Abstract
MicroRNAs (miRNAs) are endogenous oligo-ribonucleotides with exciting therapeutic potential. Early studies have established a clear role for miRNAs in leukocyte biology. The first miRNA-based therapy, Miravirsen, is now in Phase 2 clinical trials, making the reality of these therapies undeniable. The capacity for miRNAs to fine-tune inflammatory signaling make them attractive treatment targets for immunological diseases. Nonetheless, the degree of redundancy among miRNAs, coupled with promiscuity of miRNA binding sites in the transcriptome, require consideration when designing miRNA-directed interventions. Altered miRNA expression occurs across a range of inflammatory conditions including inflammatory bowel disease, arthritis and diabetes. To date, however, there have been very few studies successfully treating murine models of immunological diseases with miRNA-based approaches. While discussing recent studies targeting miRNAs to treat immunological conditions, we will also reflect on risks of miRNA-targeting and showcase some newer delivery systems that may improve the pharmacological profile of this class of therapeutics.
Introduction
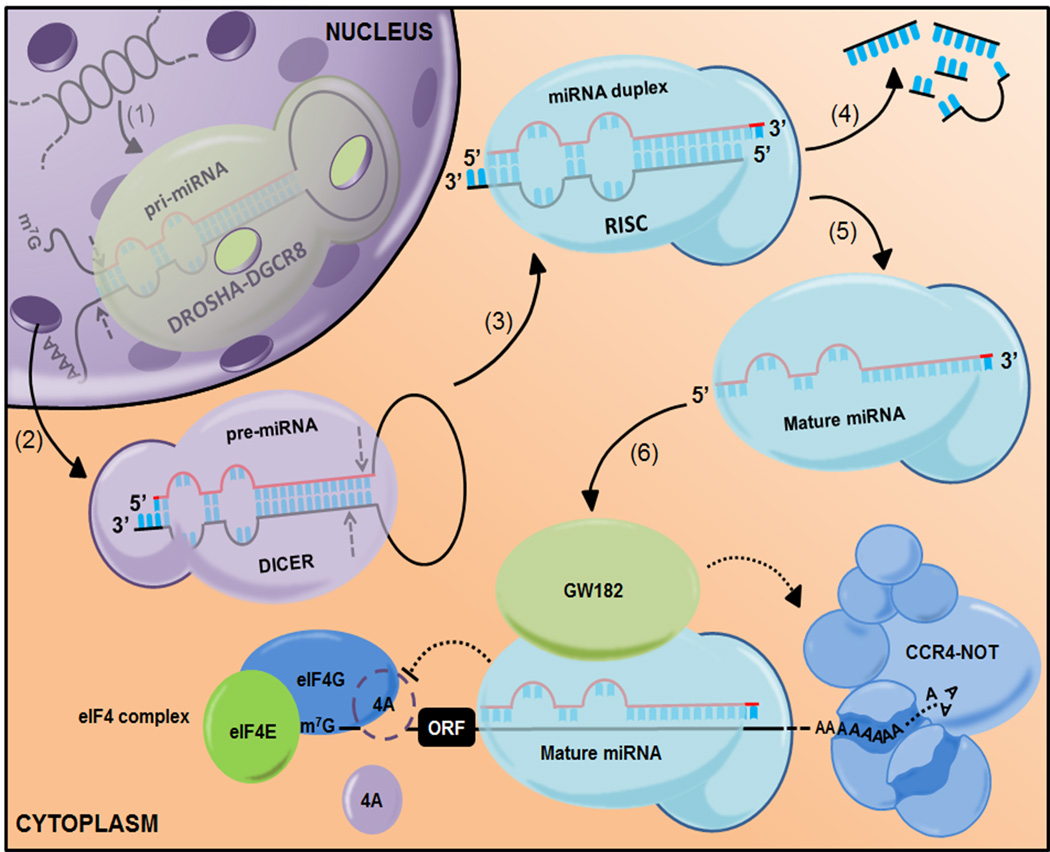
MicroRNAs (miRNAs) are short (approximately 22 nucleotides) untranslated single-stranded endogenous RNAs. They are transcribed either individually or as part of a polycistronic transcriptional unit, and the resulting primary miRNA transcripts are processed in the nucleus by the nuclear RNaseIII-like enzyme Drosha (1). The excised hairpin intermediates or precursor miRNAs (pre-miRNAs) are exported from the nucleus and processed into mature miRNAs by the cytoplasmic RNaseIII-like enzyme Dicer (2) (Figure 1). The clustering of some miRNAs into transcriptional units suggests that they may have co-evolved to perform redundant, complementary or sometimes antagonisitic functions (3), a feature that will be discussed later. Once formed, mature miRNAs are bound by Argonaute (Ago) proteins and act as guide sequences that bind to short, six to eight base pair complementary motifs on target messenger RNAs (4), generally located within the 3′ untranslated region (UTR). The six to eight base pair complementary sequence at the 5′ end of the miRNA is called the seed. Perfect complementarity between the miRNA and its target triggers RNA interference, cleavage mediated by the RNaseH-like domain of Ago2 (5, 6). However, in mammals, this has only been described for one target of miR-196 to date (7). In general, it is now believed that miRNAs promote deadenylation and mRNA decay at steady state in somatic mammalian cells (8). Under specific circumstances, miRNAs have been shown to inhibit initiation and/or elongation of protein translation (9). Recent evidence indicates that miRNA repress protein translation by blocking assembly of the eukaryotic translation initiation factor, eIF4F. Specifically, miRNAs form miRNA-induced silencing complexes with Ago proteins, which can either prevent association or actively displace the RNA helicase, eIF4A, from the eIF4F complex, thereby inhibiting the rate limiting step in translation (10, 11). Regardless of the mechanism, post-transcriptional control of gene expression by miRNAs provides a molecular rheostat to modulate specific genetic circuits, offering an additional layer to fine-tune the degree of protein synthesis.
Figure 1.
MicroRNA biogenesis pathway. The microRNA gene is transcribed into primary miRNA within the nucleus and cleaved by the Drosha-Dgcr8 Microprocessor complex. Dicer then converts the resulting hairpin intermediate, a pre-miRNA, to mature miRNA. This then binds to Argonaute proteins, core components of RNA-induced silencing complexes, leading to mRNA deaddenylation and decay as well as inhibition of mRNA translation in what is now thought to be an eIF4-dependent manner.
Current immunosuppressive therapies such as steroids induce broad-spectrum suppression of immune responses, which leads to increased susceptibility to infection. Targeting miRNAs may be of greater benefit for immunological diseases by limiting their action to a gentle nudge to restore the complex balance of our immune system between protecting and injuring the host. The feasibility of targeting miRNAs clinically has been given an enormous boost by the recent success of the pioneering miRNA-based therapeutic, Miravirsen (SPC-3649). A locked nucleic acid (LNA)-based inhibitor of miR-122, Miravirsen is currently undergoing clinical trials for the treatment of hepatitis C viral (HCV) infections (12). The role of the liver-specific miR-122 is somewhat unusual in that instead of targeting the 3′ UTR and causing signal repression, it binds to the 5′ UTR of HCV RNA and promotes its replication. Miravirsen’s success thus far paves the way for a new generation of miRNA-based therapeutics. Nevertheless, it must be noted that to date, there have been no approaches targeting miRNAs in the immune system that have advanced to clinical trials. Effective targeting of hepatocytes by Miravirsen does not guarantee the feasibility of targeting immune cells by any means. In fact, previous studies using unconjugated LNA-based miR-122 inhibitors demonstrated that significant accumulation of the LNA in hepatocytes occurred in an entirely passive manner upon intravenous injection (13). Therefore, as we shall discuss in greater detail, targeting of the immune system with miR-based therapeutics offers significantly greater challenges.
Indispensable role of miRNAs in immune cell development
Our understanding of the importance of miRNAs as a whole in immunocyte development and function initially came from genetically engineered mice carrying conditional mutant “floxed” alleles of the gene encoding Dicer, an enzyme critical for mature miRNA formation. Studies of T cell development demonstrate that early Dicer1 deletion in mice by the Lck-cre transgene impaired thymocyte proliferation and survival (14). Interestingly, restricting deletion of Dicer1 to a later stage of thymic maturation, by using CD4-cre had significantly less impact on CD4+ T cell development (14). CD4-cre-dependent deletion of Dicer1 did, however, lead to the development of colitis, due to a defect in T regulatory (Treg) cells (15). Consistent with that concept, restricting Dicer1 deletion in CD4+ FoxP3+ cells resulted in the failure of Treg development, leading to lethal multi-organ autoimmunity, similar to that seen in the Foxp3-deficient scurfy mouse (16, 17). Therefore while mature miRNAs might be partially dispensable for effector CD4+ T cell function, they are critical for development of Treg cells. Deletion of Dicer1 in CD8+ T cells results in more rapid T cell activation and impaired tissue egress (18) with both CD4 and CD8 T cell subsets displaying increased susceptibility to apoptosis (19). Furthermore, in the absence of Dicer, IFNγ is aberrantly expressed upon Th2 cell differentiation suggesting that miRNAs are essential for regulating Th2 cell plasticity. A similar phenomenon has been observed in vivo during viral infection (20), and miRNAs may provide one molecular mechanism that can control cell fate and ultimately shape immune responses. Cell-specific Dicer1 deletion has also identified important roles for miRNAs in the development and differentiation of B cells (21, 22), dendritic cells (23) and mast cells (24).
While these studies attest to the critical role of miRNAs in the maturation of the murine immune system, the global nature of the intervention, stopping production of all mature miRNA make it difficult to determine if a single miRNA (or many miRNAs) may be responsible for the resulting phenotypes. Although there are miRNA target prediction algorithms such as TargetScan (4) and PicTar (25), it is essential for the field to be able to identify and validate direct miRNA targets experimentally in a high throughput fashion. Invaluable protocols such as PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) though it requires metabolic labeling and HITS-CLIP (High-throughput Sequencing after UV-Crosslinking and Immunoprecipitation), which suffers from high background, have been developed (26, 27) for target identification. However, a good antibody or affinity tag to enrich for Ago protein and large numbers of cells are required for these approaches, which can limit feasibility of experimental designs. For CLIP, it is essential to have a negative control such as a miRNA-deficient control to help determine which Ago-binding events are miRNA-dependent (28). Moreover, the field needs a better way to validate miRNA targets rather than highly artificial luciferase reporter constructs in a heterologous transformed cell line. Although, it would be burdensome to mutate endogenous miRNA-binding sites, perhaps the field could propose agreeable alternate solutions. Nonetheless, the importance of miRNAs in immune cell development make them exciting targets for novel diagnostics and therapeutic development. Later in this article we will discuss a number of the more critical hurdles associated with the successful targeting of specific miRNA for therapeutic intervention. However, we will first discuss examples in which targeting of a specific miRNA has a beneficial effect on immunopathology.
Therapeutic targeting of miRNAs for the treatment of immune diseases
Ongoing research has progressed to the elucidation of specific miRNA actions. For example, one of the first studies demonstrated that miR-155 is required for germinal center reaction upon immunization (29, 30). Deletion of miR-155 curbed autoimmunity in a mouse model of lupus (31). Subsequently, it was shown that Th17 cells require miR-155 for development, and deletion of miR-155 confers resistance to experimental autoimmune encephalomyelitis (EAE) and uveitis (32–34). Similarly, miR-155 has been proposed as a potential therapeutic target for arthritis (35, 36). This body of work has been invaluable in furthering our understanding of how an individual miRNA contributes to the regulation of disease mechanisms. It also alerts us to the possibility that mutations in miRNAs or targets thereof (3′ UTRs) may result in human genetic disease. Certainly, miR-155 loss-of-function mutations in humans would be predicted to cause immunodeficiency.
An exciting proof of principle has come from studies in which miRNAs have been successfully targeted using synthetic miRNA inhibitors (33). One type called antagomirs are antisense 2′-O-methyl-modified, phosphorothioate-linked oligo-ribonucleotides conjugated to cholesterol, designed to enter cells and anneal to the miRNA of interest (37). These must be used with caution, however, as they have been recently identified as potent platelet activators (38). Intranasal administration of miR-126 or miR-145 antagomirs in mice inhibited Th2-mediated allergic airway disease induced by house dust mite (39, 40). Separate studies have demonstrated that this airway Th2 response is promoted by miR-106a and negatively regulated by let-7a (41, 42) and that modulation of the levels of these two miRNA in mice attenuated lung inflammation, reduced airway hyper-responsiveness and decreased Th2 cytokine IL-13 production. Recently, a study of miRNA expression in CD4+ T cells from lungs of asthmatic patients revealed elevation of miR-19a. They further show that miR-19 promotes development of Th2 cells in mice and humans, and suggest that inhibiting miR-19 might be useful for treating Th2-mediated immunopathology (43). However, it is important to understand the mechanism of action when targeting miRNAs to avoid unforeseen consequences. In many cases, the downstream targets of miRNAs have not been systematically determined in vivo.
An early example of successful inhibition of a single miRNA in a murine inflammatory model was directed against miR-326, a miRNA associated with Th17 fate determination (44). This study first demonstrated that miR-326 was significantly elevated in peripheral blood leukocytes of patients with multiple sclerosis (MS). Lentiviral approaches were used to overexpress miR-326 and accelerate EAE onset or inhibit miR-326 and attenuate EAE development. It must be stressed that a lentiviral-based approach is not currently applicable to human disease. While a number of clinical trials have been initiated using lentiviral approaches to treat immune disorders including chronic granulomatous disease and Wiskott-Aldrich Syndrome, these have yet to reach phase III trial stage and are, by design, limited to in vitro transduction. It may be necessary to include cell-specific promoters in lentivector design to improve their selectivity. Subsequent studies demonstrated a role for miR-326 in the pathogenesis of diabetes mellitus, with a similar upregulation of miR-326 in peripheral blood lymphocytes from patients with ongoing islet inflammation (45). The role played by miR-326 across a range of diseases may have implications for investment in new therapeutics. Therapies applicable to multiple diseases offer increased patient population sizes, providing greater return on investment, thereby making the cost of their research and development significantly more attractive. Traditional anti-inflammatories that have proven beneficial for the treatment of one immunological disease have then been successfully adopted for treatment of others e.g. anti-TNFα antibodies for rheumatoid arthritis (46) extended for use in Crohn’s disease (47). However, certain miRNA-based therapies may actually exacerbate other inflammatory conditions. For example miR-20b, which is down-regulated in MS and can be targeted for the treatment of EAE (48), is actually up-regulated in ulcerative colitis and may represent a potential biomarker of disease (49). Another consequence of targeting miR-326 is that it has been shown to exert an oncogenic role in glioma patients (50). Therefore chronic inhibition of miR-326 for the treatment of immunological diseases may come with increased risk of oncogenesis. Thus, it will be important to generate miR-326 deficient mice and figure out its functions in vivo to assess this risk.
Another miRNA, miR-10a has also proven protective in EAE (51). This study demonstrated that the combination of retinoic acid and TGFβ induced expression of miR-10a in naïve CD4+ T cells, and that miR-10a overexpression in transferred CD4+ T cells limited disease onset. As with the previous study, this approach attenuated, but failed to completely abrogate disease, consistent with the redundancy between miRNA regulatory pathways. It would be interesting to know if the combination of miR-10a supplementation and miR-326 inhibition would yield an additive effect. Once again, increased expression of miR-10a must be approached with caution as it too promotes cancer cell growth, migration, and invasion (52).
While a brief review of the literature might suggest that targeting anti-inflammatory miRNA would lead to increased oncogenesis, it may simply be an artifact of the cancer-heavy focus of miRNA research to date (at time of writing, a Pubmed search of “cancer” + “miRNA” yielded 15563 entries whereas “immune” + “miRNA” yielded 1884). Nevertheless, inadvertently increasing susceptibility to oncogenesis is already a feature of current anti-inflammatories such as infliximab, the gold standard anti-TNFα antibody for the treatment of inflammatory bowel disease (53). Moreover, a small increase may represent an acceptable risk especially given the association between malignancy and unchecked chronic inflammation (54). This is not a new concern for immunosuppressive therapies (55) but stresses the need to target miRNA with caution.
Importance of targeted delivery of miRNA-based therapies
As the aforementioned studies demonstrate, the risk of undesirable effects with miRNA targeting is significant. Successful approaches may depend on maximizing delivery to the target organ and thereby limiting off-target effects. While systemic delivery might be adequate for a filtering organ such as the liver, more localized delivery might improve positive outcomes in other tissues. Possibly the easiest way to restrict delivery might be to use traditional targeted delivery approaches such as rectal enema for the treatment of colitis (56) or topical delivery for dermatitis (57). In a similar way, it is not surprising that the successful miRNA therapeutic studies in lung inflammation models described previously all involved intranasal administration of antagomirs (39–42) as a means of localizing delivery. In reality, however, even organ-specific delivery of miRNA-based therapeutics may often be unsuitable as is demonstrated by driving expression of miR-141 to alleviate murine colitis (58). Despite the success of these studies, colonic delivery might be inappropriate in this situation because of the established association of miR-141 expression with colorectal cancer (59).
A small number of miRNAs provide their own solution to this issue via their intrinsic restriction of distribution. Site or cell-type specific miRNAs such as the endothelial-restricted miR-126 (60) or miR-935, whose expression is thought to be limited to eosinophils (61) are very much the exception rather than the rule. Even with cell-type specific expression of miRNA, their pleiotropic effects may make them unsuitable candidates for targeting. This has been best shown for miR-223 (62), a hematopoietic-specific miRNA that has a crucial function in control of myeloid lineage development. The multiple targets of miR-223 have caused controversy among rheumatology researchers who have separately identified both beneficial and deleterious roles for miR-223 in arthritis (63).
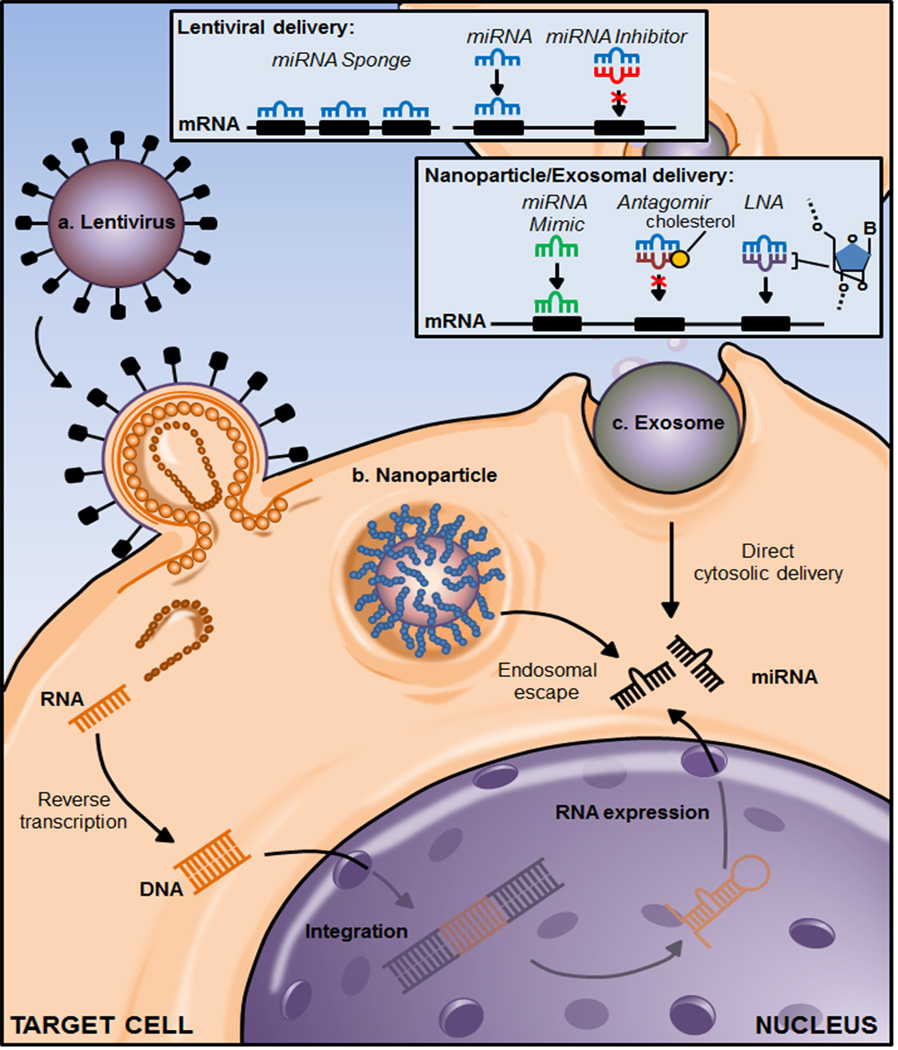
The aforementioned studies highlight the need for suitable delivery mechanisms for new miRNA-based therapeutics, to be included as an integral part of any therapeutic design (Figure 2). To that end a number of novel platforms, mostly adapted from small interfering RNA (siRNA) based or gene-delivery approaches, are being explored, such as stable nucleic acid lipid particles (SNALP) and short interfering ribonucleic neutrals (siRNN) specifically for targeting miRNAs (64, 65).
Figure 2.
Approaches to target miRNAs. Lentiviral delivery of expression vectors for miRNA over-expression or miRNA sponges and inhibitors that can block endogenous miRNA function. Synthetic miRNA mimics can be delivered into cells using nanoparticles or exosomes (89, 90). Nanoparticle and exosomal delivery can also be used for locked nucleic acids or antagomirs that bind and inhibit miRNAs. Diagrams are not drawn to scale.
Difficulties associated with miRNA targeting
Redundancy: the capacity of multiple miRNAs to regulate expression of a protein. Redundancy is thought to explain the failure of studies aimed at blocking miR-92a (66), a miRNA known to repress a number of pro-angiogenic factors including integrin α5 (67). While the study successfully demonstrated up to a fourteen-fold suppression of miR-92a in target organs in response to treatment, inhibition of miR-92a failed to have a pro-angiogenic effect (66), consistent with a possible redundant regulatory mechanism. There is significant redundancy within miRNA networks such that it may be prudent to block paralogous miRNAs, or indeed entire families of miRNAs as demonstrated with miRNA sponges (68). These false substrates act as competitive inhibitors by mimicking 3′ UTRs and stably binding complementary miRNA. This allows the simultaneous inhibition of multiple miRNAs (69, 70).
Efficiency: much like the therapeutic index of conventional therapies, understanding how much miRNA should be delivered/inhibited in a target cell is critical to the development of safe therapies. Over-expression of a miRNA can result in greater extraneous effects (71, 72) or have lethal consequences as the endogenous regulatory systems are overwhelmed (73). Thus, targeting of a miRNA must be done with a considerable degree of caution.
Immunogenicity: inhibition of miRNA activity can be achieved using the antagonistic effect of synthetic RNA that are complementary to the miRNA of interest, called antagomirs or aptamers. Naked delivery of miRNA antagomirs, aptamers or locked nucleic acids (LNA, containing an additional bond between oxygen and carbon to improve stability) offer both benefits and disadvantages for the treatment of immune diseases. Unlike current therapies such as anti-TNFα antibodies, which lose efficacy because of their capacity to elicit an antibody response (74), short nucleic acid sequences can be designed to avoid induction of an immune response to non-self RNA (75), making them significantly more attractive. Moreover, LNA have considerably longer serum half-lives (76, 77) when compared to current pharmacological or biological treatments. The downside of these approaches is the pleiotropic effects that may result.
These pitfalls have led to the development of encapsulation technologies that can then be modified to selectively target specific tissues or cell types. The use of nanoparticles to selectively target immune cells is increasingly common (78). These approaches have proven successful in the selective delivery of siRNA for the treatment of immune diseases (79) and therefore offer considerable promise for future miRNA-based studies. Surface conjugation with targeting antibodies may offer improved selectivity of the nanoparticles. One limitation of well-established liposomal delivery systems is that they are often charged nanoparticles that can have immunostimulatory effects due to their electrostatic interaction with cell surface molecules. While cationic nanoparticles have long been known to induce pro-inflammatory signaling pathways and stimulate chemokine release (80, 81), this phenomenon is actually polarity-independent as anionic nanoparticles can also augment immune responses. This side-effect may actually produce a desirable additive effect in the treatment of immunological diseases, particularly those that benefit from additional Treg induction (82), such as mouse models of Crohn’s disease (83). Another technique that is gaining traction involves highjack endogenous exosomes. This pre-existing miRNA delivery system offers some very exciting potential benefits.
Exosomal delivery of miRNA mimics
While exosomes have been proposed as a novel means of miRNA delivery for therapeutic applications, this field is still very much in its infancy. Considerable strides are still necessary to fully understand what role exosomes may play, if any, in miRNA trafficking under physiological conditions. Recent studies have suggested that during inflammatory conditions, CD4+ Tregs, which produce relatively large numbers of exosomes, deliver miRNA to other T cell subsets via exosomes in vivo as part of their suppressive function (84). This and other studies have delivered miR-155 mimics using lymphocyte-derived artificially-transduced exosomes as these are thought to express appropriate trafficking ligands on their surface (85). Exosome-like nanovesicles transfected with miR-150 and injected into recipient mice have also been shown to suppress contact sensitivity dermatitis (86). Nonetheless, important concerns remain regarding the absolute numbers of miRNA molecules contained within exosomes in vivo and hence the potential for a functional role (87). As our understanding of exosome function expands (88), their potential therapeutic applications may also.
Conclusions
While it is clear from studies outlined above that there are still hurdles to the use of miRNA-targeting approaches for clinical applications, these challenges must be weighed against the possible benefits that targeting miRNAs may have. Moreover, the rapid expansion occurring in this field currently suggests that miRNA-based therapeutics that can dampen inappropriate immune responses, without the unwanted immunosuppression seen with current therapies, are close at hand. The main concerns to be considered when designing a miRNA-based approach include understanding the selectivity of the miRNA, redundancy of the miRNA, and specificity of the delivery approach. So long as researchers treat each of these issues as important components in the design process, the prospects of miRNAs as therapeutic targets remain promising.
Acknowledgements
We wish to thank Joselyn N. Allen, B.S. (Pennsylvania State University) for creating the scientific illustrations within this review. We also thank Kelley Brodsky, Sandra Hoegl and Douglas Kominsky for helpful suggestions in writing this review. We apologize to those whose work could not be discussed because of space constraints.
Support
S.A.M. is supported by the NIH Intramural Research Program of the NIAID. C.B.C is supported by NIDDK K01DK099403-01 and CCFA CDA 253596.
Abbreviations used in this paper
- miRNAs
microRNAs
- pre-miRNAs
precursor miRNAs
- RNase
ribonuclease
- Ago
Argonaute
- UTR
untranslated region
- miR
microRNA
- eIF
eukaryotic initiation factor
- LNA
locked nucleic acid
- HCV
hepatitis C virus
- FoxP3
forkhead P3
- Treg
regulatory T cell
- IFN
interferon
- Th2
T helper type 2
- PAR-CLIP
Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation
- HITS-CLIP
High-throughput Sequencing after UV-Crosslinking and Immunoprecipitation
- Th17
T helper type 17
- TGFβ
transforming growth factor beta
- MS
multiple sclerosis
- EAE
experimental autoimmune encephalomyelitis
- siRNA
small interfering RNA
- SNALP
stable nucleic acid lipid particles
- siRNN
short interfering ribonucleic neutrals
Contributor Information
Stefan A. Muljo, Email: Stefan.Muljo@nih.gov.
Colm B. Collins, Email: Colm.Collins@ucdenver.edu.
References
- 1.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 2.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 3.Olive V, Sabio E, Bennett MJ, De Jong CS, Biton A, McGann JC, Greaney SK, Sodir NM, Zhou AY, Balakrishnan A, Foth M, Luftig MA, Goga A, Speed TP, Xuan Z, Evan GI, Wan Y, Minella AC, He L. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. eLife. 2013;2:e00822. doi: 10.7554/eLife.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 6.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 7.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Molecular cell. 2014;56:79–89. doi: 10.1016/j.molcel.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Fukaya T, Iwakawa HO, Tomari Y. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Molecular cell. 2014;56:67–78. doi: 10.1016/j.molcel.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Gebert LFR, Rebhan MAE, Crivelli SEM, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 14.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. The Journal of experimental medicine. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. The Journal of experimental medicine. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 17.Liston A, Lu L-F, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Lohning M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Xu S, Guo K, Zeng Q, Huo J, Lam K-P. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers H, Schnorfeil FM, Fehling H-J, Bartels H, Brocker T. Dicer-dependent microRNAs control maturation, function, and maintenance of Langerhans cells in vivo. J. Immunol. 2010;185:400–409. doi: 10.4049/jimmunol.0903912. [DOI] [PubMed] [Google Scholar]
- 24.Oh SY, Brandal S, Roers A, Zhu Z, et al. Global microRNA expression is essential for mast cell development in vivo. The Journal of. 2012 doi: 10.1016/j.exphem.2014.07.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nature genetics. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 26.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Molecular cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thai TH, Patterson HC, Pham DH, Kis-Toth K, Kaminski DA, Tsokos GC. Deletion of microRNA-155 reduces autoantibody responses and alleviates lupus-like disease in the Fas(lpr) mouse. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20194–20199. doi: 10.1073/pnas.1317632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escobar T, Yu CR, Muljo SA, Egwuagu CE. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Investigative ophthalmology & visual science. 2013;54:4017–4025. doi: 10.1167/iovs.13-11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. Journal of immunology. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bluml S, Bonelli M, Niederreiter B, Puchner A, Mayr G, Hayer S, Koenders MI, van den Berg WB, Smolen J, Redlich K. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis and rheumatism. 2011;63:1281–1288. doi: 10.1002/art.30281. [DOI] [PubMed] [Google Scholar]
- 37.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 38.Flierl U, Nero TL, Lim B, Arthur JF, Yao Y, Jung SM, Gitz E, Pollitt AY, Zaldivia MT, Jandrot-Perrus M, Schafer A, Nieswandt B, Andrews RK, Parker MW, Gardiner EE, Peter K. Phosphorothioate backbone modifications of nucleotide-based drugs are potent platelet activators. The Journal of experimental medicine. 2015 doi: 10.1084/jem.20140391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. The Journal of allergy and clinical immunology. 2011;128:160–167. e164. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A, Kumar M, Ahmad T, Mabalirajan U, Aich J, Agrawal A, Ghosh B. Antagonism of mmu-mir-106a attenuates asthma features in allergic murine model. Journal of applied physiology. 2012;113:459–464. doi: 10.1152/japplphysiol.00001.2012. [DOI] [PubMed] [Google Scholar]
- 42.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, Ghosh B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. The Journal of allergy and clinical immunology. 2011;128:1077–1085. e1071–e1010. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 43.Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, Wang Y, Pua HH, Baumjohann D, Montoya MM, Panduro M, Remedios KA, Huang X, Fahy JV, Arron JR, Woodruff PG, Ansel KM. A microRNA upregulated in asthma airway T cells promotes T2 cytokine production. Nature immunology. 2014 doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 45.Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes. Metab. Res. Rev. 2011;27:862–866. doi: 10.1002/dmrr.1262. [DOI] [PubMed] [Google Scholar]
- 46.Elliott MJ, Maini RN, Feldmann M, Kalden JR, Antoni C, Smolen JS, Leeb B, Breedveld FC, Macfarlane JD, Bijl JA, Woody JN. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α (cA2) versus placebo in rheumatoid arthritis. Lancet. 1994;344:1105–1110. doi: 10.1016/s0140-6736(94)90628-9. [DOI] [PubMed] [Google Scholar]
- 47.Targan SR, Hanauer SB, van Deventer SJ, Mayer L, Present DH, Braakman T, DeWoody KL, Schaible TF, Rutgeerts PJ. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn's disease. Crohn's Disease cA2 Study Group. N. Engl. J. Med. 1997;337:1029–1035. doi: 10.1056/NEJM199710093371502. [DOI] [PubMed] [Google Scholar]
- 48.Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S, Zhao Q, Zhao L, Wu Z, Yin Z. miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORγt and STAT3. J. Immunol. 2014;192:5599–5609. doi: 10.4049/jimmunol.1303488. [DOI] [PubMed] [Google Scholar]
- 49.Coskun M, Bjerrum JT, Seidelin JB, Troelsen JT, Olsen J, Nielsen OH. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J. Gastroenterol. 2013;19:4289–4299. doi: 10.3748/wjg.v19.i27.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Xu T, Yan Y, Qin R, Wang H, Zhang X, Huang Y, Wang Y, Lu Y, Fu D, Chen J. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1) PLoS One. 2013;8:e68469. doi: 10.1371/journal.pone.0068469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciumè G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O'Shea JJ. TGF-[beta] and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long M-J, Wu F-X, Li P, Liu M, Li X, Tang H. MicroRNA-10a targets CHL1 and promotes cell growth, migration and invasion in human cervical cancer cells. Cancer Lett. 2012;324:186–196. doi: 10.1016/j.canlet.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Aithal GP, Mansfield JC. Review article: the risk of lymphoma associated with inflammatory bowel disease and immunosuppressive treatment. Aliment. Pharmacol. Ther. 2001;15:1101–1108. doi: 10.1046/j.1365-2036.2001.01023.x. [DOI] [PubMed] [Google Scholar]
- 54.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 55.López-Serrano P, Pérez-Calle JL, Sánchez-Tembleque MD. Hepatitis B and inflammatory bowel disease: role of antiviral prophylaxis. World J. Gastroenterol. 2013;19:1342–1348. doi: 10.3748/wjg.v19.i9.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miner PB, Jr, Wedel MK, Xia S, Baker BF. Safety and efficacy of two dose formulations of alicaforsen enema compared with mesalazine enema for treatment of mild to moderate left-sided ulcerative colitis: a randomized, double-blind, active-controlled trial. Alimentary pharmacology & therapeutics. 2006;23:1403–1413. doi: 10.1111/j.1365-2036.2006.02837.x. [DOI] [PubMed] [Google Scholar]
- 57.Kigasawa K, Kajimoto K, Hama S, Saito A, Kanamura K, Kogure K. Noninvasive delivery of siRNA into the epidermis by iontophoresis using an atopic dermatitis-like model rat. International journal of pharmaceutics. 2010;383:157–160. doi: 10.1016/j.ijpharm.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 58.Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi H, Dong L, Zhang C, Zeng K, Chen J, Zhang J. miR-141 Regulates colonic leukocytic trafficking by targeting CXCL12β during murine colitis and human Crohn's disease. Gut. 2014;63:1247–1257. doi: 10.1136/gutjnl-2012-304213. [DOI] [PubMed] [Google Scholar]
- 59.Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D'Asaro M, Chiappe A, Sridhar S, Pacheco GD, Burczynski ME, Hochstrasser D, Vonderscher J, Matthes T. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One. 2012;7:e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y-T, Chen S-Y, Wang C-R, Liu M-F, Lin C-C, Jou IM, Shiau A-L, Wu C-L. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 2012;64:3240–3245. doi: 10.1002/art.34550. [DOI] [PubMed] [Google Scholar]
- 63.Chen S-Y. MicroRNA-223: a double-edged sword in rheumatoid arthritis. Rheumatol. Int. 2014;34:285–286. doi: 10.1007/s00296-013-2720-5. [DOI] [PubMed] [Google Scholar]
- 64.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 65.Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, Berg AV, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, Dowdy CF, Presente A, Lonn P, Kaulich M, Yoshioka N, Gros E, Cui XS, Dowdy SF. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nature biotechnology. 2014 doi: 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sengul A, Santisuk R, Xing W, Kesavan C. Systemic administration of an antagomir designed to inhibit miR-92, a regulator of angiogenesis, failed to modulate skeletal anabolic response to mechanical loading. Physiol. Res. 2013;62:221–226. doi: 10.33549/physiolres.932410. [DOI] [PubMed] [Google Scholar]
- 67.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 68.Dylla L, Jedlicka P. Growth-promoting role of the miR-106a~363 cluster in Ewing sarcoma. PLoS One. 2013;8:e63032. doi: 10.1371/journal.pone.0063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Näär AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci. Transl. Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai Z, Wu R, Zhao Y-C, Wang K-K, Huang Y-Y, Yang X, Xie Z-C, Tu C-C, Ouyang H-S, Wang T-D, Pang D-X. Early lethality of shRNA-transgenic pigs due to saturation of microRNA pathways. J. Zhejiang Univ. Sci. B. 2014;15:466–473. doi: 10.1631/jzus.B1400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frederiksen MT, Ainsworth MA, Brynskov J, Thomsen OO, Bendtzen K, Steenholdt C. Antibodies Against Infliximab Are Associated with De Novo Development of Antibodies to Adalimumab and Therapeutic Failure in Infliximab-to-Adalimumab Switchers with IBD. Inflamm. Bowel Dis. 2014 doi: 10.1097/MIB.0000000000000138. [DOI] [PubMed] [Google Scholar]
- 75.Yu D, Wang D, Zhu F-G, Bhagat L, Dai M, Kandimalla ER, Agrawal S. Modifications incorporated in CpG motifs of oligodeoxynucleotides lead to antagonist activity of toll-like receptors 7 and 9. J. Med. Chem. 2009;52:5108–5114. doi: 10.1021/jm900730r. [DOI] [PubMed] [Google Scholar]
- 76.Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C, McCauley TG. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004;21:2234–2246. doi: 10.1007/s11095-004-7676-4. [DOI] [PubMed] [Google Scholar]
- 77.Shi H, He X, Cui W, Wang K, Deng K, Li D, Xu F. Locked nucleic acid/DNA chimeric aptamer probe for tumor diagnosis with improved serum stability and extended imaging window in vivo. Anal. Chim. Acta. 2014;812:138–144. doi: 10.1016/j.aca.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 78.Laroui H, Geem D, Xiao B, Viennois E, Rakhya P, Denning T, Merlin D. Targeting Intestinal Inflammation With CD98 siRNA/PEI–loaded Nanoparticles. Mol. Ther. 2013;22:69–80. doi: 10.1038/mt.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scheinman RI, Trivedi R, Vermillion S, Kompella UB. Functionalized STAT1 siRNA nanoparticles regress rheumatoid arthritis in a mouse model. Nanomedicine. 2011;6:1669–1682. doi: 10.2217/nnm.11.90. [DOI] [PubMed] [Google Scholar]
- 80.Tanaka T, Legat A, Adam E, Steuve J, Gatot J-S, Vandenbranden M, Ulianov L, Lonez C, Ruysschaert J-M, Muraille E, Tuynder M, Goldman M, Jacquet A. DiC14-amidine cationic liposomes stimulate myeloid dendritic cells through Toll-like receptor 4. Eur. J. Immunol. 2008;38:1351–1357. doi: 10.1002/eji.200737998. [DOI] [PubMed] [Google Scholar]
- 81.Yan W, Chen W, Huang L. Mechanism of adjuvant activity of cationic liposome: phosphorylation of a MAP kinase, ERK and induction of chemokines. Mol. Immunol. 2007;44:3672–3681. doi: 10.1016/j.molimm.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 82.Keijzer C, Spiering R, Silva AL, van Eden W, Jiskoot W, Vervelde L, Broere F. PLGA nanoparticles enhance the expression of retinaldehyde dehydrogenase enzymes in dendritic cells and induce FoxP3(+) T-cells in vitro. J. Control. Release. 2013;168:35–40. doi: 10.1016/j.jconrel.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 83.Collins CB, Aherne CM, Kominsky D, McNamee EN, Lebsack MDP, Eltzschig H, Jedlicka P, Rivera-Nieves J. Retinoic acid attenuates ileitis by restoring the balance between T-helper 17 and T regulatory cells. Gastroenterology. 2011;141:1821–1831. doi: 10.1053/j.gastro.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, Seabra MC, Wilson MS. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity. 2014;41:89–103. doi: 10.1016/j.immuni.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Momen-Heravi F, Bala S, Bukong T, Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine. 2014 doi: 10.1016/j.nano.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bryniarski K, Ptak W, Jayakumar A, Püllmann K, Caplan MJ, Chairoungdua A, Lu J, Adams BD, Sikora E, Nazimek K, Marquez S, Kleinstein SH, Sangwung P, Iwakiri Y, Delgato E, Redegeld F, Blokhuis BR, Wojcikowski J, Daniel AW, Groot Kormelink T, Askenase PW. Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. J. Allergy Clin. Immunol. 2013;132:170–181. doi: 10.1016/j.jaci.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian T, Zhu YL, Hu FH, Wang YY, Huang NP, Xiao ZD. Dynamics of exosome internalization and trafficking. Journal of cellular physiology. 2013;228:1487–1495. doi: 10.1002/jcp.24304. [DOI] [PubMed] [Google Scholar]
- 89.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nature medicine. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 90.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]