SUMMARY
Live cell fluorescence microscopy is a powerful tool for characterizing aberrant mitotic phenotypes resulting from exposure to chemical inhibitors or after depletion of protein targets by RNA interference or other methods. Live imaging of cultured cells during mitotic progression presents challenges in maintaining optimal health of cells while achieving the temporal and spatial resolution to accomplish the goals of the study. Herein are strategies to monitor and analyze mammalian cell mitosis with standard, inverted, fluorescence microscopy systems that are widely available.
Keywords: Fluorescence microscopy, imaging, mitosis, cell division, fluorescent protein, microscope, RNAi
1. Introduction
1.1 Overview of Fluorescence Imaging And Mitosis
In the late 1800's Flemming, Van Beneden and their contemporaries carried out the first studies of chromosome behavior in dividing cells in both living and fixed tissue using the available microscope methodology (reviewed in [1]). Since that time, tremendous advances in microscope technology and fluorescence imaging have greatly enhanced our ability to visualize mitotic progression with high spatial and temporal resolution. Modern fluorescence imaging and molecular biology techniques allow tracking of subcellular structures such as chromosomes, kinetochores, centrosomes, or microtubules during cell division with great detail. Specific proteins of interest may be fused with fluorescent proteins to enable study of their intracellular dynamics and localization throughout the cell cycle. By creating fluorescent fusion proteins varying in wavelength emission, one can further determine if proteins of interest co-localize or concentrate in specific subcellular compartments during mitosis. For the purposes of the assay described herein a cell line was employed that expresses Histone H2B fused to green fluorescent protein (GFP). Histone H2B is a component of nucleosomes, a repeating multi-protein unit of eukaryotic chromatin that localizes exclusively and stably to DNA. Thus, cells expressing Histone H2B-GFP permit observation of chromatin and condensed chromosomes throughout interphase and mitosis by fluorescence microscopy [2].
Mitosis in vertebrate cells proceeds through multiple stages, in which chromosomes exhibit differing physical states and undergo characteristic movements. In prophase, the first stage of mitosis, the duplicated sister chromatids condense to form the mitotic chromosomes within the nucleus (Figure 1). During this time the spindle poles, also termed centrosomes, separate and begin to form the mitotic spindle composed of microtubules and accessory proteins. The chromosomes are released into the cytoplasm by the dissolution of the nuclear envelope, an event that marks the beginning of prometaphase. In the cytoplasm, the chromosomes interact with microtubules of the mitotic spindle through pairs of specialized multi-subunit substructures called the kinetochores, which power chromosome movements on the mitotic spindle. The sister kinetochores are arranged back to back on each chromosome and attach to spindle microtubules that emanate from opposite poles. The kinetochore interactions with the dynamic spindle microtubules move the chromosomes to the spindle equator. When all chromosomes have congressed to the spindle equator, the cell has reached metaphase. At the next stage, anaphase, the cohesions between sister chromatids are synchronously dissolved, and the separated chromatids move in opposite directions toward the spindle poles. In most cells the spindle poles also move apart. In the last stage of mitosis, called telophase, the mitotic spindle disassembles, the sister chromatids decondense, and nuclear envelopes form around the spatially separated chromatin masses. Cytokinesis, initiated at anaphase onset, cleaves the cytoplasm until only a small bridge termed the midbody remains between the two daughter cells. With time, the midbody thins and is severed in a process called abscission, resulting in complete separation of the two daughter cells.
Figure 1.

Stages of mitosis are depicted by landmark events. In prophase the chromosomes condense within the nucleus. At prometaphase, the nuclear envelope breaks down and the kinetochores of the chromosomes interact with spindle microtubules. The chromosomes move to the spindle equator. At metaphase, the chromosomes are aligned at the spindle equator. At anaphase the sister chromatids separate and move to the spindle poles. The spindle poles also move apart. Cytokinesis is initiated. At telophase the mitotic spindle is disassembled, the chromatids decondense and the nuclear envelope reforms. Cytokinesis cleaves the cytoplasm in two until the daughter cells are connected only by the midbody. (see Color Plates).
1.2 Live-Cell Imaging Considerations
1.2.1 Environment
Maintaining cell health is critical in live-imaging experiments. Mammalian cells must be viable and healthy during the imaging process to ensure that they will progress normally through the cell cycle. Important parameters that must be maintained include temperature, osmotic pressure and pH. Most human cells grow optimally in medium at 37°C with an osmotic pressure range between 260–320 mOsm/kg and a pH range between 7.2 and 7.4. Temperature can be maintained by various strategies including objective heaters, stage heaters, and thermo-regulated forced air. Evaporation must be avoided lest the medium to become hypertonic. Typical bicarbonate-balanced media require proper control of CO2 in the environment to maintain pH. Deviations from these optimum parameters may compromise cell viability and lead to arrest of cell cycle progression and cell division.
Use of one or a combination of methods is necessary to control these parameters. To control the incubation environment, the microscope or the microscope stage may be enclosed in commercially available or laboratory-constructed environmental chambers. This method is advantageous because the temperature, humidity, and CO2 levels can be maintained using a single system. However this strategy limits ready access to the cells. In addition, enclosures often expose microscope components to elevated humidity and temperature, which may be detrimental to the equipment. Alternatively, individual methods for the maintenance of temperature, pH, and osmotic pressure may be used. An objective heater (necessary for oil immersion objectives), a stage heater, or a system that maintains temperature by moving heated air across the culture vessel can be employed singly or in combination to maintain appropriate culture temperature. In order to maintain stable pH commonly used DMEM-based or other medium requiring 5% CO2 levels may be replaced with Leibovitz's L15-based media. Leibovitz's L15 is buffered by phosphates and free base amino acids instead of sodium bicarbonate and is suitable for culture of many cell lines. It is designed to maintain pH and support cell growth in environments without CO2 equilibration. Both DMEM- and L15-based media provide an appropriate osmotic pressure near 290 mOsm/kg. To inhibit media evaporation and ensuing changes in osmotic pressure, sealed imaging chambers can be used. Alternatively, the culture media in imaging chambers can be overlaid with mineral oil.
1.2.2 Illumination And Fluorophores
Mammalian cells expressing fluorescent proteins are extremely sensitive to photo-damage from high intensity fluorescence light sources; minimization of illumination light helps maintain favorable growth and reduce fluorophore photobleaching. Settings for illumination intensity, duration, and intervals between image capture to achieve optimal results will vary with the light efficiency of the microscope system, fluorescent protein type and abundance, cell line, and the length of the experiment. It is necessary to carry out empirical trials to determine optimal settings for each parameter. If imaging parameters result in arrest or delay at any stage of the cell cycle, it is a strong indication that cell health has been compromised. Phenol red, a pH indicator that absorbs light and adds background fluorescence upon illumination, is commonly used in tissue culture medium but it also interferes with the observation of certain fluorophores. Using medium lacking phenol red is recommended during fluorescence imaging to permit a reduction in the amount of illumination required to visualize the fluorescent protein of interest. One approach to minimize photo-damage and unnecessary photobleaching of target fluorophores is, wherever possible, to use transmitted light for focusing on cells and setting up experiments. Transmitted light can be used to identify cell or culture regions for image acquisition and for automated focusing protocols, thus reducing a cell culture's exposure to high intensity fluorescence illumination. In addition, it is often informative to monitor cytokinesis and other morphological changes by transmitted light as they may not be revealed by fluorescent Histone H2B-GFP or other fluorescent fusion proteins as cells progress through mitosis. Common bright-field methods include phase contrast and Nomarski Differential Interference Contrast (DIC). Although less phototoxic than fluorescence illumination, exposure to light commonly used for transmitted illumination should also be minimized to maintain cell health.
The constitutive expression and incorporation of H2B-GFP into the DNA of HeLa cells permits visualization of chromatin and condensed chromosomes for long periods without perturbing the cell cycle when environmental culture conditions are maintained and minimization of damaging light exposure is achieved. HeLa cells, commonly used in scientific research, are an immortal human cell line derived from cervical cancer cells. Histone H2B-GFP allows effective monitoring of the various stages of mitosis by providing a clear indication of chromosome location, movements and condensation state. Kinetochore proteins, tubulin subunits of microtubules, and other subcellular structures have also been expressed as fluorescent protein fusions and can be useful in studying mitotic progression. Although GFP remains one of the most commonly utilized fluorescent proteins, many other fluorescent proteins are available including photo-activatable and photo-convertible varieties. Co-expression of two or more proteins, each tagged with fluorescent proteins of differing emission wavelengths, allows simultaneous tracking of multiple structures (e.g. chromosomes and microtubules) within the same cell. For simplicity in this example method, we examine mitotic progression by monitoring the status of a single fluorescent protein, Histone H2B-GFP, in HeLa cells.
In this section we describe a live-cell imaging approach to examine vertebrate mitosis using an automated, inverted fluorescence microscope. This method is particularly useful for identification and characterization of aberrant mitotic phenotypes. Under experimental conditions, these errors may be induced by exposure to chemical inhibitors or from protein depletion using RNAi or other methods to target suspected mitotic regulators.
2. Materials
The following materials are used to monitor mitotic progression using HeLa cells expressing Histone H2B-GFP fusion proteins. This cell line has been described in a previous study [2].
Automated inverted fluorescence microscope system (see Note 1).
Air Stream Stage Incubator (Model ASI400, NEVTEK) (see Note 2).
HeLa cell line expressing Histone H2B-GFP fluorescent protein fusion.
Leibovitz's L15 phenol red-free media supplemented with 10% fetal bovine serum and antibiotics (penicillin and streptomycin solution).
Mineral Oil.
Culture chambers suitable for fluorescence imaging (see Note 3).
Taxol/paclitaxel (see Note 4).
3. Methods
The following procedure describes techniques to monitor chromosome location and analyze mitotic progression in HeLa H2B-GFP cells by live imaging using an inverted fluorescence microscope. In the examples we review, HeLa H2B-GFP cells were exposed to varying concentrations of Taxol, or depleted of either SKA3 (Spindle and Kinetochore Associated 3) [3] or MAD2 (Mitotic Arrest Deficient 2) [4] protein by RNAi (see Note 5). SKA3 and MAD2 are proteins that regulate proper mitotic progression and depletion of these proteins emphasizes the detection and characterization of mitotic aberrations distinct from those induced by Taxol.
3.1 Cell Culture In Chambered Coverglasses
If RNAi experiments are to be done, transfect Hela H2B-GFP cells with the appropriate siRNA one or more days before the planned experiment (see Note 6).
The day before the planned experiment transfer cells into the coverglass chambers. The cells should cover approximately 40% of the available surface prior to initiating image acquisition (see Note 7).
3.2 Time-Lapse Microscopy
Turn on the computer, microscope, and accessory devices. Accessory devices may include fluorescence illumination sources, shutters, filter wheels, and stage controllers.
Turn on the Air Stream Incubator and monitor temperature above the objective until a constant temperature of 37°C is achieved (see Note 8).
Launch the software responsible for controlling the microscope system and define known parameters such as illumination wavelengths, time-lapse imaging frequency and duration, and the location for storage of the image acquisition files (see Notes 9 and 10).
Exchange the growth medium in the chambered coverglass using 37°C Leibovitz's L15 phenol red-free supplemented media (see Note 11).
If chemical inhibitors are to be used, add to chambers immediately prior to imaging.
Overlay the L15 media with 37°C mineral oil (see Note 12).
Mount the coverglass chamber on the stage and ensure that it is firmly secured such that it does not shift with respect to the stage during movement of the stage.
Using transmitted light, select regions (imaging fields) with appropriate cell densities within coverglass chambers. Instruct the microscope system's software to retain the selected field coordinates and to monitor correct focus at each position (see Note 13).
Define parameters for both transmitted light and GFP fluorescence imaging by experimenting with exposure times and light intensities at a few locations that will not be recorded during the experiment (see Note 14).
Once proper imaging and environmental parameters for the assay have been defined or achieved, initiate image acquisition.
At the end of the total imaging period, shut down the system and turn off all accessories.
3.3 ANALYSIS
Captured images are examined to determine if perturbations in mitotic progression have occurred under experimental conditions and that control cells progressed through mitosis normally. Images can be assembled for analysis in convenient sequential series or time-lapse video records that display chronological events within single or multiple image fields. Several automated approaches have been described and applied to analyze fluorescence time lapse images of cells proceeding through mitosis that rely upon detection of various fluorescent fusion proteins. Many tools devoted to these goals are included in commercial microscopy software. However, it is important to analyze some of the results of this type of assay manually to ensure that criteria employed in automated analyses yield accurate results. Since automated systems and parameters vary widely and are specific to individual products, we describe a process for manual analysis that characterizes some general features of mitotic progression. We recommend this procedure be applied and results compared to those derived from automated data processing prior to relying upon the output from automated analyses.
Using the images acquired from this example assay, we measure multiple aspects of normal and perturbed mitotic progression. Failure to enter mitosis or aberrations that cause catastrophic errors in mitotic progression will be readily apparent and can be described accordingly. Phenotypic aberrations that are not as prevalent or penetrant can be revealed by careful examination of chromosome location and movements over time. For example, mitotic stressors can cause a normally bipolar spindle to become irregular or multipolar; these spindle alterations can often be inferred by chromosome movements and positioning during prometaphase congression. Less obvious but still measurable are perturbations that minimally alter the progression rate through mitotic stages. Convenient temporal landmarks include the release of condensed chromosomes into the cytoplasm at prometaphase, successful alignment of chromosomes at the spindle equator at metaphase, and the separation of sister chromatids as anaphase commences. Variances in the duration between periods marked by these landmarks can be compared to determine if experimental conditions have perturbed mitotic progression. Results are based upon examination of both phase contrast and fluorescence images, but the analysis described below emphasizes data obtained from Histone H2B-GFP fluorescence.
Prepare sets of individual image fields in sequential order so that cellular Histone H2B-GFP fluorescence can be examined as cells progress into and through mitosis.
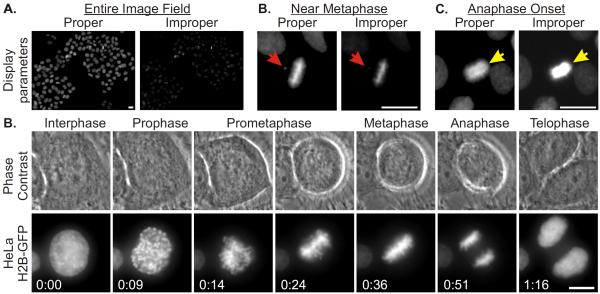
Adjust the contrast and brightness, sometimes referred to as `scale' or `scaling', within the microscopy software so that captured fluorescence signals are displayed in a manner that applies the gray levels within the images (0 to 4095 for 12 bit images) properly to the computer monitor. Details within nuclei and mitotic chromosome characteristics should be discernible (Figure 2).
Beginning with a control sample, proceed sequentially through the images and record when individual cells enter prometaphase, reach metaphase, and initiate anaphase (see Note 15).
Make notations if abnormalities are observed, such as a failure of a subset of chromosomes to congress to the metaphase plate, an inferred multipolar spindle, or a failure of chromosomes to properly segregate during anaphase.
Process data from the various stage locations and experimental conditions. Image panels revealing examples of normal mitotic progression are shown in Figure 2 (see Note 16).
Prepare graphical representations of the data to assess variances in mitotic progression under control conditions.
To determine if the culture conditions and imaging process negatively affect culture health and mitotic progression, create a plot that indicates the frequency of mitotic errors and variances in the time taken from prometaphase onset to the initiation of anaphase throughout the experimental period (Figure 3) (see Note 17).
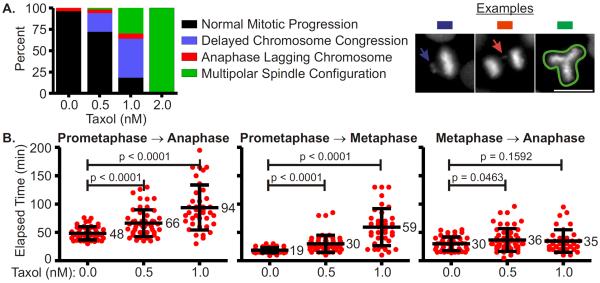
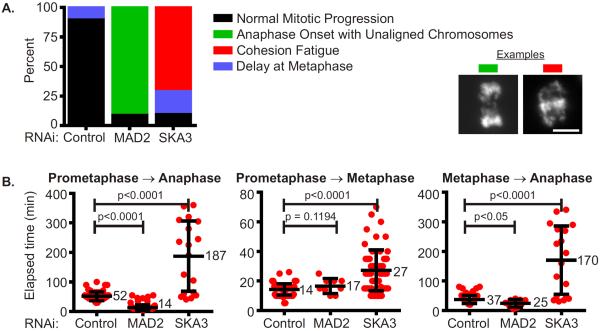
Prepare graphs using data from control and experimental conditions illustrating the type and frequency of mitotic errors. Since it is unlikely that the numbers of cells examined under each condition are equal, it is useful to display the frequency of mitotic errors as a percentage (Figure 4A and 5A).
Prepare graphical representations of the data to assess and describe variances in mitotic progression between control and experimental conditions. Prepare scatter plots illustrating the time taken for cells to progress from prometaphase to anaphase, prometaphase to metaphase, and metaphase to anaphase (Figure 4B and 5B) (see Note 18).
Figure 2.
Image display parameters and representative images from HeLa Histone H2B-GFP cell mitotic progression. (A) HeLa Histone H2B-GFP fluorescence images are presented with
varying display characteristics demonstrating proper and improper scaling parameters. An entire image field acquired using a 20X objective is shown; proper scaling parameters reveal fluorescence signals within the DNA of all interphase and mitotic cells. (B) Properly displayed Histone H2B-GFP fluorescence reveals that the cell at the center of the image has not achieved complete metaphase; the arrow indicates the position of a chromosome that is not aligned with the majority of the chromosomes at the metaphase plate. (C) Proper display of the fluorescence signal from the mitotic cell reveals that it has entered anaphase (arrow); improper scaling limits the ability to resolve the two separating masses of chromatids. (D) Image panels showing phase contrast and GFP-fluorescence images of a HeLa-H2B GFP cell progressing through mitosis. Phase contrast and GFP-fluorescence images were acquired at 5 minute intervals. Images representative of mitotic stages are shown. Scale bars = 10 μm
Figure 3.

Mitotic duration during the live cell imaging process remains constant. HeLa H2BGFP cells were imaged using an inverted fluorescence microscope system for 16 hours. After image acquisition, the images were analyzed; onset of prometaphase and anaphase for every cell was established and mitotic duration was calculated by determining the difference between these two values. These durations were plotted against the time elapsed post initiation of the live cell imaging process. Mitotic durations for the period of the assay were found to be consistent indicating that the culture was healthy throughout the experiment.
Figure 4.
Analysis of mitotic phenotypes upon Taxol treatment. HeLa Histone H2B-GFP cells were exposed to 0, 0.5, 1.0 or 2.0 nM Taxol and imaged with an inverted fluorescence microscope for 18 hours. After image acquisition, the images were examined; mitotic defects and variances in mitotic duration were determined. (A) The percentage of cells exhibiting normal mitotic progression or mitotic defects is depicted in a column graph. Example images of defects are shown. With increasing concentrations of Taxol, the percentage of cells progressing normally through mitosis decreased and the percentage of mitotic defects increased. Scale bars = 10 μm. (B) The elapsed time from prometaphase to anaphase onset, prometaphase to metaphase, and metaphase to anaphase onset was computed for cells progressing through mitosis in varying concentrations of Taxol. Results are plotted in a category scatter plot where data from each cell is depicted as a red circle. For each category the mean is labeled and the standard deviation is depicted by error bars. Statistical significance was calculated by performing an unpaired Student's t-test using data from control and experimental conditions; p values are indicated. With increasing concentrations of Taxol, the average time required to progress from prometaphase to anaphase increased. However, this increase was largely due to the increased duration taken from prometaphase to metaphase, as transition periods between metaphase and anaphase onset were largely unaffected. (see Color Plates).
Figure 5.
Analysis of mitotic aberrations caused by MAD2 or SKA3 depletion by RNAi. HeLa H2B-GFP cells were transfected with control (non-targeting), MAD2 or SKA3 siRNA duplexes at 25 nM. Cells depleted of these proteins were imaged for 15 hours. After image acquisition, the images were examined; mitotic defects and variances in mitotic duration were recorded. (A) Mitotic defects induced by different protein depletions were grouped into categories and are depicted in the column graph. Examples of observed defects are shown. The majority of control cells progressed normally through mitosis but a large proportion of MAD2-depleted cells initiated anaphase with unaligned chromosomes. In contrast a majority of SKA3-depleted cells delayed at metaphase or underwent cohesion fatigue, a phenomenon whereby extended metaphase arrest causes asynchronous separation of sister chromatids without normal anaphase onset or mitotic exit cells due to persistent microtubule pulling forces [5]. Scale bars = 10 μm. (B) Mitotic duration from prometaphase to anaphase onset, prometaphase to metaphase, and metaphase to anaphase onset was determined for every observed cell in the different protein depletions. Results were plotted in a category scatter plot where cells are depicted as red circles. For each category the mean is labeled and the error bars represent standard deviation. Statistical significance was calculated by performing an unpaired Student's t-test using data from control and experimental conditions; p values are indicated. The analysis indicates that MAD2-depletion decreases mitotic duration while SKA3-depletion increases mitotic duration. Time spent at metaphase decreases in MAD2-depleted cells as compared to control cells. The time taken to align chromosomes at the spindle equator and the metaphase duration increased in SKA3-depleted cells as compared to control cells. (see Color Plates).
NOTES
The system should include software that coordinates movements to and from assigned stage positions, that maintains focus, that regulates parameters of transmitted light and fluorescence illumination light exposures, and that controls digital image acquisition. The microscope should be designed for fluorescence microcopy with few optical elements in the light path in order to maximize the use of light; similarly, objectives should have a high numerical aperture (NA). In addition, the camera must be suitable for fluorescence microscopy. The more sensitive the detector of the camera, the lower the intensity of required illumination light. We use a Zeiss Axiovert 200M microscope equipped with a Zeiss 20X Plan-NEOFLUAR infinity-corrected 0.50 NA objective, an appropriate excitation/dichroic/emission filter set for GFP fluorescence, and a Zeiss MCU28 motorized stage controller. The filter set is a BrightLine Basic™ single-band filter set, optimized for Green Fluorescent Protein and other like fluorophores (GFP-A-Basic-ZHE, Semrock). The camera is a Hamamatsu Orca ER. Illumination light is provided by a wide-field fluorescence microscope excitation light source (X-Cite 120Q, Lumen Dynamics). Microscope system components and digital image acquisition are managed by Metamorph software (Molecular Devices).
The Air Stream Stage Incubator has a regulated heating system that moves a curtain of thermo-regulated air across the stage and culture. To ensure appropriate temperature regulation, a temperature probe is positioned adjacent to the culture chamber (digital thermometer model HH11, Omega).
We use Nunc Lab-Tek Chambered Coverglasses. These are polystyrene media chambers secured to a 1.0 Borosilicate chambered coverglass. One, 2, 4, or 8-well chambered coverglasses are available which permit up to 8 assay conditions per experiment.
Taxol/paclitaxel is a chemotherapeutic agent that inhibits normal microtubule dynamics by hyperstabilizing microtubules. Various other small molecular inhibitors that perturb mitotic progression are readily available. Inhibitors may interfere with microtubule dynamics, thus affecting the mitotic spindle, or inhibit kinases, phosphatases and other proteins integral to mitotic progression.
RNA interference (RNAi) methods can be used to target message and prevent translation of known or proposed mitotic regulators. Mitotic proteins SKA3 and MAD2 were depleted by RNAi and effects on mitotic progression were determined.
The parameters for RNAi treatment will vary with the characteristics of the target protein. For instance, siRNA treatments for stable proteins may require 2 – 3 days for adequate depletion while other proteins that turnover rapidly or that reveal phenotypes when partially depleted may require that RNAi be carried out the day before imaging. Depletions that compromise mitosis often result in cell death, and therefore parameters for RNAi-mediated depletion must be accurately established in preliminary experiments.
Ideally the cell culture will be at a density that accommodates the projected growth that will occur during image acquisition. Beginning an experiment at an appropriate cell density will help ensure the collection of interpretable images and provide sufficient space and nutrients for optimal cell proliferation.
Maintain this temperature for at least 30 minutes before imaging is initiated to ensure that the microscope stage and surrounding area has reached a temperature equilibrium that will be sustained by the Air Stream Incubator.
Usually we collect phase contrast and single plane GFP-fluorescence images at each stage location. For the 20X objective used in the above examples, a single image plane often provides sufficient depth for analysis. With the 20X objective each image filed contains many cells that can be analyzed as they enter and progress through mitosis. Acquisition of multiple focal planes at each location can be implemented to obtain additional information but this approach must be balanced with the necessity to avoid the phototoxic effects of fluorescence illumination.
The interval between image capture will determine the temporal resolution of the assay; the shorter the interval the greater the resolution. However increase in the frequency of image capture and therefore exposure to fluorescence illumination will increase the chance for phototoxic effects. For most purposes, images of the HeLa H2B-GFP cells are collected at 5 minute intervals. We find that acquisition frequencies near 5 minute intervals are sufficient to detect most errors that may occur during mitotic progression without compromising the health of cells through phototoxicity.
Medium exchange can be achieved by aspiration of the existing medium and replacement with previously prepared and warmed replacement medium. To analyze effects of Taxol on HeLa cells, the Leibovitz's L15 medium was supplemented with Taxol ranging in concentrations between 0 and 2 nM during the imaging assay.
The L15 replacement medium volume and the mineral oil volume should be approximately 50% and 40% respectively of the maximum volume recommended per chamber. The culture chamber's plastic cover may be absent during imaging; removing the cover often improves phase contrast or DIC images.
If the system relies upon the camera to set and test focus, it is preferable to use transmitted light rather than the fluorescence illumination.
The exposure times should be as short as is practical to produce an image with a reasonable signal to noise ratio and yet prevent photo-damage to the cell culture. These settings must be empirically determined based upon factors such as the efficiency of the microscope system and the fluorescent protein in use. In our system the camera contains a 12-bit digitizer that provides 4096 gray levels. For phase contrast images we typically expose the cells for 150 milliseconds to an intensity of transmitted light that results in an average intensity peak between 800 and 1200; this is sufficient to visualize the location and shape of cells within the selected field. For Histone H2B-GFP fluorescence acquisition, we minimize illumination intensity and exposure duration to obtain images that enable both detection and characterization of interphase nuclei. Histone H2B-GFP fluorescence emission in condensed mitotic chromosomes is far greater per unit area than the intensities obtained from the chromatin present in interphase nuclei, so we set our parameters to suitably visualize these dimmer objects that are required for analysis within the experiment. In our system, the fluorescence illumination light source is limited to 12.5% of maximum output and the exposure time is typically 40 milliseconds. We aim to achieve average gray levels for interphase nuclei to be approximately twice that of the average background values, those areas lacking GFP fluorescence. This representative setting results in average Histone H2B-GFP nuclear fluorescence grayscale values between 300 and 400 with average background intensities near 200. With the same parameters, Histone H2B-GFP fluorescence from condensed mitotic chromosomes ranges between 800 and 2000 gray levels. These intensities provide signal to noise capabilities sufficient for detecting single chromosomes within mitotic cells and observing cells for the duration of the experiment without causing phototoxicity.
The imaging software will have the capability to display the time at which images were acquired and/or display the elapsed time between images. The software may include functions to easily record these data points or the data may be entered manually into a separate spreadsheet application.
It is often advantageous to initially scan both control and experimental conditions to develop a catalog of frequent aberrations (e.g. tripolar spindles, lagging chromosomes in anaphase) prior to analyzing the control and experimental samples in full.
Mitosis in HeLa cells is fallible so one should expect a certain frequency of mitotic errors and variability in the duration of mitosis. However these should not significantly increase with time on the microscope under control conditions. If significant increases in these categories occur as time progresses through the image acquisition period, then the overall health of the culture was likely challenged and therefore conclusions attributed to experimental conditions may be erroneous.
Numerical analyses can be applied to the data to determine if differences are statistically significant. Herein, a commonly applied statistical hypothesis test, the unpaired t-test, is applied to establish statistically relevant differences between experimental conditions. A “p” value of less than 0.05 returned by the test indicates a likely statistically significant variation. It indicates that there is a 95% probability that control and experimental differences did not occur by chance.
REFERENCES
- 1.Paweletz N. Walther Flemming: pioneer of mitosis research. Nat Rev Mol Cell Biol. 2001;2(1):72–5. doi: 10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- 2.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8(7):377–85. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 3.Daum JR, et al. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19(17):1467–72. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 5.Daum JR, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21(12):1018–24. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]