Abstract
The prediction of protein-protein interactions and kinase-specific phosphorylation sites on individual proteins is critical for correctly placing proteins within signaling pathways and networks. The importance of this type of annotation continues to increase with the continued explosion of genomic and proteomic data, particularly with emerging data categorizing posttranslational modifications on a large scale. A variety of computational tools are available for this purpose. In this chapter, we review the general methodologies for these types of computational predictions and present a detailed user-focused tutorial of one such method and computational tool, Scansite, which is freely available to the entire scientific community over the Internet.
Keywords: Scansite, Protein-protein interaction prediction, Sequence motif, PSSM, Binding motif, Phosphorylation sites, Bioinformatics
1 Introduction
Decades of research in molecular biology have resulted in the availability of vast amounts of data, including genomic sequences, protein sequences, structural data, and protein metadata including functional domain information and interaction data. Unfortunately, the availability of these data types does not necessarily result in a clear understanding of what all the data means in a broader context. The bulk of the available data is single molecule-centric, limiting our ability to understand how molecules are integrated into pathways and networks. With the advent of new experimental techniques, it is possible to enrich these pieces of data with additional information related to protein-protein interactions and enzyme–substrate relationships. One of the most important breakthroughs in this context was the rise of experimental techniques that allowed the rapid and large-scale detection of protein-protein interactions [1]. Since the molecular apparatus of a cell is mainly controlled by protein–protein and protein–nucleic acid interactions, detecting and understanding such events, particularly direct interactions, is the first step to a broader view of biological systems. Interaction information has been collected in a number of different public databases [2, 3], and the information stored in these databases mostly contains experimentally verified information, i.e., data from in vivo or in vitro experiments. Unfortunately, given the current trend towards large-scale proteome wide analyses and the fact that these databases are far from complete, this information often proves insufficient for many analyses. The missing pieces in the puzzle that elucidates a more complete view of the cell interactome can be provided by interaction prediction tools. These tools create in silico predictions of protein-protein interactions and kinase–substrate relationships, are typically inexpensive and fast compared to conventional time- and resource-intensive experimental methods, and can provide a focused list of predictions that can then be verified or refuted by further focused experimental testing.
Over the past years, a number of different computational approaches for predicting protein-protein interactions have been developed. These techniques can be divided into those that are based on a single biological feature and those that attempt to use a range of different features and data types and can therefore be categorized based on the types of data that they use. A detailed overview of how each of these approaches works, and what the shortcomings of these methods are, can be found elsewhere [4, 5], but a short summary is provided here, focusing on which general features are used to predict protein-protein interactions.
At one extreme are methods based on a protein's three-dimensional structure, generally referred to as “protein docking” techniques. Given a 3D model (usually based on high-resolution data from X-ray crystallography or NMR experiments, deposited in the Protein Data Bank [1]) for two potentially interacting proteins, the best fit for each potential interaction interface on the surface of these models can be searched for and scored [6]. However, finding low-energy fits is very challenging, often due to the static nature of the PDB structures and the dynamic plasticity that can occur at protein-protein interfaces. Thus, conformational changes, the arrangements of side chains, and the energy levels of a potential conformation combination and interaction, potential posttranslational modifications that may or may not be included in a model, and a number of other factors have to be considered. Because of the large variance in the quality of the prediction of different methods in this field, CAPRI (Critical Assessment of Protein Interactions), a community-based program that regularly evaluates the algorithms to predict protein-protein interactions based on structures in a double-blind manner, has been initiated [7]. Alternatively, machine learning methods can be developed, usually based on databases of experimentally verified interactions and a number of additional biological properties. These points of data are then used to train a prediction engine based on known data [8]. The problem with this approach—as with any other machine learning approach used in this manner—is that the resulting predictor does not provide easily decipherable information about why the proteins are likely to interact. This means that, although it may yield useful results, it is hard to reconstruct and understand exactly why a prediction is made by this black-box predictor. A very specific type of machine learning method tries to find a pattern based on features at the interaction interface of the proteins involved. A method closely related to this approach is described later in this chapter. Other prediction methods are based on genomics. Gene fusion methods predict that discrete proteins are likely to interact if their homologues are fused into single genomic entities in other species. Other techniques based on gene neighborhood conservation are built on the hypothesis that gene pairs within such neighborhoods that are evolutionary conserved across different species are likely to interact.
No matter which method is used, it is important to keep the caveats of the method in mind. First, no prediction can guarantee either biological correctness or relevance. This is especially important if prediction tools are used to design and plan further experiments without first confirming the initial prediction. Failure of a method to predict an interaction may not reflect a fundamental problem with the method but may instead reflect limitations of the data that the method is based on. The data, be it experimentally verified interaction sites, 3D data of proteins, or other information, originates from experiments which are all error-prone, though in some cases the extent of the error may be difficult to estimate. This also applies to methods that use machine learning to train a predictor, as these methods are highly dependent on the quality of the underlying training dataset. Obviously, a large set of training data is necessary to create a good predictor and, indeed, large databases of experimentally verified protein-protein interactions are now available. However, training a predictor also requires a negative dataset that provides information of what interactions are very unlikely to happen. Experimental data of this type are typically not published, at least in part due to difficulties in distinguishing whether the lack of an observed interaction is the result of a technically failed experiment or because there is no biologically relevant interaction [9]. The end result is a lack of reliable negative training data for computational method development. The quality and nature of the training data should therefore be one important consideration in the user's choice of whether to trust a predictor trained on these kinds of data types, including the species of the proteins in the training dataset, the type of experiments used to verify the sites, and of course the number of sites and proteins included. Thus, it is very important for prediction tools to explicitly (1) give information about how the method works and what information it uses, (2) provide some type of quantitative measure that allows users to compare different results and distinguish between good and not-as-good predictions, and (3) provide any additional information that helps the user decide whether to trust the predictions. This information could be incorporated into the prediction method itself but is also very helpful if this type of metadata is simply presented for the user to examine independently of whether this information is explicitly used in the prediction algorithm.
One of the most important features in describing protein-protein interactions is elucidating the exact sites on the proteins where the interaction occurs, either at the detailed atomic/structural level or at the level of specific amino acid sequences. That is, on the surface of the protein, which part of the amino acid sequence directly contacts or indirectly influences the interaction partner? By focusing solely on sequence information, the complexities required for interaction prediction by docking-type simulations (conformational states of side chains, energetic contributions, etc.) are radically reduced.
Since protein-protein interactions are mediated by attractive forces based on the physicochemical properties of amino acids, in many cases it is sufficient to describe potential interaction partners by amino acid sequence patterns alone. This can be clearly shown by considering kinase–substrate interactions: kinases generally only phosphorylate serine (S), threonine (T), or tyrosine (Y) residues based on the ability of their phosphate acceptor hydroxyl groups to nucleophilically attack the γ-phosphate of ATP. However, more than 500 different kinases are known alone in humans, each of which targets a different set of substrates [10]. The specific site of phosphorylation is therefore not the only amino acid that plays a role in substrate recognition. Instead, 4–12 amino acid residues on the substrate flanking the phospho-acceptor likely physically contact a kinase's active site [11] and help to position the substrate for an in-line attack on the phosphate while simultaneously optimizing the geometry of the kinase's catalytic machinery to facilitate stabilization of the resulting transition state. This indicates that this part of the substrate's primary structure may be sufficient to determine whether an acceptor residue is likely to be phosphorylated by a given kinase. Although sequence patterns are only one piece of information in a puzzle of many (secondary structure, tertiary structure, surface accessibility, etc.), an abundance of data suggests that this is one of the most distinguishing factors in describing a kinase–substrate interaction and in many cases it is a sufficient predictive feature [12, 13]. Obviously, this idea does not apply only to kinases but can be used to describe other protein-protein interactions mediated by other types of modular protein domains that recognize short linear sequence motifs on their binding partners in a phospho-dependent or -independent manner, such as SH2 and SH3 domains, FHA and BRCT domains, 14-3-3 proteins, etc.
Specific amino acid preferences can be described in two ways. One is to describe them in a strict combinatorial regular expression-like pattern (Boolean matching model). This approach was originally used in PROSITE [14] to search for patterns in a sequence database. However, these patterns are very inflexible and do not allow for including differently weighted preferences for amino acids. A more flexible and powerful approach is the use of position-specific scoring matrices (PSSMs) to describe patterns/ motifs in this form. This approach was implemented in Scansite [15, 16], an application to predict short linear sequence motif sites. A PSSM matrix like this contains a probability value for each amino acid (columns) at each position of a sequence window of certain size (rows), where each value in a column and row of the matrix describes the binding partner's preference for that amino acid at that position in the motif. Scansite is a web application that uses PSSMs to predict interaction sites that are important in cellular signaling and includes more than 120 kinases and proteins that recognize specific short linear binding motifs. It can be used to show all potential sites in a given protein or all proteins in a database that contain sites for one or more motifs. Directions for both uses are provided in the following sections.
2 Materials
Scansite 3 (http://scansite3.mit.edu/) requires nothing more than a computer with an Internet connection and a modern web browser. Although it works with all popular web browsers, the recommended options are Mozilla Firefox, Google Chrome, and Opera. On some pages that display search results, Scansite will show content in pop-up windows so that results can be viewed side by side. Therefore it is recommended that you allow pop-ups in your browser for these pages to work properly. Wherever Scansite allows you to choose a sequence database (e.g., when selecting proteins or for searching a database), you can choose from these resources: SwissProt [17], SGD (yeast) [18], Ensembl (human and mouse) [19], NCBI Protein (GenPept) [20], and TrEmbl [21]. Scansite uses local mirrors of these databases in order to allow fast queries. Over the past years Scansite has also become popular for analyses of whole proteomes or subsets thereof. These are generally not done using the web interface, but computationally. If you are interested in using Scansite for this purpose, please see Note 1 for information about Scansite's web service.
2.1 Scanning a Protein for Motifs
To perform Protein Scans you need either a protein sequence or a protein identifier (accession number or ID) for the protein you are interested in from one of Scansite's protein sequence database mirrors.
2.2 Searching a Sequence Database for Motifs
Database Searches only require information about the motif that is searched for in a particular sequence database. All of the standard Scansite matrices for kinases and modular binding domains are available. In addition, you may enter more specific information to restrict the search to a smaller number of proteins.
3 Methods
Scansite's two most important interaction prediction searches will be described in detail in this section: Protein Scans that search for motif matches in a given protein and Database Searches that find proteins that contain one or more motifs in a protein sequence database. A short overview of Scansite's other features is given in Note 2. In the following, you will be guided through the steps necessary to use these features properly. Furthermore, some guidance on how to interpret these searches’ results will be given.
3.1 Scanning a Protein for Motifs
The key feature of Scansite is the prediction of motif-relevant sites in a given protein. This feature is referred to as Protein Scan or Scan Proteins for Motifs and allows a range of different inputs.
1. Navigate to Input Page
To get to the Protein Scan input screen from anywhere in Scansite, click the “Scan Proteins for Motifs” button in the navigation section on the left-hand side of the web page.
2. Choose the Protein to Scan
There are two different ways of choosing proteins in Scansite: by protein identifier (default option) and by sequence.
To choose a protein by accession number, select “Protein Accession” from the “Choose Protein by. . .” drop-down list. Below, select a protein sequence database and enter a protein ID. Links on the right-hand side of the text boxes refer to the different sequence databases that Scansite currently supports and where you can search for protein identifiers. After entering at least three characters in the text box entitled “Protein Accession”, Scansite searches for protein IDs that start with these characters and presents a list of options below the text box. The same happens when the “Check!” button next to the text box is clicked or the Enter key is pressed. You can either continue typing or select an ID from the list. The text box turns green for valid and red for invalid protein identifiers.
In order to enter a peptide sequence, select “Input Sequence” from the drop-down list. The area below this menu will change accordingly. Then, enter or paste a name and an amino acid sequence. Invalid characters (punctuation marks, white space, digits, etc.) are stripped from the sequence automatically. This means that you can just paste a sequence that is formatted with spaces and line breaks or annotated with numbers. If you paste a FASTA-formatted sequence, make sure not to copy the FASTA header (“>. . .”) with the sequence. Otherwise all possible amino acid one letter codes in the header will also become part of the sequence.
3. Choose Motifs to Consider
It is possible to search for all motifs of a motif class, for only a selected subset of motifs or motif groups or both, or for a user-defined motif (instructions on how to create your own motif can be found in Note 3). You can choose from these options in the drop-down menu entitled “Look for”. Again, the area below this menu will change accordingly dependent on your choice, offering a number of additional choices. Begin by selecting a motif class (mammalian or yeast). By default, Scansite's mammalian motifs are displayed. To select more than one motif or motif group, hold down the control key on your keyboard and make selections using your mouse. If you are not sure which motifs belong to which groups, you can either click the link below the list of groups (“Show Group Definitions”) or follow the instructions in Note 4. When using your own motif, select the motif file from your computer. After the file is uploaded (this happens automatically after you selected a file), you get a chance to make changes to affinity values if you wish to do so.
4. Select a Stringency Level
This measure defines how high sites have to score in order to be displayed as results. The setting high only displays the very best sites, i.e., the top 0.2 % of sites (sites that have a score less than or equal to the top 0.2 % of motif-specific scores in the reference proteome). Medium stringency displays the top 1 %, low the top 5 %, and minimum displays the top 15 %. These settings apply only for motifs from the Scansite database. Since no precompiled reference proteome score distribution (see Note 5) is available for user-defined motifs, these always display all sites with a score ≤5.
5. Additional Options
The two additional options that users are given are to decide whether to show predicted domains in the result as supporting information (see Note 6) and whether to use an alternative reference proteome. At the moment users can use either SwissProt's Vertebrate proteins as a reference (default) or all of SGD's proteins (default for scans using yeast motifs). Domains can also be requested later on from the result page.
6. Click the Submit Button
As an example for a Protein Scan result page, the results of a high stringency protein scan are shown in Fig. 1. The result page is split in seven sections (divided by grey bars): Protein Overview, Scan Overview, Protein Plot, Predicted Motif Sites (Table), Repeat Scan, Download Results, and Additional Analyses. Each of these sections is collapsible by clicking on the grey title areas. This allows the user to quickly get to the bottom of the page if a long list of predicted sites is displayed.
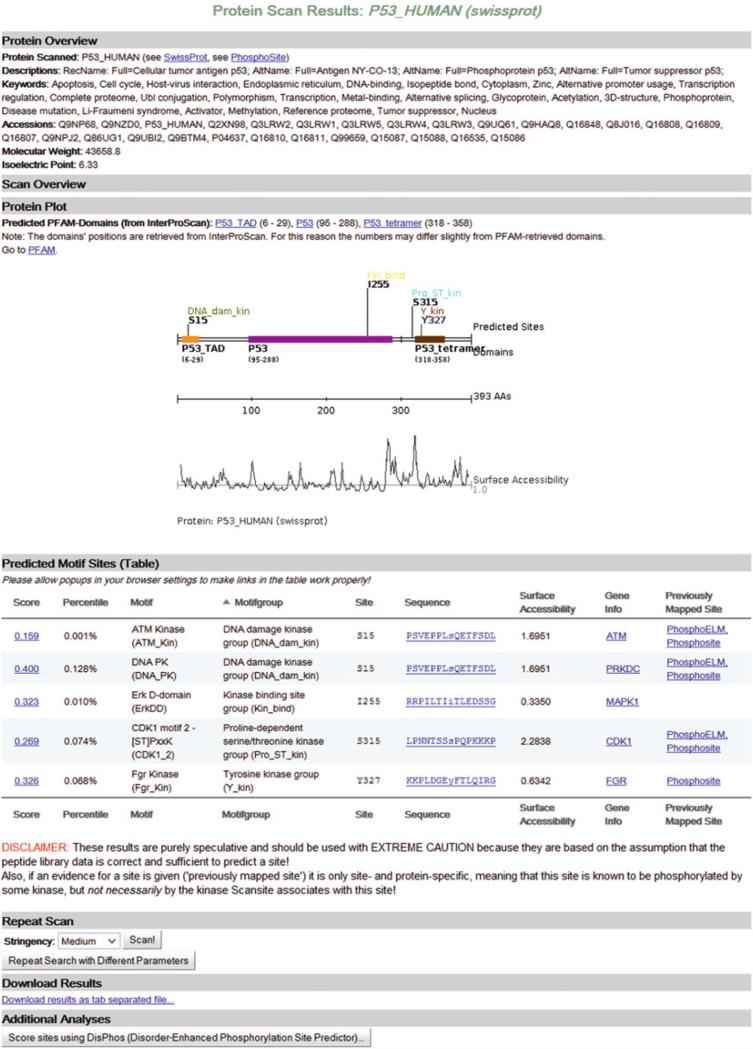
Fig. 1.
The results of a high stringency protein scan for all mammalian motifs using the SwissProt protein P53_HUMAN and the default reference proteome are shown. The section entitled “Scan Overview” which summarizes the parameters of the scan of the page is collapsed to better fit this figure on the page
In the “Protein Overview” section, some information about the input protein is listed, including alternative identifiers and keywords (only for proteins from Scansite's databases), and the protein's molecular weight and isoelectric point (calculated according to ref. 22). The “Scan Overview” summarizes the input parameters of the search and displays the number of sites that have been detected using these settings. In the next part of the page (“Protein Plot”), a plot of the protein gives a visual overview of the search results displaying the protein sequence as a straight line annotated with some additional information. If domain information about the query protein was requested to be displayed, the predicted domains are listed above the image. The plot displays the predicted sites (annotated with the position and motif group), the protein's domains (if requested) along with their names and positions, and a surface accessibility plot that shows which parts of the protein are likely to be exposed to the surface and which ones are likely to be buried. If domains have not been requested earlier, a button will be displayed below the image that allows the user to request domain prediction at this point. The links in the list of displayed domains refer to these domains’ PFAM pages (see Note 6).
The sites that are outlined in the protein plot are listed in more detail in the table view below (“Predicted Motif Sites”). Most columns can be sorted by clicking on the label in the table's header. Here, each site that was found is displayed along with some motif information (motif, motif group, hyperlink to motif's gene information page), its score and percentile, and the surrounding sequence. In addition, Scansite-3 offers hyperlinks to PhosphoSite [23], PhosphoELM [24], and Phosida [25] if a site was reported in one of these databases before (for more information about “Previously Mapped Sites” see Note 7). The other links displayed in the table, more specifically the columns “Score” and “Sequence,” refer to a histogram view of a site in the reference proteome and to a view that shows a site's sequence highlighted in the protein's sequence, respectively. The latter view also offers a link that directly submits the site's sequence (15 amino acids) to NCBI's basic local alignment search tool (BLAST) [26]. More information on BLASTing sites in Scansite can be found in Note 8.
In the “Repeat Scan” section of the result page, it is possible to either directly rerun the scan with a different stringency setting or to go back to the input page to change other search parameters. This is especially helpful if your search did not return any results. The next part in the page (“Download Results”) offers a link to a downloadable version of the table shown above (tabulator-separated file). At the bottom of the result page (“Additional Analyses”) users can directly submit the current protein's sequence to DisPhos [27], a Disorder-Enhanced Phosphorylation Site Predictor (see Note 9).
3.2 Searching a Sequence Database for Motifs
The Scansite feature Search Sequence Database for Motifs or short Database Search performs a broader search than single protein scans. Given a motif (or a set of motifs) and a sequence database, it searches the database for sequences that contain motif-relevant sites. One of the most powerful parts of this tool is the option of targeting a search to specific experimental requirements by restricting searches to proteins of a specific organism class, species, molecular weight and isoelectric point range, annotation, and sequence property. For example, this tool can be used to help identify unknown bands in two-dimensional (2D) gel electrophoresis experiments.
1. Navigate to Input Page
To get to the Database Search input screen from anywhere in Scansite, click the button “Search a Sequence Database for Motifs” in the navigation section on the left-hand side of the web page.
2. Choose the Search Method
The area below this drop-down list will change dependent on what you select. Searches for single “Database motifs” from the Scansite database are the easiest option to choose. Alternatively, you can search for your own motifs (see Note 3) or so-called “Quick Motifs” (see Note 10). It is also possible to search for sequences that match up to five motifs. These searches can include either database motifs, user-defined motifs, or a combination of both. The score of a multi-motif site is the mean (average) of all the scores of the sites involved. Co-occurrences of different motif sites in proteins can be filtered in different ways. First of all, it is possible to penalize gaps between sites of different motifs. Gap penalty settings are either high, medium, low, or none. Penalties p are then added to the score according to the maximum distance dmax between the involved sites (i.e., position of site closest to C-terminus minus position of site closest to N-terminus). The penalty values are calculated as follows: plow = 0.001 × dmax; pmedium = 0.01 × dmax; phigh = 0.1 × dmax. Secondly, it is possible to define up to three strict minimum and maximum distance bounds between motif-specific sites. This can be used if you know which motifs to expect and how far apart you expect them to be in the protein sequence. If you just want to get an overview of peptides that have multiple motif sites, it is recommended to use a gap penalty. Using distance bounds is the better option for very specific searches.
3. Select Database to Search
from the drop-down menu.
4. Restrict Search
It is recommended that you exclude as many proteins from the search as possible to both target your search as much as possible to what you are looking for and to decrease the runtime of the search. Database searches can take several minutes and the runtime of a search mostly depends on the number of proteins that are searched. You will find useful hints on what kinds of restrictions you can apply in Note 11.
5. Select Number of On-Screen Results
Since Database Searches may find a very high number of results and visual exploration of a table of thousands of results generally is avoided, the number of sites that are displayed in the web browser is limited. By default, the size of the output list is limited to 50, but users can also choose sizes 100, 200, 500, 1,000, and 2,000. Please note that these are just the numbers of sites that are displayed in the table on the result page. A file containing all the hits that were found in this search can be downloaded from the result page as well.
6. Click the Submit Button
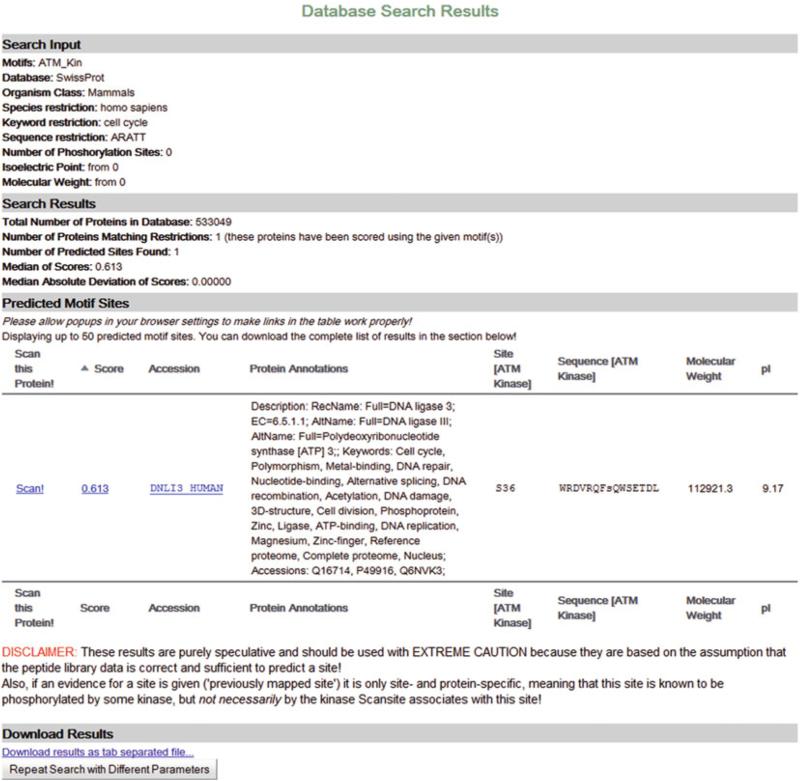
A result page of a Database Search is displayed in Fig. 2. Four sections can be distinguished within the Database Search result page. The “Search Input” section at the top of the page summarizes the preferences defined in the input page. “Search Results” gives an overview of the number of proteins in the entire sequence database, the number of proteins found that matched the given restrictions, and the number of sites found in these proteins. In addition, the median and MAD (median absolute deviation) of these sites’ scores is displayed. This part is followed by a table view of the sites found (“Predicted Motif Sites”). The table shows the (combined) site score, some information about the protein that was found (including MW and pI), and displays some site-specific information (site and surrounding sequence). For multi-motif searches a site and sequence column for each motif in the motif's site is given. The first column in the table allows to directly scan the protein for other motifs. This is useful if you want to know what other motifs are found in that protein, if a site has been reported before (previously mapped) in another database, and how the protein is generally composed (domains, surface accessibility). The link in the column labeled “Accession” takes the user to the protein's page in its primary database. The score column links to a histogram that shows the site's score in comparison to all scores found in that search. At the bottom of the page, options for downloading the entire result set and for repeating the search are given.
Fig. 2.
The results of a Database Search for ATM in human proteins of SwissProt that are annotated with “cell cycle” and contain the sequence “ARATT”. Here, only one protein matched the given restrictions and this protein also contains the motif that was searched for
Footnotes
Accessing Scansite Computationally. The current era of genomics and proteomics often requires analyses of large numbers of proteins. To make tasks like this easier it is now possible to access Scansite computationally using a web service. The parameters of protein scans, database searches, and other utility functions are sent to Scansite using a URI. The results are then returned in XML format. Detailed instructions and examples are available online at http://scansite3.mit.edu/Scansite3Webservice/. This link can also be found in Scansite's FAQ online.
Getting the Most out of Scansite. In addition to the features described in detail above, Scansite offers some more useful tools. To start with, you can search Scansite's sequence databases for simple wildcard-based sequence patterns or regular expressions. Another tool calculates a sequences molecular weight and isoelectric point for a given number of putative phosphorylations. Last, a tool called “Calculate Amino Acid Composition” visualizes a protein sequence's amino acid composition by highlighting selected sites and displaying the relative abundance of sites (e.g., all tyrosines in a sequence that are followed by leucines two residues downstream). In addition, this tool displays a protein's domain information as calculated by InterProScan [28]. One can also use one of these tools to analyze a protein sequence, copy/paste it to make changes (e.g., introduce mutations), and then use it as an input for protein scans.
Creating Scansite Motifs. Both main search options in Scansite allow the use of user-defined motifs. These motifs have to be in a Scansite-specific tabulator-separated file format. All user-defined motifs that are uploaded to Scansite are only used for the user's searches and are deleted as soon as the user leaves the site. If you have a clear idea of what motif you want to look for, use the information below to specify your own Scansite-specific motif file.
PSSMs in Scansite describe amino acid-specific affinity values for a sequence window of 15 residues. Lines correspond to positions in the sequence window, columns (separated by tabulators) to amino acids. It is not necessary to define values for every single amino acid—default values are used for omitted residues. The first line (row 1, header) defines the residue-to-column assignments using amino acid one letter codes. Those amino acids can be in any order. The following lines (rows 2–16) define affinity values for the respective residues; rows 2–8 and 10–16 define the N- and the C-terminal side of the motif, respectively. Scansite's search for sites in a peptide sequence highly depends on the PSSM's central residue (row 9). At least one site in this position needs to be invariant in the motif sequence. For example, the fixed residue should be a Y for motifs recognized by tyrosine-kinases and S and T for serine-/threonine-kinases. To mark a position as invariant, the value 21 has to be used.
In addition to columns of standard amino acids (default values of 1), it is also possible to incorporate special requirements. A motif's preference for a protein sequence's N- or C-terminus can be incorporated by using a column labeled “$” (dollar sign) or “*” (asterisk), respectively. These positions are assigned values of 0 by default. Scansite 3 also recognizes the rarely occurring amino acids selenocysteine (U) and pyrrolysine (O), which can be added by their one letter code as well. Due to their similar chemical structure, the default numbers for these residues are the values of cysteines and lysines, respectively. Lastly, some wildcard values can be used for very special cases: B (aspartate/asparagine), Z (glutamate/glutamine), J (leucine/isoleucine), and X (any residue). These symbols are included because they occur rarely in public protein databases. Generally speaking, they have no relevance for actual research purposes. The default values for these wildcards are the mean values of the amino acids they encode.
Now that the general structure and default values of motif files were defined, you may wonder what values to use to define affinities. Scansite's scoring system ranges from 0 to roughly 21. Giving an individual amino acid a score of 1 at one position in the motif indicates that no preference exists, positive or negative, for that particular amino acid in that position. Giving all amino acids in one position of the motif a score of 1 (i.e., making all values in a single row of the matrix equal to one) indicates no preference exists for any particular residue type at that position in the motif. The value 21 defines that the amino acid that is given this value in a position is required in this position for the motif to find a match. Values higher than 21 are permitted to indicate very strong affinities. However, negative values are not permitted for defining a strong disfavoring of amino acids. Instead, values between zero and one should be used for that purpose. Beware that the scoring function uses logarithms, so values less than 1, particularly those less than 0.5, strongly penalize for that particular residue in a motif.
- Is there at least one amino acid with value 21 in the central position?
- Is there a header line defining the columns using amino acid one letter codes?
- Are there 16 lines (1 header and 15 lines with values) in the file?
- Are all column separators in the file tabulators (and not spaces or other characters)?
Learn more about Scansite's Data. In a section called “Databases and Motifs” in the navigation section of the web page (left-hand side), an overview of Scansite's database mirrors (release dates and sizes), motifs, and motif group definitions is presented. In the motifs section you can select a motif and click “Get Info!.” Clicking this button will visualize the motif as a sequence logo [29] and display a link that takes you to a web page that gives information about the gene that recognizes this motif. Mammalian motifs and yeast-specific motifs are supported by information from GeneCards [30] and SGD, respectively.
Interpreting Scansite Scores. Scansite's scores range from 0 to (theoretically) ∞. However, you will never see scores higher than 5 because sites with scores that high are discarded in the scoring process. Please be aware that scores in Scansite are always motif-dependent. This means that scores for different motifs should not be directly compared to each other. For example, knowing that one motif's optimal score is 0.001 and another motif's best score is 0.4 it is easy to say that these are the best possible scores, so hits with these scores are equally good. However, the only way to extend this knowledge to slightly poorer scores is to know how likely other scores are to occur. To make this possible and allow a comparison among motifs, Scansite offers percentile values. The percentiles used in Scansite are calculated from the so-called reference proteomes which are proteomes that are commonly used in research. In the process of adding a motif to Scansite, it is scored against every single peptide in the reference proteome and the scores are stored to create a score distribution. This distribution is then used to calculate percentile values from scores calculated when users run certain searches. Using these values it is possible to rank sites from different motifs.
Domains in Scansite 3. Scansite uses InterProScan [28] to predict a protein's PFAM domains [31]. Therefore the domain positions displayed in Scansite may vary by a few amino acids from the positional assignments seen on the PFAM homepage. This is mentioned because these variations may cause confusion but do not pose a problem since all these positions are predictions and there is no way to tell which numbers are more correct in the absence of clear structural data from crystallographic or NMR experiments.
Previously Mapped Sites in Scansite. Displaying previously mapped sites in Scansite is only possible for proteins from public protein databases and works best with proteins from SwissProt. Please note that these references are only site-specific but not motif-specific. This means that if a previously mapped site shows up in the list, the site is reported in the linked databases; however, this does not imply that the Scansite motif that was found at this site is related to the site reported in the database. It could be that a completely different gene is responsible for this site. Wherever possible, the hyperlinks refer directly to the external databases page about this site. If a database does not support direct linking, the link just takes you to the database's homepage.
BLASTing of Sites. Scansite allows to directly submit the 15-mers around identified sites to NCBI's BLAST. This is a simple approach to see if a site is conserved in organisms that are expected to be physiologically similar to the one at hand. If the site is also found in similar proteins in other species, the site is more likely to be biologically relevant.
Intrinsically Disordered Proteins. Disordered regions in proteins are stretches of amino acids that do not have a rigid tertiary structure and are therefore enabled to change conformation. Disordered Proteins are proteins with disordered regions. It has been shown [32] that many posttranslational modifications and binding sites occur in disordered regions because these regions make a protein more flexible, which facilitates binding and interaction processes. DisPhos is a disorder-prediction engine that focuses on potential phosphorylation sites. The results of DisPhos searches can therefore be used as supporting information for phosphorylation sites predicted by Scansite.
Using Quick Motifs. Creating a custom motif only makes sense if enough information about the affinities of the kinase or binding domain is known. This, however, requires a very specific idea about the motif. Often, only very little detail about a motif is known. In cases like these, creating a “Quick Motif” to search a database is the best option. For defining a quick motif, the user can enter a set of primary and secondary preferences for each position of a 15-mer. These preferences are then used to calculate a simple Scansite motif. As for actual Scansite motifs, the center position needs to be fixed, so it is not possible to enter secondary preferences there. The web page describes a number of wildcards that can be used in this process to easily describe amino acid subsets by their physicochemical properties (e.g., hydrophobic or positive residues). A simplified regular expression-like version of the motif that is entered is displayed below the text boxes (with resolved wildcards) as soon as values in the text boxes are changed.
- For many searches, you may only be interested in matches from humans or a particular model organism. Searches can be restricted this way by entering the species’ name in the text box labeled “Single Species.” This feature supports many MySQL-style wildcards (regular expressions) to match species names. For example, if you are tired of writing out “Caenorhabditis elegans”, you can use “C.* elegans” instead. In a regular expression, the period (.) matches any single character, and the asterisk extends that match to multiple characters (or even zero characters). This also allows for genus-wide searches, by entering just “Rattus” for example. However, this may yield unexpected results when trying to search for all kinds of mice with “Mus.” This expression will accidentally match “Thermus aquaticus” as well, but you can avoid that by entering “^Mus.” The caret symbol (^) requires the text to match at the beginning of the entered name. One of the most common pitfalls is when the species entered does not match the organism class specified above (e.g., a search for “yeast” when “Mammals” is selected). Please note that Scansite's organism classes are not taxonomic “classes” in the conventional sense (except for Mammals) but groups of species frequently used for research purposes.
- The molecular weight, isoelectric point, and phosphorylation options are intended for use in conjunction with 2D gel electrophoresis experiments. When you find a few spots appearing reproducibly on a 2D gel under a particular test condition and not under the control, you could use Scansite to find what proteins are expected to be in that region of the 2D gel by putting in ranges for molecular weights and isoelectric points. You could simultaneously constrain the species to match the cell line you used in the experiment. If it is an experiment involving possible phosphorylation events, you can see how much a putative phosphorylation would move the peptides on the gel.
- Matches for “Keywords” are searched in a protein's annotations and are therefore primarily useful for searching well-annotated databases like SwissProt. For example, proteins involved in the cell cycle can be easily identified by entering “cell cycle,” novel proteins in GenPept by searching for “hypothetical.”
- The “Sequence Contains” text field is a quick way to restrict your search to proteins containing a consensus sequence. It is important to note that the consensus sequence entered here is not required to be part of the motif being searched for. It is merely required to show up somewhere in the sequence. Also, note that regular expressions have to be used here instead of the protein wildcard signs (“.” instead of “X”, “[ND]” instead of “B”, etc.). For example, the sequence “PXXP” is represented as “P..P” in regular expression syntax. More information on regular expressions and how they can be used in Scansite is available in Scansite's frequently asked questions (FAQ) section online.
References
- 1.Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathivanan S, Periaswamy B, Gandhi T, et al. An evaluation of human proteinprotein interaction data in the public domain. BMC Bioinformatics. 2006;7:S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turinsky A, Razick S, Turner B, et al. Literature curation of protein interactions: measuring agreement across major public databases. Database (Oxford) 2010:baq026. doi: 10.1093/database/baq026. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker B, Panchenko A. Deciphering protein-protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLoS Comput Biol. 2007;3:e43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitre S, Alamgir M, Green J, et al. Advances in biochemical engineering/biotechnology: protein-protein interaction. Springer; Heidelberg: 2008. Computational methods for predicting protein–protein interactions. pp. 247–267. [DOI] [PubMed] [Google Scholar]
- 6.Andrusier N, Mashiach E, Nussinov R, et al. Principles of flexible protein–protein docking. Proteins. 2008;73:271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janin J. Welcome to CAPRI: a Critical Assessment of PRedicted Interactions. Proteins Struct Funct Genet. 2002;47:257. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 8.Rhodes DR, Tomlins SA, Varambally S, et al. Probabilistic model of the human protein-protein interaction network. Nat Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 9.Trost B, Kusalik A. Computational prediction of eukaryotic phosphorylation sites. Bioinformatics. 2011;27:2927–2935. doi: 10.1093/bioinformatics/btr525. [DOI] [PubMed] [Google Scholar]
- 10.Hutti J, Jarrell E, Chang J, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 11.Songyang Z, Blechner S, Hoagland N, et al. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 12.Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- 13.Pinna LA, Maria Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;13143:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 14.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20(Suppl):2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaffe M, Leparc G, Lai J, et al. A motifbased profile scanning approach for genomewide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 16.Obenauer J, Cantley L, Yaffe M. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. M, UniProt-consortium UniProt Knowledgebase: a hub of integrated protein data. Database. 2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry J, Hong E, Amundsen C, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2011;40(Database issue):D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flicek P, Amode M, Barrell D, et al. Ensembl. 2011. Nucleic Acids Res. 2011;39(Suppl 1):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burks C, Cassidy M, Cinkosky MJ, et al. GenBank. Nucleic Acids Res. 1991;19:221–225. doi: 10.1093/nar/19.suppl.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeckmann B, Bairoch A, Apweiler R, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in. 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjellqvist B, Hughes G, Pasquali C, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 23.Hornbeck P, Kornhauser J, Tkachev S, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinkel H, Chica C, Via A, et al. Phospho. ELM: a database of phosphorylation sites–update. 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gnad F, Ren S, Cox J, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iakoucheva L, Radivojac P, Brown C, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter S, Apweiler R, Attwood T, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider T, Stephens R. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stelzer G, Dalah I, Stein T, et al. In-silico human genomics with GeneCards. Hum Genomics. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punta M, Coggill P, Eberhardt R, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uversky V, Dunker A. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]