SUMMARY
We have investigated the role of pairing centers (PCs), cis-acting sites required for accurate segregation of homologous chromosomes during meiosis in C. elegans. We find that these sites play two distinct roles that contribute to proper segregation. Chromosomes lacking PCs usually fail to synapse and also lack a synapsis-independent stabilization activity. The presence of a PC on just one copy of a chromosome pair promotes synapsis but does not support synapsis-independent pairing stabilization, indicating that these functions are separable. Once initiated, synapsis is highly processive, even between nonhomologous chromosomes of disparate lengths, elucidating how translocations suppress meiotic recombination in C. elegans. These findings suggest a multistep pathway for chromosome synapsis in which PCs impart selectivity and efficiency through a “kinetic proofreading” mechanism. We speculate that concentration of these activities at one region per chromosome may have coevolved with the loss of a point centromere to safeguard karyotype stability.
INTRODUCTION
Accurate segregation of chromosomes during meiosis is essential to reestablish a normal diploid genome at fertilization. The universal and defining event of meiosis is the segregation of homologous chromosomes, which in most species occurs during the first of two meiotic divisions. The ability of homologs to separate, or disjoin, from each other hinges on their ability to contact and recognize their partners and to form pairwise linkages that persist until chromosomes orient at the metaphase plate. The physical pairing of homologous chromosomes is usually stabilized during meiotic prophase by a protein scaffold known as the synaptonemal complex (SC), which polymerizes between homologous chromosomes. In most organisms, the more enduring physical links that connect homologs until metaphase also require the formation of meiotic crossovers, or chiasmata.
Perhaps the most mysterious aspect of meiosis is the mechanism by which each chromosome selectively synapses with its unique homologous partner (reviewed by Page and Hawley, 2003). Polymerization of the central element of the SC does not intrinsically require sequence homology and can occur inappropriately between nonhomologous regions under a variety of perturbed circumstances (reviewed by Zickler and Kleckner, 1998). However, synapsis is normally coupled with chromosome pairing so that only homologous regions ultimately synapse.
In C. elegans, genetic analysis of chromosome translocations, inversions, and deficiencies has revealed the existence of a particular region on each chromosome that determines its choice of meiotic segregation partner. Reciprocal-translocation chromosomes that share homology with two potential partners usually recombine with only one of the partial homologs (McKim et al., 1988). Furthermore, duplications of the left end of the X chromosome, but not the right end, will recombine with intact X chromosomes (Herman and Kari, 1989), and deletion of a region at the left end results in markedly reduced recombination between the Xs (Villeneuve, 1994). These and other observations have led to the idea that special sites, known as “homolog recognition regions,” or “pairing centers” (PCs), somehow govern meiotic interactions in C. elegans. These sites have been mapped toward one end of each chromosome by observing the segregation patterns of chromosome rearrangements (Figure 1A). Although PCs (as we refer to them) have been proposed to mediate homolog pairing, recognition, synapsis, or loading of the recombination machinery, these ideas have not been tested directly.
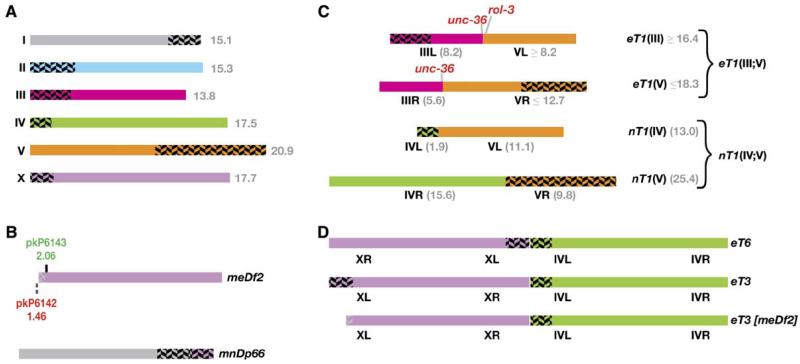
Figure 1. Normal and Rearranged Chromosomes Examined in This Study.
(A) The wild-type haploid karyotype of C. elegans. The physical interval that contains pairing-center function on each chromosome is indicated with herringbone shading. Left and right arms of each chromosome were arbitrarily designated in early genetic studies. This diagram is similar one in Albertson et al. (1997) but has been updated to include new mapping information (this study; Edgley and Riddle, 2001; Koh et al., 2004). All gray numbers indicate megabases of DNA.
(B) The X chromosome pairing-center deficiency meDf2(X) and the attached duplication mnDp66(X;I). meDf2 is a terminal or near-terminal deficiency; our SNP analysis revealed that its internal breakpoint is located between two SNP markers 1.46 and 2.06 Mb from the left end, and this region of ambiguity is indicated with light shading.
(C) Reciprocal-translocation chromosomes eT1(IV;V) and nT1(IV;V). For each half-translocation chromosome, the autosome that behaves as its meiotic segregation partner is indicated in parenthesis—i.e., eT1(III) recombines with and segregates away from chromosome III and thus (by definition) carries the chromosome III pairing center. Segment lengths shown in gray are deduced from the literature: the chromosome III breakpoint is within or very close to the unc-36 locus located 8.2 Mb from the left end. The chromosome V breakpoint is not precisely known; minimal and maximal physical lengths for each segment are based on the position of intervals known to be crossover suppressed or not suppressed. The precise breakpoints of nT1 were recently mapped by Koh et al. (2004).
(D) Fusion chromosomes eT6 and eT3 indicating the orientation of the PCs. Both of these are fusions between nearly intact copies of chromosomes IV and X. eT3[meDf2] is the product of exchange between eT3 and meDf2; it thus contains only the chromosome IV PC. All chromosome segments in (A)—(D) are drawn to scale based on physical coordinates from the WS140 freeze of Wormbase data from March 2005 (http://ws140.wormbase.org/).
Here we have used cytological methods to investigate the roles of PCs during meiosis. Our results reveal that these sites play at least two essential and separable roles in the process of homologous pairing and synapsis. Prior work has shown that pairing between homologous chromosomes is partially stabilized in SC-deficient mutants of C. elegans (Colaiácovo et al., 2003; MacQueen et al., 2002). Here we show that PCs mediate this synapsis-independent stabilization of pairing. We also show that—independent of this role in pairing—these sites promote the polymerization of the SC and are likely to be initiation sites for SC formation. We further demonstrate that PC activity is not obligately located near a chromosome end.
Our results illuminate additional key aspects of chromosome pairing and synapsis. Analysis of reciprocal translocations reveals that chromosome pairs that share a common PC undergo complete synapsis even if they are largely non-homologous and are of disparate physical lengths. This indicates that synapsis is highly processive and also implies the action of an adjustment mechanism that somehow equalizes the lengths of synapsed axial elements.
RESULTS
Pairing Centers Are Required for Normal Interactions between X Chromosomes
To evaluate the role of the pairing center during meiosis, we first analyzed the dynamics of interactions between X chromosomes deficient for these regions. meDf2 is a terminal deficiency that removes a large region from the left end of the X chromosome (Figure 1B). It is one of three similar deletions of the X PC recovered after exposure of worms to ionizing radiation (Villeneuve, 1994). X chromosome missegregation during meiosis in C. elegans hermaphrodites results in elevated frequencies of male (XO) progeny, known as the high incidence of males (Him) phenotype. Villeneuve (1994) found that hermaphrodites carrying two X chromosomes deleted for their PC regions display a very strong Him phenotype. However, a more modest segregation defect is seen when one X chromosome lacks the PC but its homolog is intact. This demonstrates that recognition of homology does not occur exclusively at these sites and also that a single, unpaired PC still contributes to homolog segregation.
We mapped three independent PC deletions using SNPs (see Experimental Procedures). All three remove at least 1.46 Mb from the left end of the chromosome but do not delete a site located 2.06 Mb from the left telomere. Because these deletions remove essential genes, homozygous or hemizygous animals must also carry a duplication of this region. Most duplications of the PC region of the X chromosome induce some detectable nondisjunction of normal X chromosomes (Herman and Kari, 1989; Herman et al., 1982). However, mnDp66 fuses approximately the same portion of the X chromosome deleted from meDf2 to the right end of chromosome I but does not recombine with normal X chromosomes or interfere with their segregation and thus seems to have a nonfunctional or noncompetitive X PC. This is further supported by experiments reported here and in the accompanying paper (Phillips et al., 2005 [this issue of Cell]), which indicate that mnDp66 does not show frequent physical associations with X chromosomes during meiosis.
To investigate the dynamics of X chromosome associations, we hybridized fluorescent probes to chromosomes in whole gonads (Figure 2; see also Table S1 in the Supplemental Data available with this article online), each of which contains a complete progression of meiotic prophase. Gonads were divided into five regions of equal size to evaluate the steady-state level of pairing as a function of meiotic progression. As demonstrated in previous work, loci on both ends of normal X chromosomes were predominantly unpaired in the premeiotic region of the gonad (zone 1) but paired rapidly after meiotic entry. Homologous pairing is normally stabilized by synapsis, such that all loci in wild-type animals remained paired through zone 5 of the time course, which corresponds to late pachytene.
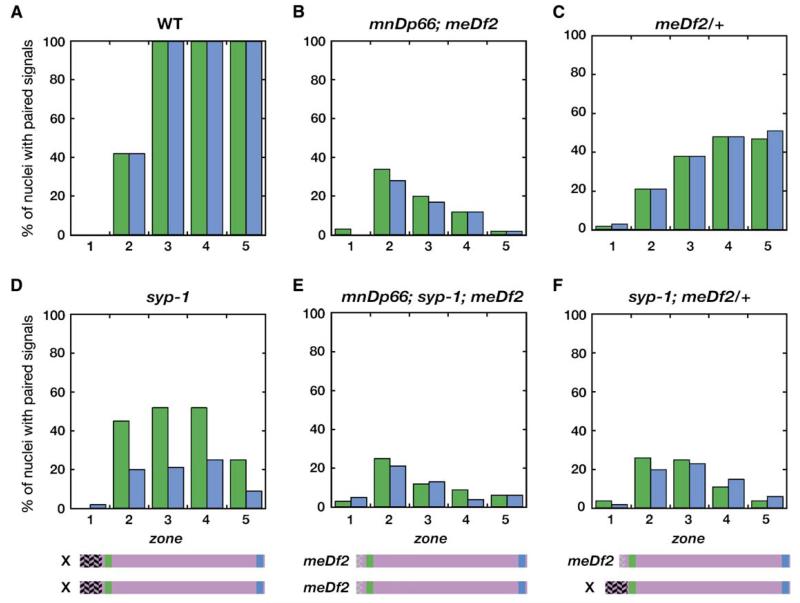
Figure 2. Quantification of X Chromosome Associations in Pairing-Center Mutants.
Probes from the left and right arms of the X chromosomes were hybridized to hermaphrodites of the indicated genotypes. The percent of nuclei with paired signals for each probe was measured for each of five temporal zones, as described by MacQueen and Villeneuve (2001). Zone 1 contains exclusively premeiotic nuclei, zone 2 contains both premeiotic and leptotene/zygotene stages, zone 3 represents early- to mid-pachytene stages, and zones 4 and 5 contain mid- to late-pachytene-stage nuclei. The genotypes in the top panels correspond to those in the bottom panels except that the bottom three panels show data from animals also homozygous for a syp-1 mutation, which eliminates an essential SC component. Corresponding numerical data are presented in Table S1.
X chromosome loci in meDf2 homozygotes displayed a transient increase in association during early meiotic prophase. However, in contrast to wild-type animals, these associations were not stabilized. The frequency of paired X chromosomes in meDf2 hermaphrodites reached a maximum in zone 2, which corresponds to the leptotene/zygotene stages of meiosis (the “transition zone”), and fell there-after. In meDf2/+ heterozygotes, homologous interactions between X chromosomes showed different dynamics (Figure 2C), consistent with their milder segregation defects. Pairing between X chromosome loci rose in early meiosis, but, compared to meDf2 homozygotes, both ends of the chromosome remained more highly associated through the end of pachytene (Figure 2B). Comparison between meDf2/+ (Figure 2C) and syp-1;meDf2/+ (Figure 2F) animals confirmed that these associations in zones 3-5 require syp-1 and therefore reflect synapsis between X chromosomes. Based on this analysis, 40% of meiotic nuclei in meDf2/+ heterozygotes achieve X chromosome synapsis by the end of prophase (Figure 2; Table S1). This difference between meDf2 and meDf2/+ hermaphrodites indicates that an unpaired PC can promote a substantial degree of homolog synapsis.
Pairing Centers Stabilize Homolog Pairing Even in the Absence of Synapsis
In principle, the importance of PCs during meiosis could be explained by our finding that they potently promote synapsis of the chromosomes that carry them. However, previous work has revealed that the PC end of each chromosome behaves distinctly even when synapsis does not occur. In syp-1 or syp-2 mutants, which lack essential structural components of the SC, all chromosome regions undergo transient pairing during early prophase. However, in mid- to late pachytene, the PC region of each chromosome remains more highly paired than loci from the opposite end (Colaiácovo et al., 2003; MacQueen et al., 2002). Thus, in the absence of synapsis, homologous pairing is locally stabilized near the PC region of each chromosome.
We tested whether this stabilization of pairing in the absence of SC proteins requires PC activity by combining meDf2 with a syp-1 mutation (Figures 2E and 2F). The differential behavior of the XL and XR probes observed in syp-1 hermaphrodites (Figure 2D) is not detected in syp-1;meDf2 animals (Figure 2E). Moreover, differences between XL and XR pairing were eliminated when even one X chromosome lacked a PC in syp-1 animals (Figure 2F). Fisher’s exact test or chi-square analysis confirm that XL showed significantly higher levels of pairing than the XR probe throughout meiosis in syp-1 mutants (p < 0.025 for either test in each of zones 2–5), but pairing of XL and XR was not significantly different in syp-1;meDf2 or syp-1;meDf2/+ animals. We conclude that meDf2 deletes all preferential stabilization activity from the left end of the X chromosome and that this synapsis-independent stabilization of pairing therefore requires the X chromosome PC. Further, a PC must be present on both chromosomes to promote local stabilization of pairing. Thus, meDf2/+ heterozygotes display a separation of function: the PC on the intact X chromosome retains some ability to promote synapsis in the absence of any local stabilization of homolog pairing.
Synapsis Does Not Strictly Require a Pairing Center
We directly visualized synaptonemal complex (SC) formation using antibodies to structural proteins of the SC (Figure 3). To detect the axial elements, we generated antibodies to HTP-3 (HIM three paralog 3). We observed that this protein forms axes along the chromosomes even before nuclei show other signs of meiotic entry—i.e., prior to and independently of homolog pairing (data not shown). This behavior is similar to that of a related protein, HIM-3 (Zetka et al., 1999), but HTP-3 loading occurred somewhat earlier and was independent of him-3 function (data not shown). The central region of the SC was detected using antibodies against SYP-1, which normally polymerizes along chromosomes concomitant with homolog pairing (MacQueen et al., 2002).
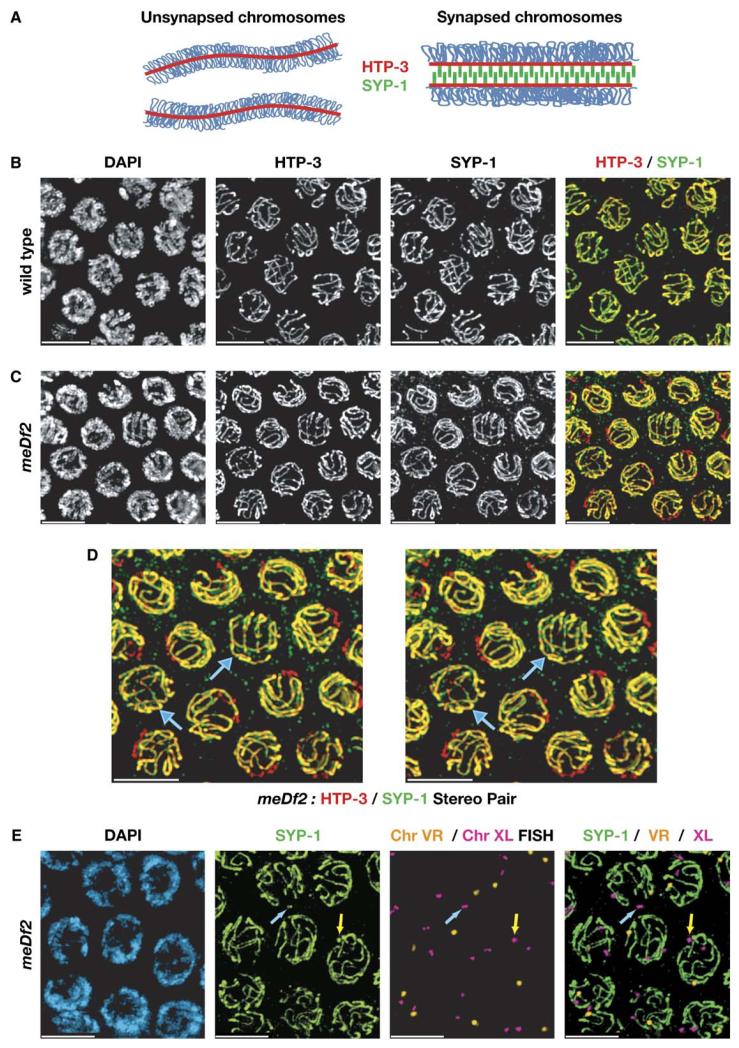
Figure 3. X Chromosomes Lacking Pairing Centers Form Axial Elements but Only Rarely Undergo Synapsis.
(A) illustrates the behavior of the proteins we have used here to visualize the axial and central elements of the SC. Axial-element proteins, including HTP-3, load onto meiotic chromosomes prior to pairing and synapsis. The presence of SYP-1, a component of the central element, defines synapsed segments. (B) and (C) show projections through fields of pachytene nuclei from gonads stained with antibodies against HTP-3 and SYP-1. (B) shows nuclei from a wild-type hermaphrodite. SYP-1 staining corresponds closely with all HTP-3 segments in the merged image, indicating an absence of unsynapsed chromosome regions. Six SCs, corresponding to the six synapsed chromosome pairs, can be detected in each nucleus. (C) shows images from a hermaphrodite homozygous for the PC deficiency meDf2. Regions of HTP-3 staining lacking SYP-1 staining can be detected in most of these nuclei, although in a small fraction of nuclei, complete synapsis with six contiguous stretches of SC is detected. Because the asynapsed regions and the difference between synapsed and asynapsed chromosomes is more easily observed in 3D images, the same region shown in (C) is displayed in (D) as a stereo pair of images. Blue arrows indicate the two nuclei in this field that are fully synapsed. In (E), in situ hybridization confirms that the X chromosomes are specifically asynapsed in meDf2 homozygotes. The X chromosome probe used here hybridizes to a region 2.2 Mb from the left end of the chromosome, which is retained on meDf2 and is not duplicated on mnDp66. In most pachytene-region nuclei, two separated X chromosome signals can be seen, in contrast to the chromosome V-derived probe, which is consistently paired. Interestingly, the X chromosome probes often colocalize with small foci of SYP-1 (two clear examples are indicated with arrows), suggesting that although SYP-1 does not polymerize along these PC-deficient chromosomes, it may load onto the chromosome end (or ends). All scale bars represent 5 μm.
By the onset of pachytene in wild-type gonads, all HTP-3 segments were associated with SYP-1 protein, indicating synapsis of all regions (Figure 3B). Six contiguous strands stained by both HTP-3 and SYP-1 antibodies could often be counted in 3D views of pachytene nuclei. By contrast, most nuclei in the pachytene region of meDf2 hermaphrodites contained segments of HTP-3 staining that lacked SYP-1, indicating regions of asynapsis (Figures 3C and 3D). Frequently, two distinct regions of HTP-3 devoid of SYP-1 could be observed in three-dimensional images of meDf2 nuclei. Immunolabeling combined with FISH confirmed that these SYP-1-negative regions corresponded to the X chromosomes (Figure 3E).
Despite their obvious synapsis defects, a small but significant fraction of nuclei in meDf2 homozygotes contained six HTP-3 and SYP-1-stained SCs of normal length, indicating complete synapsis (Figures 3C and 3D). Twenty-six fully synapsed nuclei were observed among 260 nuclei from the pachytene regions of four meDf2 hermaphrodites, or 10% of the total that we counted.
Fully synapsed nuclei comprised nearly half of the population observed at pachytene in meDf2/+ heterozygotes (Figure 4C and data not shown), consistent with our FISH-based measurement of 40% synapsis (Figure 2C; Table S1). The remainder appeared to have completely unsynapsed Xs. No indication of partial synapsis (i.e., HTP-3-staining segments partially stained with SYP-1) between X chromosomes was observed in this or any genetic background, suggesting that the SC spreads rapidly along the full length of the chromosomes whenever it initiates successfully.
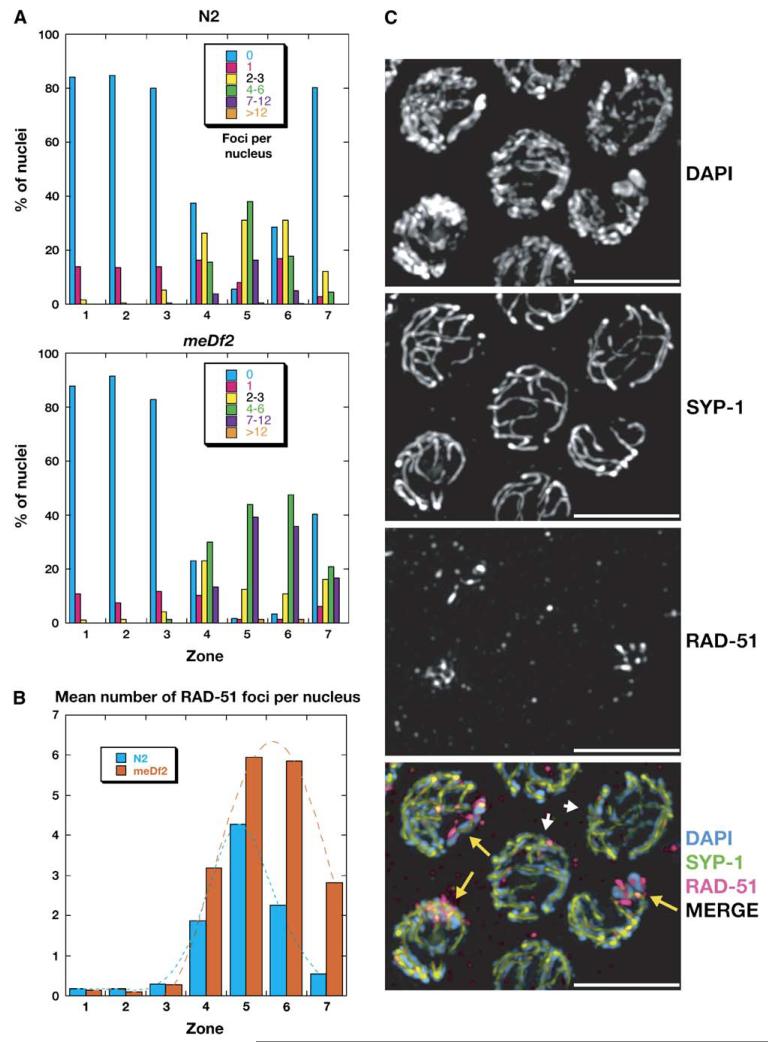
Figure 4. Recombination Intermediates Reach Higher Steady-State Levels and Persist Later in Nuclei with Defective X Chromosome Synapsis.
(A) The distribution of RAD-51 foci as a function of meiotic progression in N2 (wild-type) and meDf2 hermaphrodites was measured as in Colaiácovo et al. (2003). Each genotype is plotted independently. Gonads were divided into seven zones of equal size, which are enumerated along the x axis.
(B) To facilitate comparison between the genotypes analyzed in (A), the same data were consolidated to derive a mean number of RAD-51 foci per nucleus in each zone. For (A) and (B), the total numbers of nuclei scored for each genotype were as follows (in order by zone). N2: 344, 453, 471, 373, 327, 276, 138; total = 2382. meDf2: 257, 323, 342, 364, 296, 240, 149; total = 1971.
(C) RAD-51 foci are detected on unsynapsed X chromosomes. This projection displays late-pachytene nuclei from a meDf2/+ heterozygote. Two fully synapsed nuclei are indicated with small white arrows, and three nuclei with unsynapsed X chromosomes are indicated by yellow arrows. Nuclei containing unsynapsed chromosomes retain a more asymmetric chromosome distribution and display more numerous RAD-51 foci. Unsynapsed X chromosomes can be detected as brightly DAPI-stained regions that lack extensive SYP-1 staining. Late RAD-51 foci are abundant on the X chromosomes, perhaps most easily seen in the nucleus on the lower right. Scale bars represent 5 μm.
These observations indicate that the X chromosome PC is not absolutely required for SC polymerization or, alternatively, that meDf2 does not eliminate the synapsis-promoting activity of the PC although it deletes all local pairing stabilization activity. These possibilities are further considered in theDiscussion, below.
X Chromosomes Lacking Pairing Centers Incur Double-Strand Breaks and Can Undergo Crossing-over
To investigate the consequences of PC deletion on recombination, we used antibodies against RAD-51, a protein required to repair meiotic double-strand breaks (Rinaldo et al., 2002). In wild-type worms, RAD-51 foci begin to accumulate on chromosomes at the leptotene/zygotene stages of meiosis, peak during early pachytene, and are infrequently detected by late pachytene (Colaiácovo et al., 2003). Staining of meDf2 and meDf2/+ hermaphrodites revealed abundant RAD-51 foci at later-than-normal stages in the gonad (Figure 4). Similar delays in RAD-51 clearance have been observed in other mutants with defects in synapsis (Colaiácovo et al., 2003; Couteau et al., 2004). Interestingly, this delay appears to be cell autonomous: the subpopulation of nuclei in meDf2 and meDf2/+ hermaphrodites that does achieve synapsis also shows normal clearance of RAD-51 foci (Figure 4C). In meDf2 and meDf2/+ hermaphrodites, the unsynapsed X chromosomes showed a high concentration of persistent RAD-51 foci, indicating that PCs are not required in cis for initiation of meiotic recombination (Figure 4C and data not shown). Interestingly, many late foci are also detected on the synapsed autosomes of these nuclei, suggesting that the failure of the X chromosomes to synapse may influence recombination on the autosomes. We have explored this issue in detail elsewhere (Carlton et al., 2005).
The frequency of crossing-over between PC-deficient X chromosomes was analyzed by counting DAPI-stained bodies at diakinesis, the stage of prophase just prior to the first meiotic division. Gonads from meDf2 animals were hybridized with X chromosome FISH probes to enhance discrimination between bivalent (chiasmate) and univalent X chromosomes. Consistent with the observations of Villeneuve (1994), 23% of oocytes in meDf2 hermaphrodites showed only six bivalents, while 77% revealed seven distinct DAPI-staining bodies with separated X chromosomes (n = 100).
This observed frequency of bivalent X chromosomes in meDf2 hermaphrodites substantially exceeds the fraction of nuclei that displayed extensive X chromosome synapsis at pachytene, based on either FISH or immunostaining assays (above). A possible interpretation of this discrepancy is that some X chromosomes manage to undergo exchange without completely synapsing with their partners. However, the observation that nuclei with unsynapsed X chromosomes contain persistent RAD-51 foci suggests an alternate explanation. Unrepaired meiotic breaks can trigger a DNA-damage checkpoint, resulting in programmed cell death (Gartner et al., 2000). Apoptosis in C. elegans oocytes occurs during late pachytene, prior to diakinesis. Preferential apoptosis of nuclei with unsynapsed X chromosomes could potentially enrich the pool of oocytes at diakinesis and after division for the small fraction that have achieved X chromosome synapsis. In support of this idea, when a ced-3(n717) mutation, which reduces apoptosis, is combined with meDf2, only 12% of nuclei at diakinesis showed bivalent X chromosomes (n = 174). We believe that this lower frequency of chiasmata more accurately reflects the actual frequency of crossover formation, in part because crossovers are likely to require synapsis in C. elegans (MacQueen et al., 2002), and this frequency is similar to the 10% of meDf2 nuclei that displayed X chromosome synapsis (above). These observations have broad implications since they reveal that chromosome missegregation observed among the progeny of mutant worms is not a simple function of the frequency of defects in synapsis or crossover formation.
Evidence that Synapsis Initiates Near the Pairing-Center Region of the Chromosome and Proceeds through Nonhomologous Regions
Recognition of a special activity located near one end of each C. elegans chromosome arose from genetic studies of structurally rearranged chromosomes (Herman and Kari, 1989; McKim et al., 1988). In many organisms, heterozygosity for chromosome rearrangements such as inversions and translocations inhibits meiotic recombination near the breakpoints. This is usually attributed to local disruption of synapsis, which can sometimes be observed cytologically. In C. elegans, crossovers are suppressed in all regions distal to translocation breakpoints, where “distal” is defined in relation to the pairing centers, and not just near the breakpoints. In principle, the regions that are crossover-suppressed could be asynapsed, synapsed with nonhomologous regions, or homologously synapsed but not capable of forming crossovers, although preliminary cytological studies have indicated that the latter possibility is unlikely (Goldstein, 1986; Loidl et al., 2004).
To understand the basis for crossover suppression in translocation heterozygotes, we examined their synapsis behavior. eT1 and nT1 are two of the best characterized “balancer” rearrangements used in C. elegans genetics (Edgley et al., 1995; Koh et al., 2004; Rosenbluth and Baillie, 1981). Each is a reciprocal translocation that suppresses exchange over extensive portions of two autosomes (III and V or IV and V; Figure 1C). Crossing-over in eT1 and nT1 heterozygotes occurs only from the pairing-center end to the breakpoint on each autosome.
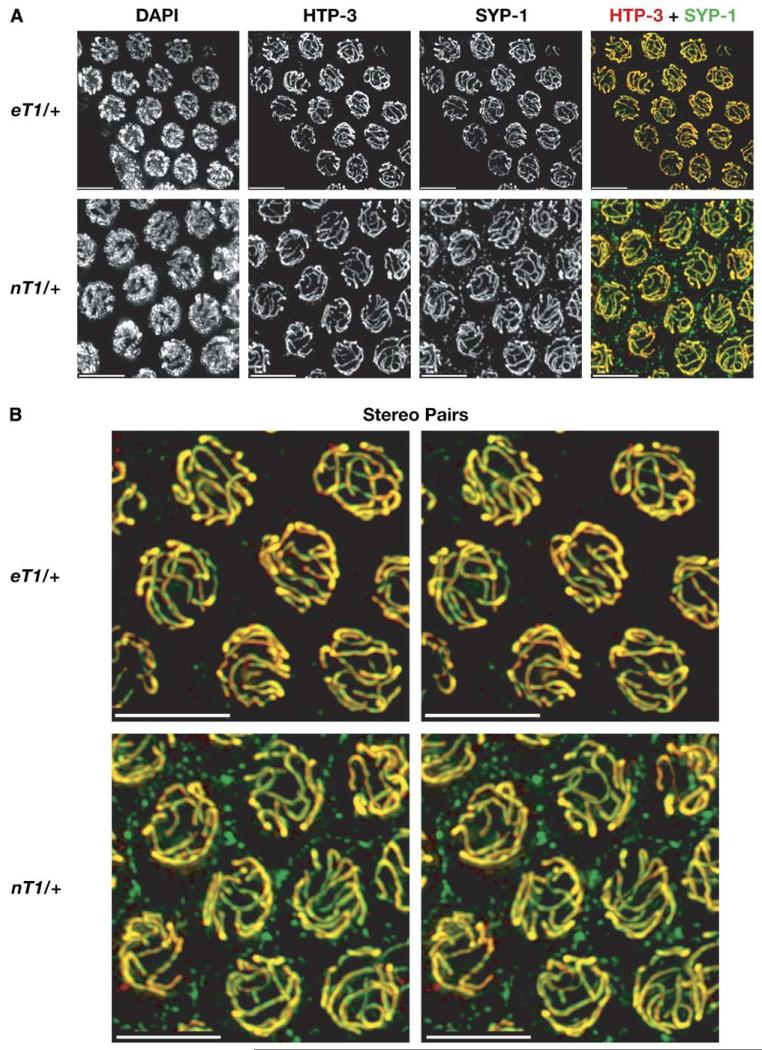
In eT1 or nT1 heterozygotes, we observed six SC segments in pachytene nuclei (Figure 5). All HTP-3 staining segments were associated with SYP-1, indicating synapsis of all chromosome regions. Strictly homologous synapsis in these animals would result in cruciform, or quadrivalent, SCs, and such structures were never detected. This suggests that the crossover-suppressed regions are nonhomologously synapsed, which would account for their inability to undergo reciprocal recombination.
Figure 5. Reciprocal Translocations that Suppress Crossing-over Do Not Cause Asynapsis.
These panels show immunostaining of the SC in hermaphrodites heterozygous for the reciprocal translocations eT1 and nT1 (diagrammed in Figure 1). As in wild-type animals, no regions of unsynapsed axial elements are observed. Stereo pairs of regions from the merged images are also shown because they make it easier to observe the six contiguous SCs in each nucleus and the absence of cruciform (quadrivalent) structures. All scale bars represent 5 μm.
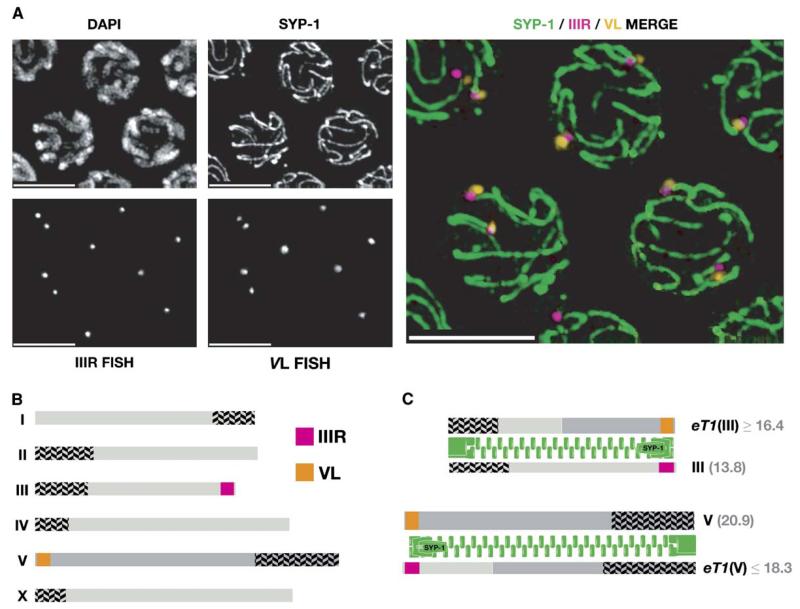
By combining immunostaining with FISH, we directly observed nonhomologous synapsis in these crossover-suppressed regions (Figure 6). Probes to the nonhomologous ends of eT1(III)/III and eT1(V)/V were closely juxtaposed at pachytene at one end of each of two linear SCs. By contrast, probes from the pairing-center regions of these chromosomes were homologously paired (data not shown). Thus, eT1 and nT1 heterozygotes undergo complete synapsis between chromosomes with matching PC regions (Figure 6C), despite the fact that they have extensive regions of nonhomology.
Figure 6. Regions that Are Crossover-Suppressed in Reciprocal-Translocation Heterozygotes Participate in Nonhomologous Synapsis.
(A) shows separate and merged images of pachytene nuclei from an eT1/+ heterozygote stained with anti-SYP-1 antibodies and hybridized with fluorescent probes to the crossover-suppressed (non-PC) ends of chromosomes III and V. As in Figure 5, six SCs are observed in each nucleus, and two of these show closely associated nonhomologous probe pairs at one end. (B) indicates the position of the probes on the wild-type karyotype. (C) illustrates the most straightforward explanation for the observations: synapsis initiates at the homologously paired PC end of the chromosomes and “zippers” up along the complete length of each chromosome pair, resulting in nonhomologous synapsis of these distal regions associated with homologous PCs. The eT1 (III)/III and eT1(V)/V pairs both involve synapsis between chromosomes of unequal length, suggesting that there is a mechanism that compacts and/or extends chromosome regions to equalize these lengths. Scale bars represent 5 μm.
Together with evidence that PCs are important for the efficient initiation of synapsis, these observations are most easily explained if we propose that (1) synapsis initiates at or near the PC and (2) polymerization of the SC is highly processive and, once initiated, will “zipper” up two chromosomes with out regard to homology distant from the PC. The idea that synapsis is very processive is also consistent with the absence of partial synapsis observed for X chromosomes in any of the mutant genotypes analyzed here.
Interestingly, both the homologous and nonhomologous ends of the translocation chromosomes were closely juxtaposed with their partners at pachytene, with no apparent “overhang” of axial-element proteins or chromatin on either end (Figures 5 and 6). This is significant in light of the discrepancy between the lengths of the synapsed chromosome pairs. The chromosome III breakpoint in eT1 is in or very near the unc-36 locus, and eT1 balances the left portion of chromosome V through at least the rol-3 gene (Barbazuk et al., 1994); thus, the pairs of synapsed chromosomes in an eT1 heterozygote differ in physical length by at least 22% (III and eT1(III)) and 14% (V and eT1(V)) (Figures 1C and 6C). The length disparity is even greater between synaptic partners in nT1 heterozygotes (Figure 1). These observations imply the existence of a mechanism that globally aligns the mismatched chromosomes and adjusts the lengths of their axes, perhaps by matching up the telomeres and condensing or decondensing the chromosomes, as necessary, to equalize their lengths at pachytene. We cannot currently determine whether this length equalization occurs before or concomitantly with synapsis since it has not been possible to measure the lengths of unsynapsed axial elements with enough precision.
These observations can explain the genetic behavior of large heterozygous insertions and deletions in C. elegans, which disrupt crossing-over in the vicinity of the insertion but are often partially permissive for crossing-over distal to the region of presumed nonhomologous synapsis (McKim et al., 1993). Similar adjustments in SC formation between chromosomes of different lengths have been observed in mammals and plants (reviewed by Zickler and Kleckner, 1999).
Pairing-Center Function Does Not Require Proximity to Chromosome Ends
Genetic mapping has revealed that PCs are located toward one end of each chromosome (Figure 1A), raising the question of whether their function requires proximity to a telomere. A possible relationship between PCs and telomeres is further suggested by our findings that PCs associate with the nuclear envelope during meiosis (Phillips et al., 2005), as do telomeres in many organisms. To test whether pairing centers can function at locations other than chromosome ends, we examined the meiotic behavior of eT3 and eT6, two different fusions between chromosomes X and IV (S. Ahmed, personal communication; Figure 1D). C. elegans hermaphrodites can transmit these and other X autosome fusion chromosomes efficiently to their progeny, although heterozygous animals show high X and autosomal nondisjunction (Herman et al., 1982; Hillers and Villeneuve, 2003; Sigurdson et al., 1986).
On eT6, the PC ends of both the X and chromosome IV are oriented internally, near the junction (Figure 1D). Consistent with their efficient segregation, we found that synapsis occurred regularly along the entire length of homozygous eT6 chromosomes (Figure S1). The behavior of eT6 argues that the PC does not need to be near an end to promote homologous synapsis, but it does not eliminate the possibility that telomeric sequences are important for PC function since eT6 does contain TTAGGC repeats at the junction between X and IV (M. Lowden and S. Ahmed, personal communication).
In principle, each PC on eT6 could act near the junction to promote synapsis directionally toward its respective end of the chromosome. To explore whether a single, internally located PC can promote synapsis toward both chromosome ends, the X chromosome PC was deleted from the X-IV fusion eT3 by recombination with meDf2. This created an X-IV fusion that has only the chromosome IV PC, located internally near the junction (diagrammed in Figure 1). Immunostaining of SC components revealed that this chromosome, like eT6 and intact eT3 (data not shown), regularly undergoes synapsis along its length in homozygous animals (Figure S1). This demonstrates that a medially located PC can promote synapsis that extends to both ends of the fusion chromosome.
DISCUSSION
Pairing Centers Are Essential for Accurate Meiotic Segregation but Are Not Strictly Required for Homolog Pairing, Synapsis, or Crossing-over
Our data have demonstrated that PCs locally stabilize homolog pairing in the absence of synapsis; they also promote synapsis even in the absence of this stabilization activity. It is not yet clear whether PCs also actively facilitate encounters between homologs. Our observations that the unpaired PC in meDf2/+ heterozygotes retains substantial synapsis-promoting activity in the absence of local synapsis-independent stabilization activity reveal that the stimulation of synapsis by the PC is not simply a consequence of increased perdurance of pairing interactions. It is not yet known whether these distinct activities originate from the same locus on the chromosome or from separate but closely linked sites. Either way, it may be advantageous to the cell to physically link these activities as a means to coordinate the processes of homolog pairing and synapsis.
One surprising conclusion is that PCs profoundly influence the probability of synapsis and crossing-over without being strictly required for their occurrence. Homolog pairing, synapsis, and crossover recombination all take place, albeit transiently or inefficiently, between chromosomes lacking pairing centers. This could be attributed to residual PC activity on the meDf2 chromosome, but our results argue against this interpretation. Specifically, we have shown here that meDf2 eliminates preferential stabilization activity from the left end of the X chromosome. Results in an accompanying paper (Phillips et al., 2005) also reveal that meDf2 removes the primary binding site for an essential PC-interacting protein. Previous work has shown that the recognition of homology is not an exclusive property of the pairing-center region of the chromosome (Villeneuve, 1994). Moreover, chromosome regions distant from PCs can cross over, which requires intimate homologous interactions, even when partner chromosomes lack contiguous homology (McKim et al., 1993). We favor the idea that, while activities that stabilize pairing and promote synapsis are highly concentrated at the PCs, these activities are also distributed elsewhere along the chromosomes, either at discrete sites or to some extent at all sequences.
A Multistep Model for PC-Mediated Pairing and Synapsis
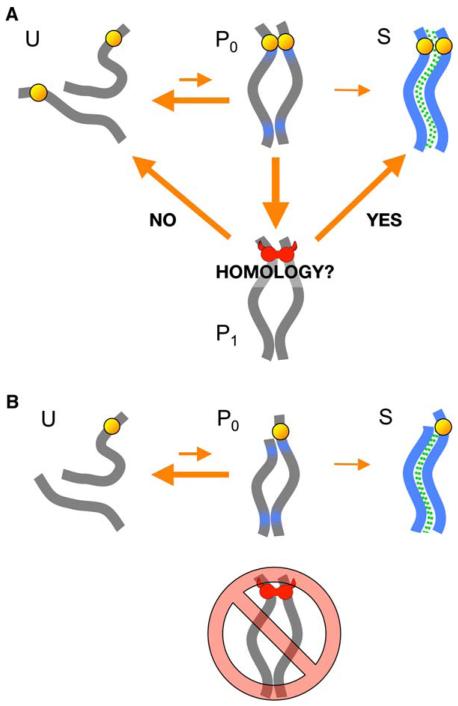
Our results suggest a model (Figure 7) describing how the PC might enable chromosomes to transition efficiently and accurately from an unpaired (U) to a homologously synapsed (S) state. We propose that the local stabilization of pairing by the PC creates a long-lived but reversible state (P1) that allows the evaluation of local homology. Initial, transient contacts between homologous or nonhomologous chromosomes are designated in the model as P0. When pairing centers interact with each other, P0 can transition to the longer-lived P1. If homology criteria are satisfied, the paired chromosomes are maintained at P1 until they transition to the S (synapsed) state. On the other hand, if homology recognition mechanisms determine that the partners are inappropriate, chromosomes transition from P1 back to U to enable a new search for the correct partner. For synapsis to preferentially occur homologously, the rate of synapsis initiation must be slow relative to the average persistence of the P0 state. When the P1 state occurs between homologous partners, it is long lived, providing an extended time window for synapsis to initiate.
Figure 7. A Model for the Role of PCs in Chromosome Pairing and Synapsis.
(A) The normal pathway leading from unpaired (U) chromosomes to homologous synapsis (S). The P0 paired state is transient, but the P1 state is stabilized by an interaction between homologous PCs. Yellow balls represent pairing centers, and the green dashed lines represent the synaptonemal complex. The red “horns” represent the Maxwellian demon-like properties of the pairing centers at the P1 state. See the text of the Discussion for explanation.
(B) An alternate pathway to synapsis when the pairing center is missing from one of two homologous chromosomes.
This model can account for the substantial levels of synapsis observed between normal and PC-deficient X chromosomes if we propose that a single PC enhances the P0→S transition rate (Figure 7B), thereby promoting synapsis initiation even in the absence of the P1 state. When both PCs are absent, the P0→S transition rate is further reduced, but our observation that meDf2 chromosomes do undergo occasional synapsis implies that this transition can still occur.
One prediction of this model is that chromosome synapsis might occur not only less frequently but also more stochastically in the absence of the stabilization activity due to ongoing collisions during meiotic prophase. This idea is supported by our FISH analysis of meDf2/+ heterozygotes, in which the fraction of synapsed X chromosomes continues to rise throughout pachytene (Figure 2C, zones 3–5). Because it sidesteps the stabilized state, which we have proposed to play an important role in homology discrimination, we predict that this route to synapsis might also be error prone relative to the normal pathway. However, this prediction has not been tested here. In meDf2/+ heterozygotes, all of the autosomes carry functional PCs, which promote their efficient synapsis and prompt removal from the pool of unsynapsed potential partners. This lack of competition may facilitate proper X chromosome partner choice even in the absence of stabilization.
The Pairing Center as a Maxwellian Demon
We suggest that the PC enables the cell to overcome a thermodynamic barrier to achieve accurate sorting and that it does so by using energy to kinetically stabilize a key transition state, the P1 state. This proposed function meets the definition of “kinetic proofreading,” as outlined by Hopfield (1974). He demonstrated that the error rate in multistep biological processes could be reduced through kinetic stabilization of transition states and recognized that this proofreading must consume energy.
In the case of PC function, energy might be expended to make the transition from the P0 to the P1 state irreversible and/or to perpetuate the P1 state for long periods relative to the rate of synapsis initiation. For example, homologous associations at PCs might be maintained through phosphorylation of protein targets or by hydrolysis of ATP or GTP by PC-associated molecular motors. Molecular dissection of pairing-center components will make it possible to test these ideas.
This proposed sorting function of the PC suggests an analogy with Maxwell’s demon, an imaginary creature proposed by physicist John Clerk Maxwell that could distinguish fast-from slow-moving molecules in a volume of gas. In our model, the stabilization of pairing facilitates sorting by allowing homology to be assessed prior to synapsis initiation. This aspect of the model is indicated by the horns on the PCs in the P1 state (Figure 7).
Pairing-Center Conservation and Evolution
The essential role of cis-acting pairing centers distinguishes C. elegans from other species in which meiosis has been extensively analyzed. In most organisms, the stabilization of pairing and initiation of synapsis are not generally thought to depend on specific chromosome regions. Telomeres may contribute to this process via formation of the meiotic bouquet, but synapsis frequently initiates at interstitial positions (reviewed by Zickler and Kleckner, 1998).
In Drosophila, there is evidence for sites that play special roles during female meiosis as well as a distinct set of sites that are essential for meiotic segregation in Drosophila males. Both differ in significant ways from the pairing centers of C. elegans. The sites that have been identified in female meiosis act as boundaries of crossover regulation, but it is not yet clear whether they contribute to either pairing or synapsis (Hawley, 1980; Sherizen et al., 2005). Male pairing sites share a major function with C. elegans PCs in that they are required to stabilize homolog association and to permit proper segregation. Unlike PCs, however, they do not contribute to synapsis initiation since Drosophila spermatocytes lack SCs.
Evidence from other model systems has indicated that the recombination machinery contributes to synapsis-independent stabilization of pairing, although such a contribution is either nonexistent or dispensable in C. elegans and Drosophila. Perhaps the most straightforward demonstration comes from real-time analysis of meiotic chromosomes in living S. pombe cells (Ding et al., 2004). In fission yeast, there is no synapsis. In the absence of double-strand breaks, homologous interactions are very transient throughout meiosis, but they are normally stabilized via a recombination-dependent mechanism. In other organisms, it has been difficult to tease apart the role of the recombination machinery in pairing and synapsis, largely because recombination and synapsis are often mutually interdependent. However, recent evidence from S. cerevisiae shows that both crossover and noncrossover recombination intermediates stabilize interhomolog interactions, and at least the noncrossover pathway’s contribution is independent of any role in initiating synapsis (Peoples-Holst and Burgess, 2005). It remains ambiguous whether the recombination machinery plays a direct role in initiating synapsis in any system or instead stimulates SC formation by stabilizing pairing, but in vivo kinetic analysis should help to resolve this issue.
It is possible that the concentration of these key meiotic functions at a unique region on C. elegans chromosomes is related to the fact that worm chromosomes are holocentric in mitosis and probably also in meiosis (reviewed by Albertson et al., 1997). The overt functions of pairing centers and centromeres are quite distinct, and there is no evidence that pairing centers contribute to the quintessential roles of the centromere in kinetochore formation or microtubule attachment during mitosis or meiosis. Nevertheless, centromeres play another, less obvious but essential role in chromosome integrity simply by virtue of their singularity. In monocentric species, where a unique site on each chromosome mediates the indispensable functions of spindle attachment and segregation, chromosome rearrangements are usually eliminated through mitotic divisions. In particular, any event that produces an acentric fragment will result in chromosome loss, usually leading to the death of that cell’s progeny. By contrast, chromosome fragments are transmissible through mitotic divisions in holocentric species (White, 1973). We propose that the presence of a unique and essential PC region on each chromosome prevents the efficient transmission of chromosome fragments through meiosis.
There are further reasons to suspect that pairing centers in C. elegans may be related to centromeres. In wheat meiosis, centromeres play a role in distinguishing homeologous from true homologous chromosomes (Martinez-Perez et al., 1999), indicating that they contribute to the recognition or enforcement of homology. Recent evidence has revealed a potentially analogous role for centromeres in budding-yeast meiosis (Tsubouchi and Roeder, 2005). Prior to synapsis, pairwise associations between centromeres are observed. These initial contacts occur irrespective of homology, and chromosome pairs are subsequently re-sorted to lead to homologous synapsis. As in our model (Figure 7), pairing between centromeres may mediate a metastable, paired state in which homology can be assessed. Centromeres are also thought to play a role in the segregation of nonexchange chromosomes in yeast (Kemp et al., 2004), which may be a manifestation of this putative stabilization function.
One possibility is that, as centromeres became delocalized in the lineage leading to C. elegans, the same sites acquired the indispensable function that they now play in meiotic pairing and synapsis. Although the existence of a unique site on each chromosome that confers centromere activity is dispensable and has been lost in many evolutionary lineages among plants and animals, an essential site that functions properly in one and only one copy per chromosome may be a universal feature of eukaryotic genomes.
EXPERIMENTAL PROCEDURES
Genetics and Mapping
All worms were raised at 20°C under standard culture conditions. Specific alleles used in this study were chromosome IV, ced-3(n717); chromosome V, syp-1(me17); chromosome X, unc-1(e528);dpy-3(e27);lon-2(e678); and chromosome aberrations eT1(III;IV), nT1[qIs51](IV;V), eT3(IV;X), eT6(IV;X), eT3[meDf2](IV;X), mnDp66(X;I), meDf2(X), meDf3(X), and meDf5(X).
To generate meDf2/ + animals lacking mnDp66, mnDp66;meDf2/0 males were mated to unc-1 dpy-3 hermaphrodites. Hermaphrodite crossprogeny were allowed to self-fertilize, and their Unc nonDpy progeny were selected. To generate the eT3[meDf2] chromosome, lon-2 was first crossed onto meDf2 to mark its left end.
The extent of the deficiencies meDf2, meDf3, and meDf5 was mapped using “snip-SNPs,” single nucleotide polymorphisms that alter a restriction site between N2 (the reference strain) and a Hawaiian isolate of C. elegans (Wicks et al., 2001). An unc-1 dpy-3 strain with Hawaiian-derived SNPs to the left of dpy-3 was generated by extensive outcrossing. mnDp66;meDf2/0 males were crossed to these Hawaiian unc-1 dpy-3 hermaphrodites; crossprogeny were allowed to self-fertilize, and Unc nonDpy hermaphrodites (which lack mnDp66) were picked and lysed for SNP analysis. Six SNPs ranging from 0.05 to 2.06 Mb from the left end of the X chromosome were examined; for SNPs within the deletion, only the Hawaiian restriction digest pattern was observed, whereas for SNPs outside the deletion, we detected both N2 and Hawaiian alleles.
Cytological Methods
FISH experiments were performed essentially as in Dernburg et al. (1998). Either formaldehyde fixation alone or a combined EGS/formaldehyde fixation procedure was used. Probes to single-copy sequences were generated using cosmids or YACs provided by the Sanger Centre. The X chromosome probes used for the pairing analysis in Figure 3 and Table S1 were synthesized from YACs Y24A9 (XL) and Y68A3 (XR). The probes for the experiment presented in Figure 6 were synthesized from cosmid pools T27A9, T25C8, T12D8, and ZK526 (for IIIR) and T01G6, D2051, and R11G11 (for VL). For experiments in which FISH was combined with immunostaining, FISH was generally performed first on samples fixed in 1% formaldehyde.
To raise antisera specific for HTP-3, the C-terminal exon of the predicted gene was amplified and cloned into pET-D/TOPO (Invitrogen), verified by sequencing, and expressed in E. coli. The recombinant His6-tagged protein was purified under denaturing conditions, separated from contaminants by SDS-PAGE, and excised from the gel. Chickens and guinea pigs were immunized, and reactive sera were used without further purification. Antisera against SYP-1 (MacQueen et al., 2002) and RAD-51 (Colaiácovo et al., 2003) have been described previously.
For immunostaining, dissected gonads were fixed in 1% formaldehyde in egg buffer (Dernburg et al., 1998) containing 0.1% Tween 20 for 5 min, freeze cracked into cold 100% dimethylformamide, and then transferred to PBS + 0.1% Tween 20 at room temperature. All primary antisera except for anti-RAD-51 were used without affinity purification, but in some experiments they were preadsorbed against formaldehyde/methanol-fixed whole worms to reduce nonspecific staining. Secondary antibodies were purchased from Jackson ImmunoResearch or Molecular Probes.
Three-dimensional images of stained gonads were collected using a DeltaVision microscope system on an Olympus IX70 platform (Applied Precision, Issaquah, WA). Most images were recorded using a 100× NA 1.35 UPlanApo objective. All images were deconvolved with a measured point-spread function. For display of all three-dimensional images presented here, projections through the full nuclear volumes were generated using a maximum-intensity algorithm.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Shawn Ahmed and Mia Lowden for generating, characterizing, and providing the chromosome fusions used in this work. This study was supported by NIH grant R01 GM53804 to A.M.V.; an NIH/Ruth L. Kirschstein Individual NRSA (GM067408) to N.B.; an NSF predoctoral fellowship to C.M.P.; and by LDRD funds from Lawrence Berkeley Lab, NIH R01 GM655591, and Burroughs Wellcome Career Award 1000950 to A.F.D.
Footnotes
Supplemental Data include one table and one figure and can be found with this article online at http://www.cell.com/cgi/content/full/123/6/1037/DC1/.
REFERENCES
- Albertson DG, Rose AM, Villeneuve AM. Chromosome organization, mitosis, and meiosis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 47–78. [PubMed] [Google Scholar]
- Barbazuk WB, Johnsen RC, Baillie DL. The generation and genetic analysis of suppressors of lethal mutations in the Caenorhabditis elegans rol-3(V) gene. Genetics. 1994;136:129–143. doi: 10.1093/genetics/136.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton PM, Farrugio AP, Dernburg AF. A link between meiotic prophase progression and crossover control. PLoS Genet. 2005 doi: 10.1371/journal.pgen.0020012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Couteau F, Nabeshima K, Villeneuve A, Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- Edgley ML, Riddle DL. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics. 2001;266:385–395. doi: 10.1007/s004380100523. [DOI] [PubMed] [Google Scholar]
- Edgley ML, Baillie DL, Riddle DL, Rose AM. Genetic balancers. Methods Cell Biol. 1995;48:147–184. [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Goldstein P. The synaptonemal complexes of Caenorhabditis elegans: the dominant him mutant mnT6 and pachytene karyotype analysis of the X-autosome translocation. Chromosoma. 1986;93:256–260. doi: 10.1007/BF00292746. [DOI] [PubMed] [Google Scholar]
- Hawley RS. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics. 1980;94:625–646. doi: 10.1093/genetics/94.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman RK, Kari CK. Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans. Genetics. 1989;121:723–737. doi: 10.1093/genetics/121.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman RK, Kari CK, Hartman PS. Dominant X-chromosome nondisjunction mutants of Caenorhabditis elegans. Genetics. 1982;102:379–400. doi: 10.1093/genetics/102.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B, Boumil RM, Stewart MN, Dawson DS. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 2004;18:1946–1951. doi: 10.1101/gad.1227304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Bernstein Y, Sundaram MV. The nT1 translocation separates vulval regulatory elements from the egl-18 and elt-6 GATA factor genes. Dev. Biol. 2004;267:252–263. doi: 10.1016/j.ydbio.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Loidl J, Pasierbek P, Rose AM. Conservation and variability of meiotic processes—lessons from the unconventional meiosis of C. elegans. In: Schmid M, Nanda I, editors. Chromosomes Today. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2004. pp. 93–101. [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Shaw P, Reader S, Aragon-Alcaide L, Miller T, Moore G. Homologous chromosome pairing in wheat. J. Cell Sci. 1999;112:1761–1769. doi: 10.1242/jcs.112.11.1761. [DOI] [PubMed] [Google Scholar]
- McKim KS, Howell AM, Rose AM. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics. 1988;120:987–1001. doi: 10.1093/genetics/120.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Peters K, Rose AM. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics. 1993;134:749–768. doi: 10.1093/genetics/134.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- Peoples-Holst TL, Burgess SM. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 2005;19:863–874. doi: 10.1101/gad.1293605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123 doi: 10.1016/j.cell.2005.09.035. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–479. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth RE, Baillie DL. The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics. 1981;99:415–428. doi: 10.1093/genetics/99.3-4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherizen D, Jang JK, Bhagat R, Kato N, McKim KS. Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics. 2005;169:767–781. doi: 10.1534/genetics.104.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson DC, Herman RK, Horton CA, Kari CK, Pratt SE. An X-autosome fusion chromosome of Caenorhabditis elegans. Mol. Gen. Genet. 1986;202:212–218. doi: 10.1007/BF00331639. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- Villeneuve AM. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics. 1994;136:887–902. doi: 10.1093/genetics/136.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. Animal Cytology and Evolution. Third Edition Cambridge University Press; Cambridge: 1973. [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.