
(B) In wild-type hermaphrodites, these antibodies show very similar localization patterns at pachytene along the entire lengths of all six pairs of chromosomes.
(C) In him-8 mutant hermaphrodites we observe one pair of unsynapsed chromosomes in each nucleus. Here these are revealed as chromosomes that stain with HTP-3 (red) but not SYP-1 (green). Examples are indicated with blue arrows.
(D and E) Oocyte nuclei at diakinesis, shortly prior to the meiotic nuclear divisions.
(D) Wild-type nuclei have six DAPI staining bodies, indicating the formation of chiasmata between all six pairs of homologous chromosomes. One of these bivalents is marked by a FISH probe specific for the X chromosomes.
(E) him-8 nuclei usually reveal seven DAPI staining bodies at diakinesis. Achiasmate, or univalent, X chromosomes marked by a FISH probe are indicated by yellow arrows.
(F) Diagram showing the location of diakinesis within the worm gonad. At this stage, the SC has largely broken down and homologs are held together by chiasmata.
All images are projections of 3D images following deconvolution. Scale bars represent 5 µm.
We examined chromosome synapsis in him-8 mutants using antibodies to components of the synaptonemal complex (SC). As described in the accompanying paper (MacQueen et al., 2005), HTP-3 antibodies were used to mark the axial elements of both unsynapsed and synapsed chromosomes. SYP-1 antibodies (MacQueen et al., 2002) were used to label the central component of the SC, thereby defining regions of synapsis. In wild-type hermaphrodites, all six pairs of chromosomes load HTP-3 in early prophase, prior to homolog pairing, and acquire SYP-1 staining along their entire lengths by pachytene (Figure 1B). In him-8 mutants, one chromosome pair failed to load SYP-1 and thus (by definition) remained unsynapsed in most nuclei (Figure 1C). FISH experiments have confirmed that this single pair of unsynapsed chromosomes is consistently the Xs (data not shown). By late pachytene, some X chromosomes in him-8 hermaphrodites did appear to load SYP-1 protein along their lengths, although this staining tended to be faint and patchy relative to autosomal SYP-1 immunofluorescence. Moreover, staining with SYP-1 antibodies combined with X chromosome FISH revealed that the homologous X chromosomes are still located at a distance from each other in most cases where SYP-1 is detected on the X chromosomes (data not shown). Thus, it appears that SYP-1 can polymerize along some unpaired X chromosomes late in prophase. This autosynapsis may occur either by folding back of an unpaired X chromosome or by loading of SYP-1 along the axis between sister chromatids.
In addition to forming axial elements, the unsynapsed X chromosomes in him-8 mutants initiate meiotic recombination events, as evidenced by RAD-51-positive foci (see Figure S3 in the Supplemental Data available with this article online). We measured the frequency of chiasmata at diakinesis in him-8 oocytes by counting univalent and bivalent X chromosomes, which were hybridized with FISH probes to facilitate scoring (Figure 1E). In him-8 mutants, only 5% of oocytes (n = 100) revealed six bivalents at diakinesis, in contrast to nearly 100% of oocytes in wild-type animals. This minor population indicates that some X chromosomes successfully form chiasmata in him-8 mutants, which is not entirely surprising in light of the fact that some crossing-over has been detected genetically in him-8(e1489) and him-8(mn253). (Broverman and Meneely, 1994; Herman and Kari, 1989). Moreover, as we have discussed in the accompanying paper (MacQueen et al., 2005), the fraction of bivalent X chromosomes at diakinesis is probably enriched relative to the actual frequency of chiasma formation earlier in meiotic prophase.
Thus, in several key respects, the cytological phenotype of him-8 mutants mirrors the effects of deletion of the X chromosome PC (MacQueen et al., 2005). Specifically, neither the PC nor HIM-8 appears to be necessary for axial-element formation or for initiation of meiotic recombination, but both the cis-acting site and trans-acting him-8 function are necessary for efficient synapsis of the X chromosome pair.
X Chromosome Pairing Analysis in him-8 Mutants
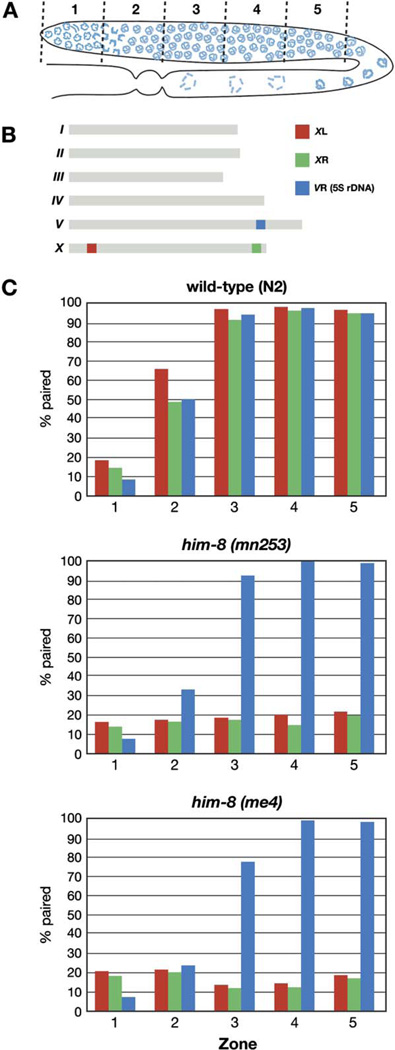
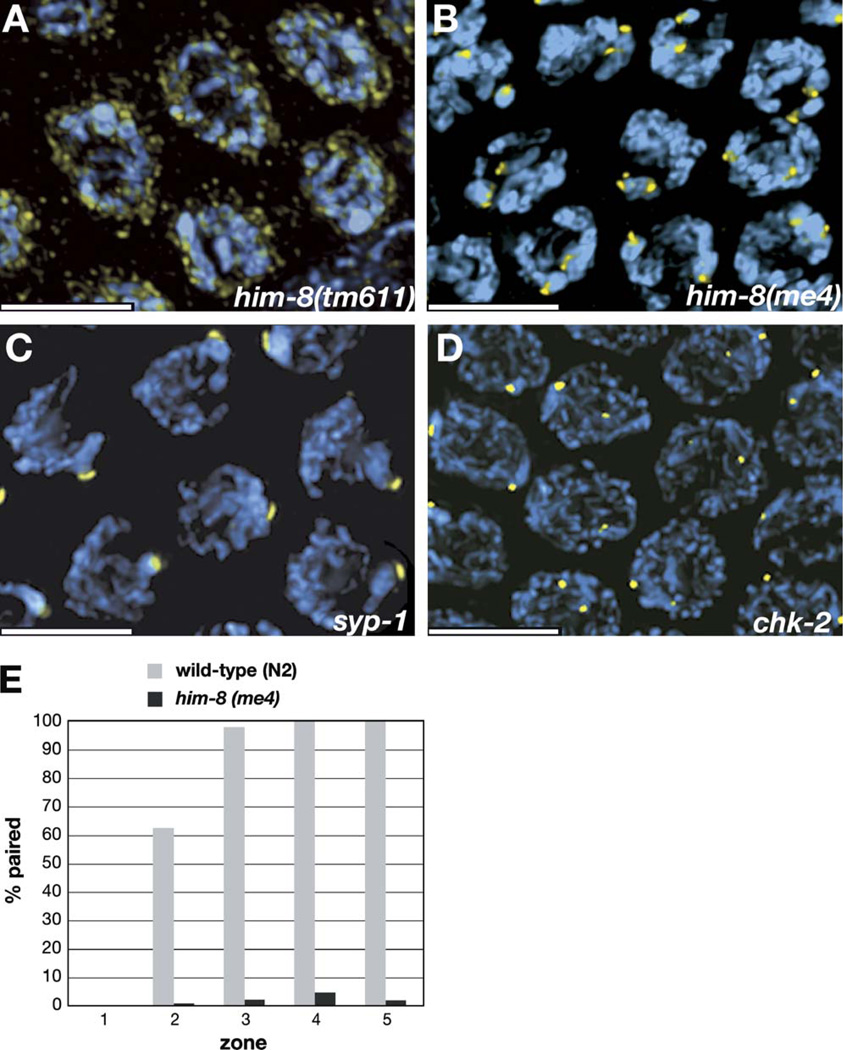
To evaluate the effects of him-8 on X chromosome interactions, we quantified associations between homologous loci in him-8 mutants (Figure 2). Each gonad in an adult animal contains a gradient of nuclei that span the progression of meiotic stages. Fluorescent probes to sequences from the left and right arms of the X chromosome were hybridized to wild-type gonads as well as samples from two mutant him-8 alleles (me4 and mn253). A probe to the 5S rDNA locus on chromosome V was included as an autosomal control (Figure 2B). Germlines were divided into five evenly spaced intervals, starting from the distal nuclei, which are undergoing premeiotic divisions, and continuing through the end of pachytene. The fraction of nuclei containing paired FISH signals was counted for each probe in each zone.
Figure 2. X Chromosome Pairing Is Defective in him-8 Mutants.
(A) Diagram of a hermaphrodite gonad, indicating the five zones in which the pairing of FISH signals was scored.
(B) Genomic localizations of the three FISH probes used to quantify homolog pairing.
(C) Graphs indicating the fraction of paired FISH signals in each zone for wild-type (N2), him-8(mn353), and him-8(me4) hermaphrodites. Three probes were scored independently: one from the left end of X chromosome (red), one from the right end of X chromosome (green), and the 5S rDNA, which marks the right arm of chromosome V (blue). In both him-8 alleles, pairing of the X chromosome probes did not rise above the baseline levels observed in the premeiotic region (zone 1), whereas Chromosome V association rates and dynamics were very similar to what we observed in wild-type hermaphrodites.
In wild-type hermaphrodites, pairing of all three probes initiated at the leptotene/zygotene stage of meiosis, represented in zone 2. By early pachytene (zone 3), pairing was stabilized by synapsis at all three loci in nearly 100 percent of nuclei. In both him-8 mutant alleles that we examined, the frequency of chromosome V pairing was very similar to that seen in wild-type throughout all zones, but pairing of the X chromosomes never rose detectably above the background level of association in the premeiotic region (zone 1) (Figure 2C; Table S1). The absence of a detectable rise in X chromosome pairing in the transition zone is subtly distinct from observations from meDf2 homozygotes (MacQueen et al., 2005). This difference suggests that loss of him-8 activity reduces the frequency or perdurance of X chromosome interactions even more severely than loss of both X pairing centers, as discussed below.
When mutations in the SC components syp-1 and syp-2 were analyzed, the PC region of each chromosome was more frequently paired than the opposite end throughout most of meiotic prophase (Colaiácovo et al., 2003; MacQueen et al., 2002, 2005). In the accompanying paper, we show that this local synapsis-independent stabilization of pairing requires the presence of a PC on both homologs. In him-8 mutants, no preferential stabilization of pairing is observed at the PC end of the X chromosomes (Figure 2). This suggests that HIM-8 is required for this local stabilization of pairing. Alternatively, him-8 lesions may cause an earlier defect than syp-1 or syp-2 mutations, such that X chromosome pairing never occurs and therefore cannot be stabilized. We believe that the first possibility, that him-8 plays a key role in stabilizing pairing, is the most likely explanation for these observations, both because it is consistent with a function we have demonstrated for the PC and also because some crossovers do occur between X chromosomes in him-8 mutants, indicating that they are still capable of homologous pairing to some degree. To fully understand the role of HIM-8 in X chromosome interactions will require real-time analysis of homolog pairing in him-8 mutant animals, which is beyond the scope of the current work.
him-8 Encodes a C2H2 Zinc-Finger Protein that Binds Specifically to the Pairing-Center Region of the X Chromosome
To better understand the role of him-8, we identified the affected gene. The genetic map position was refined by three-factor crosses, which showed that him-8 lies about 65% of the genetic distance from unc-24 to dpy-20 on chromosome IV. Candidate genes were tested by analyzing transcripts from wild-type and mutant animals. Because him-8 was implicated in chromosome behavior, our attention was drawn to a group of predicted genes from a single operon on the cosmid T07G12, each of which contain two predicted C2H2 zinc fingers. No other predicted genes with obvious chromatin- or DNA-associated motifs have been identified in this region of the genome. RT-PCR products were synthesized and sequenced for each gene within this operon using RNA isolated from either wild-type or him-8 hermaphrodites as the template. Both previously characterized alleles of him-8, e1489 and mn253, were associated with point mutations in a transcript from predicted gene T07G12.12. Each of these mutations would be expected to cause a nonconservative change in an amino acid critical to the function of one of two zinc fingers (Figure 3C). A third allele of him-8, me4, was isolated in a general screen for meiotic mutations in the laboratory of Anne Villeneuve, and this allele results in an amino acid change in the N-terminal portion of the same gene. A targeted deletion of this gene that removes a C-terminal portion of the T07G12.12 coding sequence confers an X chromosome segregation defect indistinguishable from the other three him-8 alleles (Table 1).
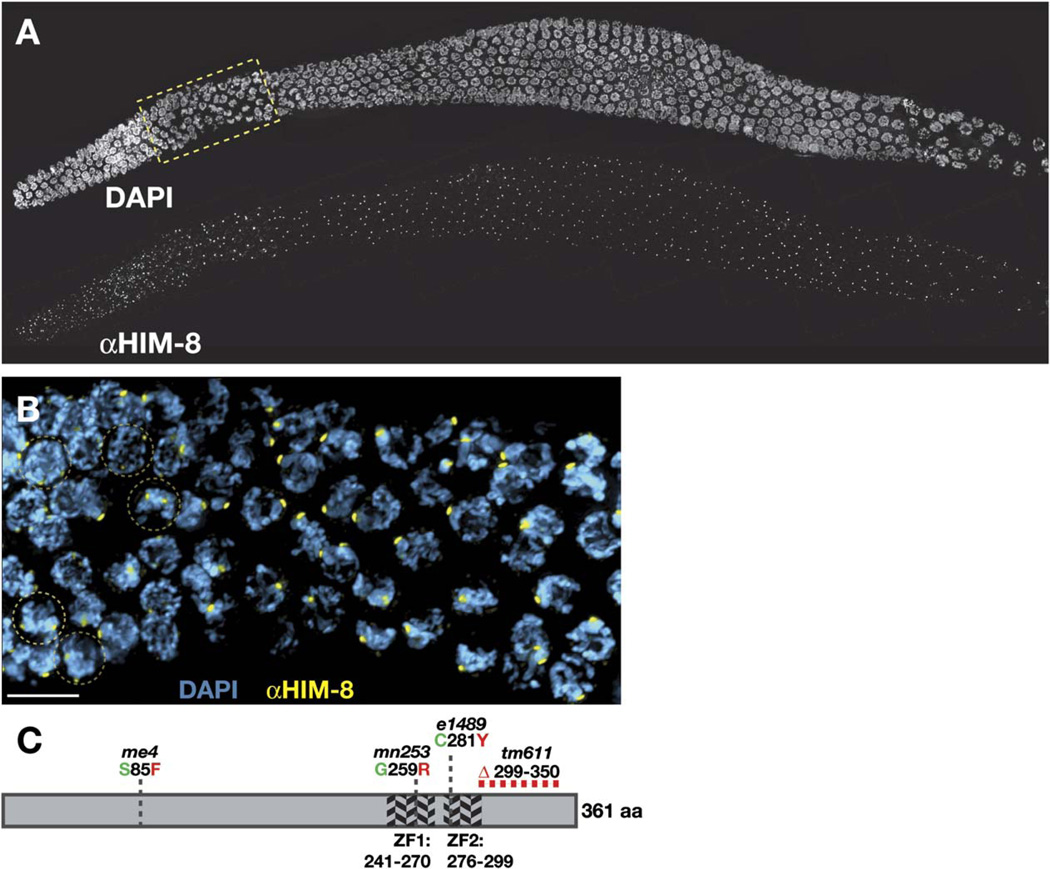
Figure 3. HIM-8 is a C2H2 Zinc-Finger Protein that Localizes to Distinct Nuclear Foci during Meiosis.
(A) Three-dimensional projection through a wild-type gonad stained with DAPI and antibodies against HIM-8. Subnuclear HIM-8 foci are present in all germ-line nuclei throughout the premeiotic, transition-zone, and pachytene region of the gonad. The transition-zone region outlined by the yellow box is magnified in (B).
(B) Prior to meiotic entry, two HIM-8 foci (yellow) are observed in each nucleus. Examples of premeiotic nuclei in which two foci can be clearly observed are outlined in brown circles. Once nuclei have entered the transition zone, representing the leptotene/zygotene stages of meiosis, they usually reveal only a single HIM-8 focus or closely spaced pair of foci. The scale bar represents 5 µm.
(C) Schematic representation of the HIM-8 protein. The diagram displays the location of two predicted zinc fingers as well as the sites of point mutations or deletions resulting from the four mutant alleles of him-8.
We conclude that him-8 corresponds to predicted gene T07G12.12, which encodes a C2H2 zinc-finger protein. Because all extant him-8 alleles have equally severe consequences for X chromosome crossing-over and segregation, they are likely to eliminate the normal function of the him-8 gene. However, immunofluorescence analysis (see below) and Western blotting (data not shown) indicate that HIM-8 protein is probably produced in all four mutant alleles.
Polyclonal antisera were raised against a partial HIM-8 fusion protein (see Experimental Procedures). Wild-type gonads stained with these antibodies showed conspicuous chromosome-associated foci from premeiotic stages through late pachytene of both males (data not shown) and hermaphrodites (Figure 3A). As nuclei condensed at diplotene in preparation for the meiosis I division, HIM-8 foci abruptly disappeared from the chromosomes.
In each nucleus within the premeiotic region of the hermaphrodite gonad, two distinct HIM-8 foci were visible. Early in the transition zone, where homologous pairing and synapsis are initiated, each nucleus displayed either one or two foci. Throughout pachytene, only a single focus, or sometimes a closely spaced doublet, was detected in each nucleus (Figure 3B). This result strongly suggests that HIM-8 binds to a particular chromosome locus that is unpaired in premeiotic nuclei but pairs at the initiation of meiotic prophase.
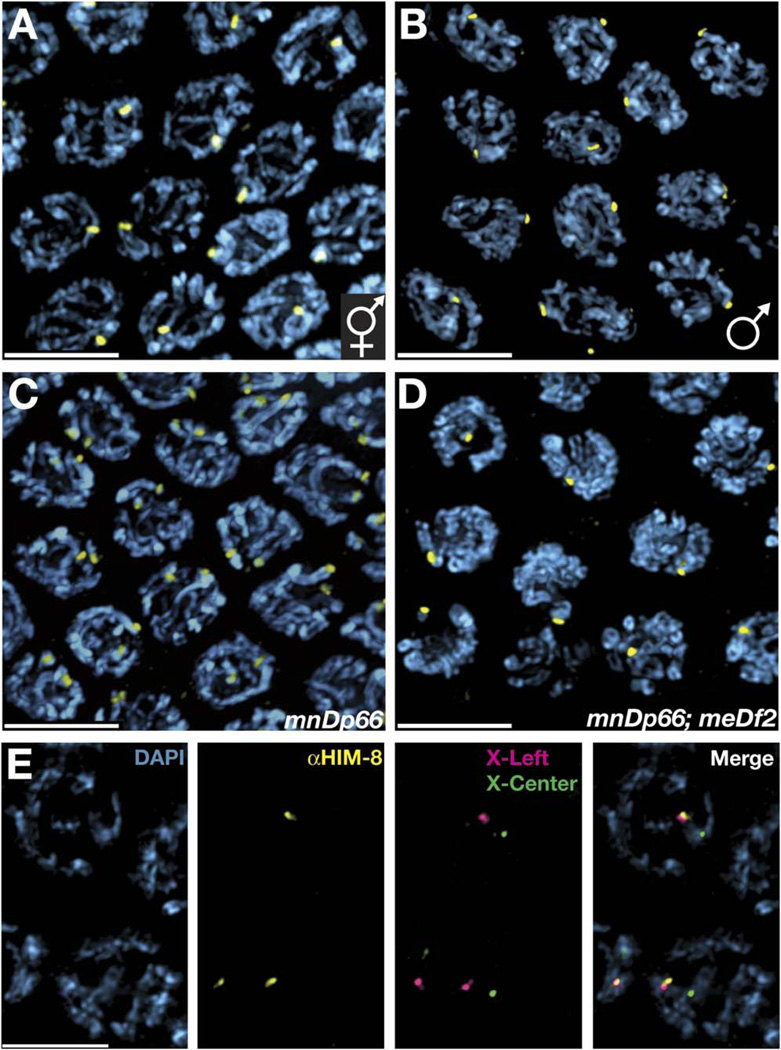
Based on the genetic interactions we had observed and the similarity of the phenotypes of PC deletions and him-8 mutations, we considered it likely that HIM-8 foci might correspond to the PC region of the X chromosomes. Currently it is not possible to generate animals that completely lack the X chromosome PC because the existing deletions of the PC are large and must be covered by duplications of the region (Villeneuve, 1994; MacQueen et al., 2005). Nevertheless, several informative genotypes were examined. In male spermatocyte nuclei, only a single HIM-8 focus was observed even prior to meiotic entry, consistent with a localization of the protein to the single male X (Figure 4B). Hermaphrodites carrying two copies of mnDp66, a duplication of the left two megabases of the X chromosome that includes the PC region (Villeneuve, 1994; Colaiácovo et al., 2003; MacQueen et al., 2002), revealed up to four distinct HIM-8 foci in premeiotic nuclei and two foci at pachytene, indicating that each copy of mnDp66 introduces an extra HIM-8 signal (Figure 4C). We also examined mnDp66;meDf2 hermaphrodites, which carry both a duplication and a deletion of the X chromosome PC, and observed the expected pair of foci in premeiotic nuclei and merged focus at pachytene, indicating that whereas mnDp66 provides an additional HIM-8 binding site, meDf2 removes this site (Figure 4D).
Figure 4. The HIM-8 Protein Localizes to the PC Region of the X Chromosome.
All images show projections through fields of pachytene-region nuclei from animals of the indicated genotypes. The entire nuclear volume is shown in each projection. All scale bars represent 5 µm.
(A) Wild-type hermaphrodite, revealing a single HIM-8 focus in each nucleus due to close association between the two binding sites detected in premeiotic and very early meiotic nuclei (Figure 3).
(B) Wild-type male, which displays a single (unpaired) HIM-8 focus in each nucleus. In contrast to hermaphrodites, males also have one HIM-8 focus per nucleus in the premeiotic region of the gonad (data not shown).
(C) Hermaphrodite carrying two copies of the mnDp66 duplication of the X chromosome PC region. An additional HIM-8 focus is present in each nucleus, corresponding to the paired and synapsed duplication.
(D) Hermaphrodite carrying both mnDp66 and the PC deficiency meDf2, which eliminates one of the two foci observed in (C).
(E) HIM-8 immunostaining was performed in conjunction with FISH to two probes on the X chromosome, one from the left arm (1.5 Mb from the telomere) and one from a more medial position (7.4 Mb from the left end). The XL probe is closely associated with HIM-8 focus, while the XC probe is clearly more distant from the HIM-8 focus. Probes to the right end of the X and autosomal loci were also tested (data not shown) and did not correlate in their position with HIM-8 foci.
As an independent test for the genomic location of the HIM-8 binding region, FISH was performed to a number of loci on both the X and autosomes in conjunction with HIM-8 immunostaining. The HIM-8 signal consistently localized very close to a probe derived from the PC region on the left arm of the X chromosome but did not spatially correspond with probes from the middle or right arm of the X or with autosomal probes. (Figure 4E and data not shown) We therefore conclude that HIM-8 associates with the X chromosome in a PC-dependent fashion and that its binding site coincides with the genetically defined PC region. Whether the PC is solely defined by its ability to recruit HIM-8 remains to be determined.
We tested whether HIM-8 localization requires the function of a number of other meiotic genes involved in chromosome pairing or synapsis by immunostaining mutant hermaphrodites. Both the expression of the HIM-8 protein and its ability to localize to the X chromosome were independent of the function of all genes we tested, including him-1(e879), him-3 (gk149), him-5(e1467 and e1490), zhp-3(jf61), htp-1(gk150) chk-2(me64), and syp-1(me17) (Figures 5C and 5D and data not shown). In chk-2 hermaphrodites, the HIM-8 foci were usually unpaired throughout meiotic prophase, which is consistent with the failure of the normal chromosome pairing process in this mutant (MacQueen and Villeneuve, 2001; Figure 5D). By contrast, syp-1 hermaphrodites revealed uniformly paired HIM-8 foci throughout pachytene (Figure 5C). This indicates that, in the absence of SC formation, the HIM-8 site is more robustly paired than other loci previously examined in syp-1 or syp-2 mutants (Colaiácovo et al., 2003; MacQueen et al., 2002). This result reinforces the idea that a HIM-8 binding site is at or very close to a site that mediates local synapsis-independent stabilization of pairing.
Figure 5. HIM-8 Localization in him-8 Mutants and Other Informative Meiotic Mutants.
All images show projections through the nuclear volumes of fields of pachytene-region nuclei from animals of the indicated genotypes. Immunofluorescence with anti-HIM-8 antibodies is shown in yellow, and DAPI staining is shown in blue.
(A) In him-8(tm611) mutant hermaphrodites, discrete foci of HIM-8 are not detected on the chromosomes. The intensity of the staining is shown more brightly here than in images of other genotypes to reveal that the residual staining is concentrated at the nuclear periphery. Similar staining is seen in him-8(e1489) and him-8(mn253) animals, which also carry mutations in the zinc-finger domain of HIM-8. This residual staining is detected using several different antisera raised against HIM-8, suggesting that it is specific rather than nonspecific background.
(B) In him-8(me4) hermaphrodites, two distinct foci are visible in each nucleus at pachytene. See also Figure 6B and Figure S1.
(C) In syp-1 hermaphrodites, a single focus of HIM-8 is detected in each nucleus in the pachytene region of the gonad, indicating that pairing of the HIM-8 binding region is stabilized despite the absence of synapsis.
(D) HIM-8 foci are detected on the X chromosomes but usually do not pair in chk-2 mutants.
(E) Immunofluorescence with the HIM-8 antibody was performed on wild-type and him-8(me4) hermaphrodites. The fraction of paired foci was scored in each of five zones of the gonad, which were defined in the same way as in our FISH analysis (Figure 2).
All scale bars represent 5 µm.
HIM-8 foci were absent from meiotic chromosomes in worms carrying three different him-8 alleles (e1489, mn253, and tm611), each of which is predicted to disrupt one of the two zinc fingers. While we do not yet know whether HIM-8 binds directly to DNA, this result indicates that the zinc fingers are necessary for chromosome association. Weaker immunostaining was still detected in these mutant animals and appeared to be diffusely associated with the nuclear envelope (Figure 5A and data not shown). This staining is likely to reflect authentic HIM-8 localization rather than nonspecific background because it was observed using polyclonal sera from three different immunized animals, two guinea pigs and one rat.
A fourth mutant allele, me4, results in a missense mutation in the N-terminal portion of the him-8 gene, distant from the zinc-finger motifs (Figure 3C). By immunofluorescence, we observed that this mutant protein not only is expressed but retains its ability to bind to the left end of the X chromosome (Figure 5B). In contrast to wild-type animals, however, the HIM-8 foci observed in him-8(me4) mutants did not pair as the nuclei entered meiosis and progressed to pachytene (Figure 5B). Functional analysis (above) has revealed no differences in the severity of pairing, synapsis, or segregation defects between me4 and other him-8 alleles, indicating that the presence of the protein on the X chromosome PC is not sufficient for its function.
Because the protein is still present at the PC region in him-8(me4) mutants, we compared the pairing behavior of the HIM-8 foci in these animals to wild-type hermaphrodites, analogously to our FISH time courses (above). The differences in X chromosome behavior between wild-type and me4 mutant animals were even more apparent by this assay (Figure 5E; Table S2), almost certainly because the higher signal-to-noise ratio and better morphological preservation provided by this immunofluorescence approach allow better discrimination between paired and unpaired loci than FISH. Qualitatively, the results are the same: throughout meiotic prophase, only baseline (premeiotic) levels of pairing of the HIM-8 foci were observed in the him-8(me4) mutant.
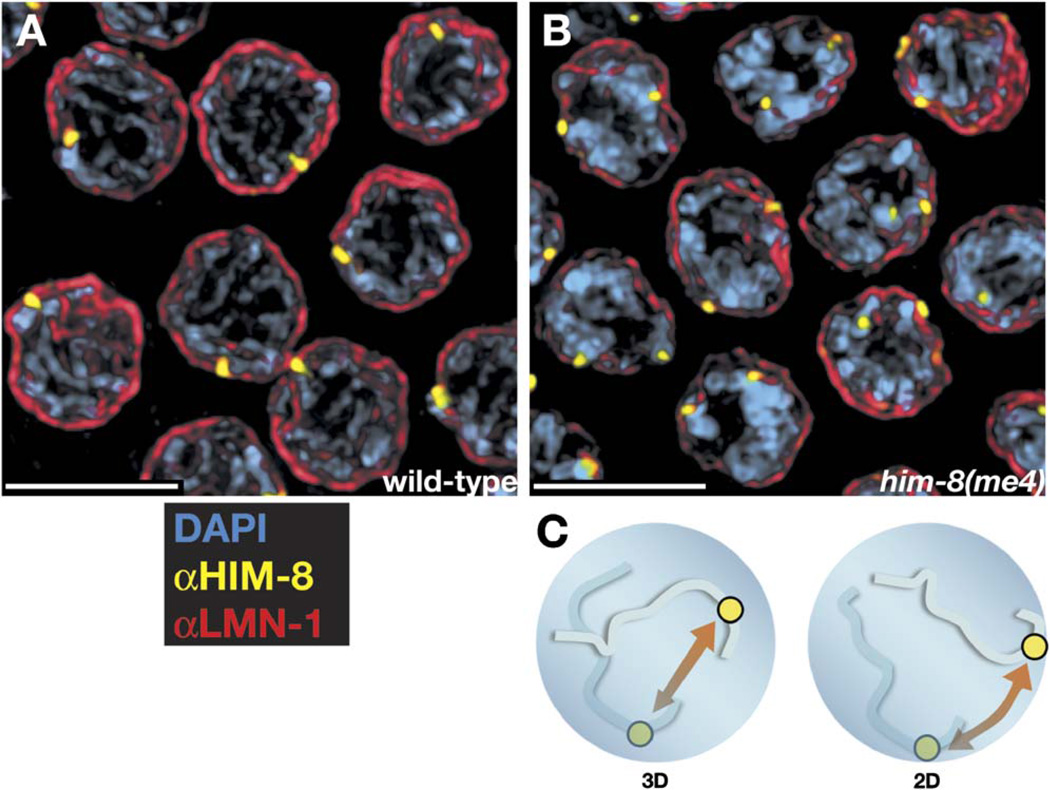
HIM-8 Associates with the Nuclear Envelope
Although associated with chromatin, HIM-8 foci are consistently observed at the extreme periphery of meiotic nuclei. This is true of both unpaired foci in premeiotic nuclei and paired HIM-8 foci throughout meiotic prophase. To better assess whether these foci might be associated with the nuclear envelope (NE), we costained dissected gonads with antibodies to HIM-8 and to the NE proteins LMN-1/lamin (Figures 6A and 6B; Figure S1) or SUN-1/matefin (data not shown). Close association between HIM-8 foci and nuclear-envelope staining is clearly evident, particularly in individual optical sections from these data sets (Figure S1). Fluorescence-intensity profiles confirm that HIM-8 foci are peripheral with respect to the mass of DAPI-stained chromatin and lie very close to or embedded within the nuclear lamina (Figure S1). Frequently, HIM-8 foci have a crescent-shaped appearance, especially in the transition zone, as if the protein localizes to a patch along the nuclear surface (cf. Figure 3B and Figure 5C). In him-8(me4), the only mutant allele in which HIM-8 foci remain associated with the X PC, the protein also retains its localization at the NE (Figure 6B and Figure S1).
Figure 6. HIM-8 Foci Associate with Nuclear-Envelope Components in Both Wild-Type and him-8(me4) Animals.
(A and B) Pachytene nuclei were stained with antibodies against HIM-8 (yellow) and anti-LMN-1 (red), which marks the nuclear lamina, or nuclear envelope. All scale bars represent 5 µm. See Figure S1 for an optical section through a similar data set, where nuclear-envelope association is somewhat more evident.
(C) A possible role for nuclear-envelope association of meiotic chromosomes is the reduction of homology search from a 3D to a 2D problem. As discussed in the text, our analysis of the him-8(me4) mutant indicates that the requirement for HIM-8 is likely to extend beyond such a role.
In all three mutant alleles of HIM-8 that disrupt the C-terminal zinc-finger domain, the protein fails to localize to prominent foci, but diffuse staining is seen at the periphery of the nuclei (Figure 5A and data not shown). This suggests that HIM-8’s association with the NE may be independent of its PC-localization activity. The protein does not include a transmembrane domain, lamin binding motif, FFAT motif, or other known NE-recruitment hallmarks. We speculate that HIM-8 is likely to be recruited to the NE by protein-protein interactions that remain to be elucidated.
We conclude that the prominent X chromosome-associated foci of HIM-8 are located at or very close to the nuclear envelope throughout most of meiotic prophase. It is not yet known if HIM-8 binding is required for association of this locus with the NE or whether NE localization is required for HIM-8’s function. However, the knowledge that the me4 allele causes defects in X chromosome pairing and segregation as equally severe as all other him-8 mutations is highly informative since it reveals that binding of HIM-8 to the X chromosome and nuclear-envelope association of this binding site are not sufficient for pairing or synapsis. Even if HIM-8 does specifically target the PC region to the NE, this cannot be its only essential function during meiosis.
DISCUSSION
Functional Relationships between HIM-8 and the Pairing Center
HIM-8 is the first example of a protein that is required for homologous synapsis of a particular chromosome pair. We have demonstrated that it binds specifically to the PC region of the X chromosome and is required for both synapsis-independent stabilization of pairing and efficient synapsis of this chromosome.
The conclusion that X chromosomes rarely, if ever, synapse in him-8 mutants is at variance with the results of Goldstein and Slaton (1982). They analyzed him-8 mutants by transmission electron microscopy (TEM) and observed six extensive synaptonemal complexes in pachytene nuclei. We suggest that this conclusion, which was based on observation of a limited number of nuclei, may be related to the apparent autosynapsis we observed in some him-8 nuclei stained with SYP-1 antibodies.
In light of our results that HIM-8 concentrates so prominently at the PC, it is perhaps surprising that all him-8 mutations result in X chromosome pairing, synapsis, and segregation defects that are subtly but reproducibly more severe than deletions of its major X chromosome binding site. This difference is apparent in segregation data (Table 1; Broverman and Meneely, 1994; Villeneuve, 1994) and is most obvious in our analysis of the frequency of chiasmate X chromosomes in oocytes at diakinesis (5% for him-8 versus 23% for meDf2 hermaphrodites; MacQueen et al., 2005). Double mutants that lack both X PCs and him-8 function are affected as severely as him-8 mutants but no worse (Table 1). We cannot yet fully explain this difference, but it implies that HIM-8 must act in a partially PC-independent fashion to promote X chromosome pairing and/or synapsis. Based on the recombination behavior of large insertions or deletions (McKim et al., 1993), as well as the consequences of PC deletions (Villeneuve, 1994; MacQueen et al., 2005), we know that other sites must mediate intimate alignment and promote synapsis between homologous chromosomes. A direct role for HIM-8 in stabilizing homolog pairing is strongly supported by our evidence that him-8 mutants lack the localized stabilization that is normally observed at the PC in the absence of synapsis. Even in the absence of synapsis, X chromosomes lacking PCs attain greater steady-state levels of pairing in early prophase than in him-8 mutants (Figure 2; MacQueen et al., 2005, Figure 2E). Taken together, we believe that our data are best explained by the idea that weak synapsis-independent stabilization of pairing activity is distributed along the X chromosome in addition to being highly concentrated at the PC and that HIM-8 contributes to this activity at all sites. It remains possible that HIM-8 also plays a direct role in initiating synapsis. To better understand its molecular mechanism, it will be useful to identify factors that interact directly with HIM-8 as well as further separation-of-function mutations in the him-8 gene or PC locus.
The Role of the Nuclear Envelope in Pairing and Synapsis
The observations that HIM-8 localizes to PC region of the X chromosomes and colocalizes with both DAPI-staining chromatin and the NE suggest that it might tether the chromosome to the NE. Such a role is consistent with the electron microscopic analysis of Goldstein and Slaton (1982), who observed that each of the six C. elegans chromosomes is attached to the NE at a single site during meiosis. Although they believed this site to be a chromosome end, their data are also consistent with each chromosome attaching to the NE via its PC.
Meiotic associations between the chromosomes and the NE have been observed in a wide variety of species, including fungi, plants, and animals (reviewed by Zickler and Kleckner, 1998). Usually these associations are thought to involve telomeres, which frequently cluster at a small region of the NE in a conformation known as the “meiotic bouquet.” It has been speculated that this tethering of chromosomes at the NE might expedite pairing simply by reducing the complexity of the homology search from a 3D to a 2D spatial problem (Figure 6C). However, this concept has not been directly supported by experimental evidence and is apparently contradicted by evidence from some organisms that bouquet formation only occurs after extensive homolog alignment.
The me4 point mutation retains both PC binding and nuclear association activities of HIM-8 but causes defects in X chromosome pairing, crossing-over, and disjunction as severe as mutations that prevent HIM-8 from binding to chromosomes. This allows us to conclude that PC-NE association is not sufficient to promote either pairing or synapsis in C. elegans, although it may be necessary. Further analysis of HIM-8 with particular attention to the domain that is altered in him-8(me4) should help to clarify how and why chromosomes associate with the nuclear envelope during meiotic prophase.
We have argued that the association of HIM-8 with the nuclear envelope may be required for stabilization of pairing, a key function of the PC. We propose that the meiotic bouquet in other species may perform an analogous role in prolonging homologous interactions to facilitate chromosome sorting and/or to promote a relatively slow process of synapsis initiation. This hypothesis is consistent with the order of events that is seen in all organisms, including those that align their homologs before bouquet formation. It also can explain why, in some species where a bouquet is observed, synapsis frequently initiates at interstitial chromosome loci (Zickler and Kleckner, 1998) if the stabilization function is not directly linked to synapsis initiation activity.
Functional Relationships between HIM-8 and Other Meiotic Factors
Sequence comparisons with known meiotic factors have not revealed any obvious orthologs of HIM-8. One of HIM-8’s zinc fingers shows unusual spacing of the cysteine and histidine residues, which seems to be shared only by other nematode proteins, including several in the same operon as the him-8 gene. However, genes involved in meiosis diverge very quickly during evolution, despite the fundamental conservation of this process. Even core structural components of the SC have been identified independently through genetic analysis or monoclonal antibodies in several model systems rather than by virtue of obvious homology. Zinc-finger proteins such as HIM-8 are also known to diverge very rapidly, and the C2H2 family in particular has undergone massive expansion in the evolution of eukaryotes (Chung et al., 2002; Englbrecht et al., 2004). The essential role played by him-8 in C. elegans may be delegated to distantly related proteins in other organisms.
A few known meiotic factors do share structural and/or functional similarities with HIM-8. Most compelling is Teflon, a C2H2 zinc-finger protein essential for accurate segregation of the autosomes during male meiosis in Drosophila (Tomkiel et al., 2001). Similar to our proposed function for HIM-8, Teflon is not necessary for homolog association but is required to stabilize pairing in Drosophila spermatocytes, which do not make SCs. Other, more distant potential HIM-8 relatives include telomere binding components that play roles in bouquet formation, such as Taz1p from fission yeast (Nimmo et al., 1998) or NDJ1p from budding yeast (Trelles-Sticken et al., 2000).
Additionally, it is noteworthy that a key component of budding-yeast axial elements, Hop1p, contains an atypical zinc-finger sequence (Hollingsworth et al., 1990). This cysteine-rich domain is absent from HIM-3 and HTP-1, −2, and −3, the four predicted Hop1 homologs in C. elegans, as well as from Hop1 homologs in plants and other animals (data not shown). However, it is the most highly conserved region among Hop1 homologs from fungi, other than the HORMA domain shared by all Hop1 homologs (Figure S3). In vitro, the zinc finger of Hop1p binds to DNA with a strong preference for G-rich sequences and can mediate interactions between double-stranded DNA molecules containing runs of Gs (Anuradha and Muniyappa, 2004a, 2004b; Kironmai et al., 1998). This activity has been speculated specifically to promote meiotic pairing at telomeres or other G-rich sites in vivo. These observations raise the intriguing possibility that fungal Hop1 proteins incorporate a domain that functions analogously to HIM-8 to stabilize homolog interactions during meiosis.
The chromosome specificity of defects caused by him-8 mutations begs the question of whether the autosomes rely on analogous factors to stabilize homolog pairing and promote synapsis. It is conceivable that him-8 has evolved to mediate the unique features of the X, which must pair and recombine to segregate accurately during meiosis in hermaphrodites yet be transmitted efficiently as a univalent in males. However, each autosome also has a PC that appears to confer local stabilization of pairing (Colaiácovo et al., 2003; MacQueen et al., 2002). The work of Goldstein and Slaton (1982) indicates that each chromosome associates with the nuclear envelope during meiosis, which also suggests the possibility of autosomal counterparts to HIM-8. We are currently investigating whether the predicted HIM-8 homologs in the same operon might contribute to meiotic alignment of the autosomes.
EXPERIMENTAL PROCEDURES
Genetics
The C. elegans wild-type strain used was N2 Bristol. All worms were cultured at 20°C according to standard conditions (Brenner, 1974). Counts of male and hermaphrodite progeny among the broods of mnDp66/+; meDf2/+ or him-8/+;meDf2/+ hermaphrodites were corrected for inviability based on the expectation that 12.5% of male progeny (those meDf2/0 males that do not inherit a copy of mnDp66) die whereas 6.25% of hermaphrodite progeny die (see Villeneuve, 1994).
FISH and Temporal Analysis of Chromosome Pairing
Pairing analysis was carried out using age-matched adult worms at 20–24 hr post-L4 larval stage. The gonads were divided into five equal sized regions, beginning at the distal tip of the gonad and progressing through the end of pachytene. Three complete germlines were scored for each genotype. The XL probe was made by PCR amplification of sequences from a single cosmid, K06A9, which is 1.5 Mb from the left end. The X center probe (Figure 4) and XR probe (Figures 2 and 4) are synthetic oligonucleotides that match short repeats enriched on the X chromosome (Lieb et al., 2000). XR has the sequence GACTCCATCCACCAGCACTGCTTCGAGTACGACAGAAAGCACTTC, which is concentrated in a small region 17.4 Mb from the left end and 340 Kb from the right end of the X chromosome. XC is TTTCGCTTAGAGCGATTCCTTACCCTTAAATGGGCGC CGG, which is repeated many times on cosmid C07D8, 7.4 Mb from the left end. The 5S rDNA probe to the right arm of chromosome V has been described elsewhere (Dernburg et al., 1998).
All FISH probes were synthesized by 3’ end labeling of DNA fragments with aminoallyl-dUTP (Sigma) followed by conjugation to Alexa 488-NHS ester (Molecular Probes) or Cy3- or Cy5-NHS-ester (Amersham). Fixation and in situ hybridization of dissected worm gonads were carried out essentially as described by Dernburg et al. (1998), except that microwave irradiation using a variable-wattage microwave with a circulating water-bath (Biowave, Ted Pella) was used to accelerate probe diffusion and annealing, and the annealing time was reduced to 1 hr. More detailed protocols for probe synthesis and FISH will be provided on request.
For pairing analysis, FISH signals were regarded as paired if they lay within 0.7 µm of each other, which (as discussed in MacQueen et al., 2002) is the maximum distance typically measured between FISH signals on synapsed homologs. This criterion probably results in a consistent overestimate of pairing frequency since two FISH signals will sometimes fall within 0.7 µm linear distance by chance when they are constrained to lie within a volume of less than 4 µm diameter. We also note that each zone analyzed here may include a somewhat different distribution of stages depending on the genotype of the sample since the temporal progression of meiosis and the number of nuclei that abort the process and undergo apoptosis are affected by defects in chromosome pairing, synapsis, and/or recombination (Alpi et al., 2003; Bhalla and Dernburg, 2005).
Antibodies, Immunofluorescence, and DAPI Staining
All immunofluorescence experiments were performed with polyclonal antisera. Rabbit anti-SYP-1 was kindly provided by Anne Villeneuve, and anti-LMN-1 and anti-SUN-1 were gifts of Yossi Gruenbaum.
To raise antibodies specific for HIM-8, a 167-residue internal segment of the protein that shares minimal homology with other predicted C. elegans proteins was cloned into an expression vector. The primers CTGAATC TTTCGGAAAAAATATCC and CGGGAAATGACATTGAATATTGTG were used to amplify the sequence from total worm RNA by RT-PCR. The resulting coding sequence was cloned into pET100-D/TOPO (Invitrogen) downstream of a His6 tag. Individual clones were sequenced, and a correct clone was selected for expression in E. coli BL21 DE3. Recombinant protein was purified using nickel chromatography under denaturing conditions. Guinea pigs and rats were immunized by Pocono Rabbit Farm and Laboratory. Affinity purification of HIM-8 antisera was not performed for these experiments, but subsequently we have found purification to have no effect on the staining patterns reported here.
DAPI staining and immunofluorescence were carried out after dissecting worms in egg buffer containing 0.1% Tween 20 (Dernburg et al., 1998) and fixation in 3.7% or 1% formaldehyde in egg buffer, respectively. Secondary antibodies were purchased from Jackson ImmunoResearch or Molecular Probes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Anne Villeneuve for contributing the me4 allele of him-8 that is characterized here. The tm611 allele was isolated and provided by Shohei Mitani and the Japanese National BioResource for C. elegans. Many of the strains we used were provided by the Caenorhabditis Genetics Center. We thank Gary Karpen and Amy MacQueen for helpful comments on the manuscript. This work was supported by a National Science Foundation (NSF) predoctoral fellowship to C.M.P.; a UC Berkeley Biology Fellows Program grant to C.W.; an NIH/Ruth Kirschstein Individual NRSA (GM067408) to N.B.; grants from the NSF to P.M.M.; and by LDRD funds from Lawrence Berkeley Lab, NIH R01 GM/CA655591, and Burroughs Wellcome Career Award 1000950 to A.F.D.
Footnotes
Supplemental Data
Supplemental Data include two tables and three figures and can be found with this article online at http://www.cell.com/cgi/content/full/123/6/1051/DC1/.
REFERENCES
- Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans . Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Anuradha S, Muniyappa K. Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Res. 2004a;32:2378–2385. doi: 10.1093/nar/gkh559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anuradha S, Muniyappa K. Saccharomyces cerevisiae Hop1 zinc finger motif is the minimal region required for its function in vitro. J. Biol. Chem. 2004b;279:28961–28969. doi: 10.1074/jbc.M403727200. [DOI] [PubMed] [Google Scholar]
- Baker BS, Carpenter AT. Genetic analysis of sex chromosomal meiotic mutants in Drosophilia melanogaster . Genetics. 1972;71:255–286. doi: 10.1093/genetics/71.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans . Science. 2005;310 doi: 10.1126/science.1117468. in press. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broverman SA, Meneely PM. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans . Genetics. 1994;136:119–127. doi: 10.1093/genetics/136.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HR, Schafer U, Jackle H, Bohm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila . EMBO Rep. 2002;3:1158–1162. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Couteau F, Nabeshima K, Villeneuve A, Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Englbrecht CC, Schoof H, Bohm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein P, Slaton DE. The synaptonemal complexes of Caenorhabditis elegans: comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma. 1982;84:585–597. doi: 10.1007/BF00292857. [DOI] [PubMed] [Google Scholar]
- Herman RK, Kari CK. Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans . Genetics. 1989;121:723–737. doi: 10.1093/genetics/121.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans . Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K, Friedman DB, Hollingsworth NM, Byers B. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1424–1435. doi: 10.1128/mcb.18.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, de Solorzano CO, Rodriguez EG, Jones A, Angelo M, Lockett S, Meyer BJ. The Caenorhabditis elegans dosage compensation machinery is recruited to X chromosome DNA attached to an autosome. Genetics. 2000;156:1603–1621. doi: 10.1093/genetics/156.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2 . Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans . Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans . Cell. 2005;123 doi: 10.1016/j.cell.2005.09.034. this issue, 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Peters K, Rose AM. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans . Genetics. 1993;134:749–768. doi: 10.1093/genetics/134.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- Tomkiel JE, Wakimoto BT, Briscoe A., Jr. The teflon gene is required for maintenance of autosomal homolog pairing at meiosis I in male Drosophila melanogaster . Genetics. 2001;157:273–281. doi: 10.1093/genetics/157.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E, Dresser ME, Scherthan H. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans . Genetics. 1994;136:887–902. doi: 10.1093/genetics/136.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Rose AM. The meiotic behavior of an inversion in Caenorhabditis elegans . Genetics. 1992;131:321–332. doi: 10.1093/genetics/131.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.