Abstract
Background
Hibernation is a survival mechanism in the winter for some animals. Fat preserved instead of glucose produced is the primary fuel during winter hibernation of mammals. Many genes involved in lipid metabolism are regulated by the peroxisome proliferator-activated receptor alpha (PPARα). The role of PPARα in hibernation of mammals remains largely unknown. Using a multidisciplinary approach, we investigated whether PPARα is adapted to hibernation in bats.
Results
Evolutionary analyses revealed that the ω value of Pparα of the ancestral lineage of hibernating bats in both Yinpterochiroptera and Yangochiroptera was lower than that of non-hibernating bats in Yinpterochiroptera, suggesting that a higher selective pressure acts on Pparα in hibernating bats. PPARα expression was found to be increased at both mRNA and protein levels in distantly related bats (Rhinolophus ferrumequinum and Hipposideros armiger in Yinpterochiroptera and Myotis ricketti in Yangochiroptera) during their torpid episodes. Transcription factors such as FOXL1, NFYA, NFYB, SP1, TBP, and ERG were bioinformatically determined to have a higher binding affinity to the potential regulatory regions of Pparα in hibernating than in non-hibernating mammals. Genome-wide bioinformatic analyses of 64 mammalian species showed that PPARα has more potential target genes and higher binding affinity to these genes in hibernating than in non-hibernating mammals.
Conclusions
We conclude that PPARα is adapted to hibernation in bats based on the observations that Pparα has a more stringent functional constraint in the ancestral lineage of hibernating bats and a higher level of expression in hibernating than in non-hibernating bats. We also conclude that PPARα plays a very important role in hibernation as hibernators have more PPARα target genes than non-hibernators, and PPARα in hibernators has a higher binding affinity for its target genes than in non-hibernators.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-015-0373-6) contains supplementary material, which is available to authorized users.
Keywords: PPARα, Hibernation, Bats, Mammals, Molecular evolution, Gene regulation
Background
Hibernation is an adaptive strategy used by some animals to survive cold winter weather when food is scarce [1, 2]. Small mammalian hibernators (e.g., squirrels and bats) undergo a series of torpor-arousal cycles during hibernation, in which a torpor bout lasts for several days or weeks but an arousal bout lasts only several hours [1, 3]. During torpor, the metabolic rate (MR) of these mammals is reduced to 2-4 % of the euthermic metabolism, and their body temperature (Tb) may fall just a few degrees above the ambient temperature; however, both their MR and Tb are rapidly restored to the euthermic levels upon arousal [2]. The body weight and fat mass of these hibernators are dramatically increased in the pre-hibernation season. The preserved fats are used as the primary energy source during hibernation [2, 4, 5].
Bats belong to the order Chiroptera in the mammalian clade Laurasiatheria [6]. They comprise almost 1-quarter of mammalian species and are the only flying mammals [7, 8]. Hibernating bats (e.g., some species in superfamilies Rhinolophoidea, Emballonuroidea, and Vespertilionoidea) are mostly distributed in subtropical or cold latitudes, whereas non-hibernating bats are mainly reside in warm subtropical or tropical latitudes [9]. Many hibernating bat species in genera Myotis and Rhinolophus are deep hibernators that stringently control their overall metabolism during torpor [9, 10]. Positive selection in the coding regions of some genes, such as Leptin and Bssl, is found in these bats, suggesting that lipid metabolism has undergone adaptive evolution in response to hibernation [11, 12].
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptors that regulate the expression of many genes (e.g., Leptin, Hmgcs1, Ucp1, and Pgc1α) involved in lipid and glucose homeostasis [13, 14] and the development of obesity, diabetes, and hypertension in mammals [15]. Ligand-dependent transcriptional regulation by PPARs depends on the heterodimerization of PPARs with their coactivators (e.g., RXR). PPARs can also be activated directly in a ligand-independent manner by phosphorylation [16, 17]. 3 different subtypes (α, β, and γ) of PPARs have been identified. PPARα is activated upon energy deprivation and is highly expressed in the liver, heart, kidney, and adipose tissues [17, 18], while subtypes β and γ, expressed ubiquitously, are associated with cell migration and adipocyte differentiation, respectively [19].
The actions of PPARα are regulated by the fibroblast growth factor 21 (FGF21), and the PPARα-FGF21 signaling cascade has been shown to induce torpor in fasting mice [20]. PPARα is recently shown to shift fuel utilization from carbohydrate to lipid during torpor of arctic ground squirrel, Urocitellus parryii [21]. A positive selection at the glycine residue of codon 96 (96G) of Pparα has been detected in members of super-clade Laurasiatheria (placentals) and suborders of Euarchontoglires (primates and rodents) [22]. In addition, increased expression of PPARγ and its co-activator PGC-1α is observed in Spermophilus tridecemlineatus squirrels during a cold-induced torpor [23]. The expression of these 2 proteins is also increased in many organs of torpid Myotis lucifugus bats [24].
Although differential expression of PPARs and their co-activators (e.g., RXR) has been investigated in some hibernating mammals during different phases of hibernation [21, 23-26], the evolution of PPARα in hibernating bats remains unknown. Since PPARα plays a significant role in metabolic regulation during torpor [27], we hypothesize that PPARα has evolved in bats in adaption to hibernation. To test this hypothesis, we studied the evolution of Pparα in fifty-six species of mammals including twelve species of bats. We also compared mRNA and protein expression of Pparα in hibernating bats between torpid and arousal states. The number of potential PPARα target genes and the binding affinity of PPARα to these genes in hibernating and non-hibernating bats and other mammalian species were also determined.
Results and Discussion
Higher Selection Pressure on Pparα in Hibernating Bats
To determine whether the coding region of Pparα is positively selected in bats in response to hibernation, we conducted a branch model test on Pparα from 9 hibernating and 3 non-hibernating species of bats by the maximum likelihood method using other mammalian species as an out-group (Fig 1 and Table 1). Although some of the branches were short and might lead to inaccurate estimation of ω (dN/dS), the ω value of the ancestral lineage of hibernating bats in both Yinpterochiroptera (Yin) and Yangochiroptera (Yang) was found to be lower than that of non-hibernating bats in Yin (Fig 1), suggesting a higher selective pressure acting on Pparα in hibernating bats. A significant lower ω value was obtained by the 2-ratio model E (P = 0.008) when the ancestral branch of hibernating bats in Yang was labeled for comparison with other branches, indicating that Pparα is much more conserved in this lineage (Table 1). It is conceivable that such conservation is required for the survival of hibernating bats, especially for those living in relatively low latitudes (see Additional file 3: Fig S1). No positive selection on any amino acid of Pparα was found in the ancestral branches of hibernating bats in both Yin and Yang using the branch-site model A test (see Additional file 1: Table S1 and Additional file 2). There was also no positively selected site detected in these bats using the site model test (see Additional file 1: Table S2 and Additional file 2). However, Pparα was found conserved across 56 mammalian species and experienced a higher selective pressure (ω = 0.057) in ancestral Chiroptera (see Additional file 1: Table S3, Additional file 2, and Additional file 3: Fig S2). These data suggest that Pparα is conserved in evolution and support the previous postulation that heterothermy is an ancestral chiropteran trait [8, 12, 27]. A positive selection on codons 96 and 195 (96G and 195 V) of Pparα was detected in these mammals by site model comparisons (M8-M7 model) (see Additional file 1: Table S4 and l file 2). This result is consistent with the positive selection on codon 96 (96G) of PPARα observed in super-clade Laurasiatheria (placentals) and suborders of Euarchontoglires (primates and rodents) [22].
Fig. 1.
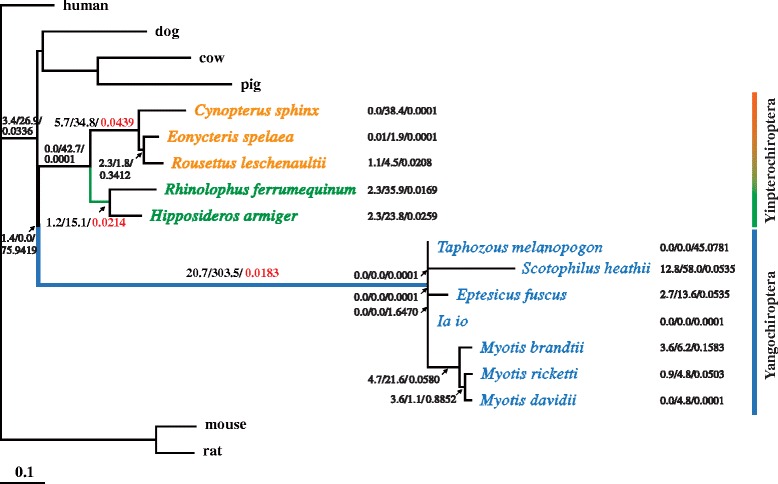
Species tree of Pparα of bats. Non-hibernating bats in Yinpterochiroptera, hibernating bats in Yinpterochiroptera, and hibernating bats in Yangochiroptera are represented by orange, green, and blue colors, respectively. The N × d N/S × d S/ω value of each tree branch of bats is shown
Table 1.
Branch model tests on Pparα in bats
| Model | npa | ℓ b | ω0 c | ωlabeled d | Model compared | 2Δℓ | P |
|---|---|---|---|---|---|---|---|
| A. One ratio: ω0 | 35 | −5047.902 | 0.033 | ω0 | |||
| B. One ratio: ω = 1 (fixed) | 34 | −6001.416 | 1.000 | ω0 | A and B | 1907.029 | 0.000 |
| C. Free ratio | 67 | −5016.255 | — | — | A and C | 63.294 | 0.001 |
| D. Two ratios: ω0 = ωH-Yan, ωH-Yin | 36 | −5047.813 | 0.033 | 0.021 | A and D | 0.179 | 0.672 |
| E. Two ratios: ω0 = ωH-Yin, ωH-Yan | 36 | −5044.408 | 0.037 | 0.018 | A and E | 6.988 | 0.008 |
| F. Two ratios: ω0, ωHN | 36 | −5047.801 | 0.032 | 0.0042 | A and F | 0.203 | 0.653 |
anp: number of parameters.
b ℓ: −Lnlikelihood ratio.
cω0: one ω ratio for all branches.
dωlabeled are ω ratios for ancestral branches of Yinpterochiroptera hibernating bats (ωH-Yin), Yangopterochiropter hibernating bats (ωH-Yan), and non-hibernating bats (ωHN).
Upregulation of Pparα Expression in Torpid Bats
We next investigated the possibility that bat Pparα is adapted to hibernation at the level of transcription. The amount of Pparα mRNA in the liver of torpid bats was compared to that of active bats, including the distantly related Rhinolophus ferrumequinum and Hipposideros armiger bats in Yin and Myotis ricketti bats in Yang (Fig 2). Results of quantitative PCR showed that Pparα mRNA levels were significantly higher in torpid states than in active states and were increased by 1.19, 1.39, and 1.71 fold in torpid R. ferrumequinum, H. armiger, and M. ricketti bats, respectively (Fig 2). These results indicate that Pparα in bats adapts to hibernation at the level of transcription.
Fig. 2.
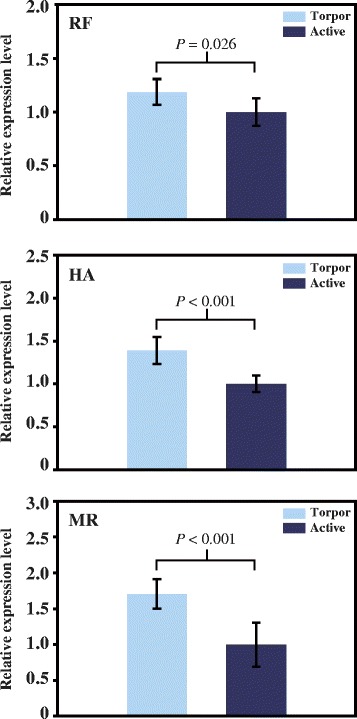
Expression levels of Pparα mRNA determined by real-time RT PCR. RF, HA, and MR represent hibernating R. ferrumequinum, H. armiger, and M. ricketti bats, respectively. Relative mRNA levels in torpid and active states are indicated with light blue and dark blue colors, respectively. The expression level of Pparα in active bats was set as 1.0. Data are presented as mean ± SD. A P value < 0.05 is considered statistically significant
The mechanisms for Pparα upregulation in torpid bats remain to be investigated. During hibernation, lipid is the main fuel source, and a complex lipid signaling exists not only on the cell membrane but also in the nucleus [28]. Lipid signaling has a close relationship with the signaling pathways of numerous biological processes, such as inflammation, immunity, and glucose and amino acid metabolism [29-32]. PPARs are known to be activated by fatty acids and their derivatives [30, 31]. As some transcription factors may also activate Pparα transcription, we used bioinformatic approaches to search for those that can bind to the 3 potential regulatory regions of Pparα, including the region around the transcription start site (TSS_Around), the region upstream from TSS (TSS_Up), and the gene body (TSS_Body) (see Additional file 1: Table S5 and Additional file 2).
We found that transcription factors FOXL1, NFYA, NFYB, SP1, TBP, and ERG can bind to various regulatory regions of Pparα in hibernating bats (FOXL1: TSS_Up 10 kb; NFYA, NFYB, and ERG: TSS_Body 7.5 kb; NFYB: TSS_Around 7.5 kb and TSS_Body 5 kb; SP1: TSS_Up 2.5 kb; TBP: TSS_Around 10 kb; ERG: TSS_Body 2.5 kb and 10 kb). In contrast, the NF-κB p65 subunit (RELA) and E2F1 were found to have the potential to bind to the regulatory regions of Pparα in non-hibernating bats (RELA: TSS_Around 7.5 kb; E2F1: TSS_Body 10 kb). FOXL1 is a forkhead box transcription factor and is crucial for liver development and function. Knockdown of Foxl1 in mice leads to defective intestinal glucose uptake [33]. NFYA, NFYB, and SP1 work cooperatively with PPARα to regulate the transcription of many lipogenic genes [34, 35]. TBP is a TATA box binding protein that interacts with PPARα to mediate gene transcription [36]. E2F1 regulates the expression of many genes involved in cell cycle control and proliferation [37]. RELA is a pleiotropic TF associated with the regulation of inflammation and immunity. Its activity is repressed by ligand-bound PPARs [37, 38].
It is known that E2F1 competes with histone deacetylase 1 (HDAC1) for binding to SP1 [39]. The activity of HDAC1 is significantly elevated in the skeletal muscle of Spermophilus tridecemlineatus ground squirrels during hibernation [40]. In these squirrels, the activation of NF-κB in skeletal muscle and intestine is positively linked to the antioxidant defense [41, 42]. NF-κB is also activated in the heart and muscle of torpid Myotis lucifugus bats [43]. These observations indicate that NF-κB and E2F1 are critical for hibernation. Since down regulation of PPARα depends on an intact signaling pathway of RELA [44] and E2F1 (an endogenous co-activator of NF-κB) [37], the potential of RELA and E2F1 to bind to the regulatory regions of Pparα in non-hibernating bats suggests their roles in transcriptional repression of Pparα. Similar binding preferences of FOXL1, NFYA, NFYB, SP1, TBP, ERG, RELA, and E2F1 on Pparα were found in many other mammalian species (see Additional file 1: Table S5 and Additional file 2).
Elevated Production of PPARα in Torpid Bats
To correlate the mRNA level of Pparα with its protein level, we determined the amounts of PPARα protein in mice, hibernating Rhinolophus ferrumequinum, Hipposideros armiger, and Myotis ricketti bats, and non-hibernating Rousettus leschenaultii bats by Western blotting. Mouse PPARα (~55 kDa) protein was used as the positive control (Fig 3A). For each hibernating bat, the amount of PPARα in the torpid state (T) was compared to that in the active state (A). The levels of PPARα were found to be increased by 1.22, 1.24, and 1.25 fold in torpid R. ferrumequinum, H. armiger, and M. ricketti bats, respectively (P < 0.05) (Fig 3B). These results were consistent with the elevated expression of Pparα mRNA in these bats during hibernation (Fig 2).
Fig. 3.
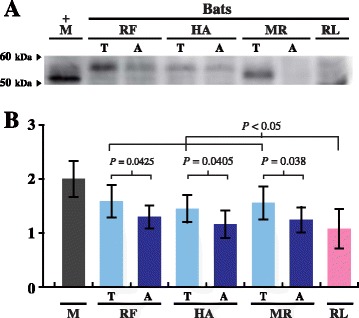
Expression levels of PPARα in bats determined by Western blotting. (A) Western blotting results of PPARα. (B) Relative levels of PPARα. The expression level of the PPARα protein in mice (M) was set as 2, and that of PPARα in each bat species was relative to it. RF, HA, and MR represent hibernating R. ferrumequinum, H. armiger, and M. ricketti bats, respectively. RL represents non-hibernating R. leschenaultii bats. T and A indicate bats in torpid and active states, respectively. Arrowheads in (A) indicate molecular weights (kDa) of protein markers. Relative levels of the PPARα protein in mice (gray), torpid (light blue) and active (dark blue) bats, and non-hibernating bats (pink) are shown in (B). Results are presented as mean ± SD. A P value < 0.05 is considered significant
Because the expression of PPARα was upregulated in distantly related hibernating bats and its expression level was higher in torpid bats than in active and non-hibernating bats (Fig 3), these results strongly suggest the adaptation of bat PPARα to hibernation. The sizes of PPARα detected in samples from R. ferrumequinum and H. armiger bats were slightly higher than expected (Fig 3A). This discrepancy may be due to variations in post-translational modifications (PTMs) of PPARα as described previously [45]. The expression of PPARα in mice was found to be higher than in all the hibernating and nonhibernating bats examined (Fig 3), suggesting a potential hibernation capability of mice. This possibility is supported by the adenosine-induced torpid mice recently established by Swaop et al. [46].
In addition to using fat as the main energy source, bats maintain their blood glucose at 2–3 mM and catabolize amino acids to generate ketone bodies for energy supply during torpor [27]. PPARs are known to be involved in lipid metabolism and other cellular processes, such as insulin sensitivity, amino acid homeostasis, adaptation to starvation, and inflammation [31]. Overexpression of PPARα activates nearly every gene associated with fatty acid catabolism [31]. Furthermore, Pparα-null mice exhibit increased glucose utilization, diminished fatty acid oxidation [15], and elevated expressions of many genes (e.g., arginase, phenylalanine hydroxylase, and spermidine synthetase) involved in amino acid metabolism [47]. Since hibernating mammals progressively suppress glucose utilization and use fat as the primary fuel [2], it is conceivable that bats produce sufficient amounts of PPARα to meet their metabolic requirements during torpor. Previous studies indicated that PPARα homeostasis is regulated by heterodimerization, recruitment of cofactors, post-translational modifications, and micro-RNAs (e.g., miR-519d) [48, 49]. Such regulation and the possible cross-talk among the various transcription factors observed in this study agree with the notion that transcriptional or translational reprogramming of genes occurs during mammalian hibernation [40].
Variation of amino acid sequences of PPARα in bats
To investigate the adaptation of PPARα to hibernation, we compared the amino acid sequences of PPARα from 9 hibernating and 3 non-hibernating species of bats. The human PPARα sequence was used as the template. In the aligned region (amino acid positions 87 to 446), 316 amino acid sites were identical (87.78 %) and 44 amino acid sites were variable (see Additional file 3: Fig S3).
5 positions including V90, Y155, I195, T200, and I382 of PPARα were conserved in hibernating bats but were different or divergent among non-hibernating bats. 38 amino acid sites of PPARα were conserved in non-hibernating bats but were different among hibernating bats. Missense mutations at V90 and Y155 of PPARα had been reported in carcinomas of the large intestine and liver, respectively [50], and missense mutation at G96 had been observed in malignant melanoma [50]. The amino acid site 155 is located in the zinc-finger motif (C4 degenerated type, CX2CX13CX2CX14–15 CX5CX9CX2C) of PPARα (see Additional file 3: Fig S3). These results suggest the important role of these conserved amino acid sites in PPARα. Taken together, these data agree with the conservation and functional constraint of PPARα toward evolution (Fig 1 and Additional file 3: Fig S3).
Potential PPARα Target genes
Genome-wide bioinformatic analyses were performed to estimate the mean ratio of the number of PPARα potential target genes to the number of all annotated genes (see Additional file 1: Table S6) in bats and to calculate the mean affinity score of PPARα to its target genes. 4 species of hibernating bats Myotis brandtii, Myotis davidii, Myotis lucifugus, and Eptesicus fuscus were found to have a higher mean target gene ratio than non-hibernating Pteropus vampyrus bats (see Additional file 3: Fig S4A), and the mean affinity score of PPARα to its target genes of these hibernating bats was higher than that of non-hibernating bats, Pteropus vampyrus and Pteropus alecto (see Additional file 3: Fig S4B). These results imply that mammalian hibernators have more PPARα target genes than non-hibernators and that PPARα has a higher binding affinity to its target genes in hibernators than in non-hibernators. To test these possibilities, the mean ratio of the number of PPARα potential target genes to the number of all genes in each of the 16 hibernating species and 48 non-hibernating species of mammals was determined; the mean affinity score of PPARα to its target genes in each of these mammalian species was also calculated. Results showed that the mean target gene ratio of the hibernators was significantly higher than that of the non-hibernators at a P value threshold of 5 x 10−7 (Fig 4A). This result was consistent across different settings of threshold P values (see Additional file 3: Fig S5 and Additional file 4). The mean affinity score of PPARα to its target genes in the hibernators was 9.68 ± 0.24, which is significantly higher than the score 9.37 ± 0.21 of the non-hibernators (Fig 4B and Additional file 1: Table S6). These data showed that hibernating mammals have more PPARα target genes and that PPARα has a higher affinity to its target genes in hibernating than in non-hibernating mammals.
Fig. 4.
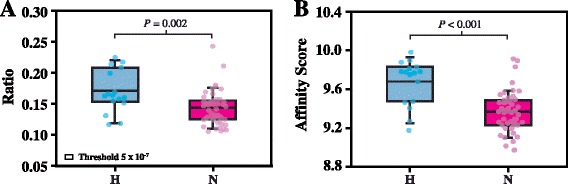
Bioinformatic analyses of PPARα in mammals. The y-axis in (A) represents the ratios of the number of PPARα potential target genes to the number of all genes of different animals. The small open rectangle indicates the threshold setting (5 x 10−7) for the matrix scan. The scores in (B) indicate the binding affinity of PPARα to its target genes in different animals. Blue and pink boxes indicate data calculated from 16 hibernating species (H) and 48 non-hibernating species (N) of mammals. Each dot represents the mean value obtained from each mammalian species. The box plot shows the median, 25/75 percentiles (box), and 10/90 percentiles (bars). A P value < 0.05 is considered significant
To investigate phylogenetic inferences [51], we constructed a phylogenetic tree of 64 mammals (see Additional file 3: Fig S6A). The correlation between mammalian hibernation and the target gene ratio, or affinity score of PPARα, was analyzed by 2 different phylogenetic comparative methods, including the quantitative genetic threshold model proposed by Felsenstein [52] and the phylogenetic ANOVA proposed by Garland et al. [53] (see Additional file 2). With both approaches, we found that the correlation between hibernation and affinity score of PPARα was significant (95 % confidence interval 0.198 to 0.261; P < 0.002; see Additional file 1: Table S7). The correlation between hibernation and target gene ratio of PPARα was also significant (P value threshold 5 x 10−7) (see Additional file 1: Table S7 and Additional file 3: Fig S6B). However, the correlation coefficients were substantially decreased if the phylogeny of these mammals was considered. Therefore, phylogenesis is an important factor in the analysis of evolutionary adaptation of mammalian hibernation.
Differential gene expression plays a key role in the evolution of morphological phenotypes and some biological traits [54]. The evolution of phenotypic fitness is influenced, in part, by the divergent pattern of TF binding sites and different affinities of TFs to their binding sites. The crosstalk among TFs regulates transcription, ultimately leading to the complex phenotypes [54, 55]. It has been shown that TF binding sites evolve rapidly in order to adapt to metabolic control and detoxification in the liver [56]. Our observations of more PPARα target genes and a higher binding affinity of PPARα to its target genes in hibernating mammals suggest an important regulatory role of PPARα in hibernation. It is probable that the genomes of hibernators have evolved to adapt to hibernation.
Mammalian hibernation is an ideal model to investigate the role of differential gene expression in adaptive evolution [57]. The coding regions of most differentially expressed genes are shaped by purifying selection [54]. A recent comparative genomic study of 3 hibernating and several non-hibernating species of mammals found no signs of positive selection on the coding regions of genes associated with hibernation [58]. However, differential expression of genes of metabolic pathways commonly shared by 4 hibernating species (Myotis brandtii, Ursus americanus, Spermophilus parryii, and Spermophilus tridecemlineatus) was found, indicating the pivotal role of regulation of gene expression in mammalian hibernation [57, 58]. Our findings together with previous reports [58] suggest that both genomic information (e.g., gene sequences) and genomic processes (e.g., transcription and translation) of Pparα have evolved toward adaptation to hibernation. More comparative studies of TF binding between torpid and active mammals or between hibernating and non-hibernating species will allow us to gain further insights into the evolution of mammalian hibernation.
Conclusions
In this study, we found PPARα upregulation at both mRNA and protein levels in bat liver during torpor, providing clear evidence of PPARα in adaptation to mammalian hibernation. Mechanisms by which bats upregulate the transcription and translation of PPARα to achieve a torpid phenotype remain to be investigated. A complex crosstalk among different TFs, such as FOXL1, NFYA, NFYB, SP1, TBP, ERG, RELA, and E2F1, may be involved in the control of PPARα expression. Results of evolutionary analyses and amino acid sequence alignments indicated that PPARα is highly conserved among hibernators. Bioinformatic analyses revealed that the genomes of mammalian hibernators have evolved to become more susceptible to PPARα regulation.
Methods
Animals and tissue acquisition
Acquisition of bats and experiments involving animals were done in accordance with ethical principles of the Animal Ethics Committee, East China Normal University (approval number AR2012/03001). 6 males each of the following hibernating bat species were captured from various locations in China: Rhinolophus ferrumequinum from Guan Ma Karst Cave (40°08′N, 126°05′E) in Jilin Province; Hipposideros armiger from Fish Cave (30°20′N, 117°50′E) in Anhui Province; Myotis ricketti from 7 Star Cave (25°16′N, 110°18′E) in Guangxi Province; Taphozous melanopogon from Ladian County (24°41′N, 108°01′E) in Guangxi Province; Scotophilus heathii from Mengla County (21°27′N, 101°33′E) in Yunnan Province; and Ia io from Feilong Cave (24°58′N, 104°53′E) in Guizhou Province (see Additional file 3: Fig 1). 3 bats of each species were sacrificed on site while they were in torpor, and the remaining bats were scarified 48hr after arousal from torpor in the laboratory. Rectal temperatures were approximately 8-10 °C for torpid and 36-37 °C for active bats. 3 males each of non-hibernating bat species Rousettus leschenaultii, Cynopterus sphinx, and Eonycteris spelaea were captured from Mashan County (23°55′N, 108°26′E) in Guangxi Province, Haikou Park (20°02′N, 110°20′E) in Hainan Province, and Xishuangbanna (22°0′N, 100°47′E) in Yunnan Province, China, respectively. Non-hibernating bats captured were sacrificed in the field. Mice were purchased from Sino-British Sippr/BK Lab Animal Ltd (Shanghai, China). These animals were sacrificed by cervical dislocation, and their liver tissues were rapidly excised, snap frozen in liquid nitrogen, and then stored in a −80 °C freezer until used.
RNA Isolation, cloning, and sequencing
Total RNA was isolated from the liver tissues using Trizol® reagent (Invitrogen, USA). A total of 5 μg RNA was reverse transcribed to cDNA using the SuperScript® III Reverse Transcriptase kit (Invitrogen, USA). Primer pairs listed in Table 2 were used to amplify the coding region of bat Pparα. PCRs were carried out under the following conditions: denaturation at 95 °C for 5 min; 32 cycles of 95 °C for 30 s, 50 - 60 °C for 30 s, and 72 °C for 2 min; and a final extension at 72 °C for 10 min. The resulting DNA fragments were isolated by electrophoresis in 1 % agarose gel and then ligated into pGEM®-T Easy Vector (Promega, USA). Recombinant plasmids were transformed into E. coli strain TOP10 (Tiangen, China). Bacterial colonies were screened by blue-white selection, and white colonies were picked for colony PCR. 6 positive clones of each PCR product were sequenced on an ABI 3730 DNA sequencer. All sequences obtained were submitted to GenBank (see Additional file 1: Table S8).
Table 2.
Primers for cloning of bat Pparα
| Name | Sequences | Amplified species | Primer used | Product length (bp) | Length proportion |
|---|---|---|---|---|---|
| Pparα-1 F | 5’-CTTGAGGCTGATGAYCTGGAAA | E. spelaea | 1 and 2 | 1361 | 96.73 % |
| Pparα-1R | 5’-TYGGGAAGAGGAAGRTGTCG | R. leschenaultii | 1 and 2 | 1361 | 96.73 % |
| Pparα-2 F | 5’-CAAYCCACCTTTYGTCAT | C. sphinx | 1 and 2 | 1361 | 96.73 % |
| Pparα-2R | 5’-ATRTCCCTGTAGATYTCCT | R. ferrumequinum | 1 and 3 | 1362 | 96.80 % |
| Pparα-3 F | 5’-TGAATAAAGACGGGATGCT | H. armiger | 1 and 3 | 1362 | 96.80 % |
| Pparα-3R | 5’-CATGTCCCTGTAGATTTCCT | T. melanopogon | 4 | 1118 | 79.46 % |
| Pparα-4 F | 5’-CGGTGTCTTACCCTGTGGT | S. heathii | 4 | 1118 | 79.46 % |
| Pparα-4R | 5’-CGCCTCGGTCCTCTTGAT | M. ricketti | 4 | 1118 | 79.46 % |
| I. io | 4 | 1118 | 79.46 % |
Sources of Pparα nucleotide sequences
Nucleotide sequences of Pparα were obtained from the following: 56 mammalian species of Plancentalia and Marsupialia [59] (see Additional file 1: Table S8 and Additional file 3: Fig S2); 5 species of Yinpterochiroptera bats including 3 non-hibernating bat species (family Pteropodidae) C. sphinx, E. spelaea, and R. leschenaultii and 2 hibernating bat species R. ferrumequinum (family Rhinolophidae) and H. armiger (family Hipposideridae); and 7 species of hibernating Yangochiroptera bats including T. melanopogon (family Emballonuridae) and 6 species of family Vespertilionidae: S. heathii, I. io, E. fuscus, M. ricketti, M. brandtii, and M. davidii. These bats were chosen because their hibernation behaviors are known [9, 12]. The DNA sequences of Pparα of E. fuscus, M. brandtii, M. davidii and non-bat mammals, such as Homo sapiens and Mus musculus, were downloaded from GenBank.
Evolutionary analysis of Pparα of bats
The nucleotide sequences of Pparα were aligned using the software Clustal X [60]. DNA sequences were translated into amino acid sequences using the software MEGA5 [61]. The program CODEML in PAML (version 4.8) was used to estimate the ω value, which is derived from dN (nonsynonymous substitution rate) divided by dS (synonymous substitution rate). Species topology was constructed as previously described (Fig 1) [6, 62]. An ω value of 1, < 1, or > 1 represents neutral evolution, negative purifying selection, and positive selection, respectively. The one-ratio model analysis, which assumes an equal ω value among all branches, was first conducted to establish the null hypothesis [63]. The free ratio model, which allows ω values to vary among branches, was used to compare with the one-ratio model to test the heterogeneity of ω across the tree. The two-ratio model, which allows ω values to vary between the labeled and other branches, was also applied to each of the ancestral branches of 2 clades of hibernating bats from Yinpterochiroptera and Yangochiroptera (Table 1).
Real-time PCR
Total RNA was extracted from the liver tissues of hibernating bats H. armiger, R. ferrumequinum, and M. ricketti. 6 bats of each species were used, including 3 active and 3 torpid bats. The mRNA was reverse transcribed into cDNA as described above. The primer pairs used are listed in Table 3. 100 nanogram of cDNA was used to determine the expression level of Pparα. Real-time PCR was performed on an ABI Prism 7300 real-time PCR system (Applied Biosystems, UK) using the SYBR® Premix Ex TaqTM (Tli RNaseH Plus) kit (Takara, Japan). The 2-ΔΔCT method was applied to normalize the level of Pparα to that of Gapdh and to calculate the relative expression levels of Pparα between torpid and active bats [64-68].
Table 3.
Primers used in RT-PCR
| Genes and bat species | Primersa |
|---|---|
| Pparα and M. ricketti | F: 5’-AAAGCGAAACTGAAAGCAGAAATCC |
| R: 5’-TCATGTTGAAGCTCCGCAGGTAG | |
| Pparα and R. ferrumequinum | F: 5’-AGCCAACAACAATCCACCTTT |
| R: 5’-AGCTCCGTGACAGTCTCCACA | |
| Pparα and H. armiger | F: 5’-TTTCACAAGTGCCTTTCGGTTGG |
| R: 5’-GATTTGAGGTCCGCCGTTTCG | |
| Gapdh (internal control for all bat species) | F: 5’-ATGGGTGTGAACCAYGASAAGT |
| R: 5’-GGTCATGAGTCCYTCCACRAT |
aY = C + T, S = G + C, R = A + G.
Western blotting
Liver tissues (100 mg) from bats and mice were homogenized separately in 2-ml Eppendorf tubes, each containing 1.7 ml of lysis buffer (10 % glycerol, 2 % SDS, 1.25 % β-mercaptoethanol, 25 mM Tris–HCl, pH 6.8) and ceramic beads (0.17 g, 1 mm in diameter), using a Precellys® 24 grinder (Bertin Technologies, France) at 4 °C. The homogenates were centrifuged at 13,400 xg for 10 min at 4 °C. Supernatants were boiled at 100 °C for 10 min and then centrifuged at 13,400 xg for 10 min at 4 °C to remove insoluble cell debris. Each of the clarified supernatants was divided into small aliquots and stored at −80 °C until used. Protein concentration of each sample was determined using the Quick StartTM Bradford protein assay kit (Bio-Rad, USA) according to manufacturer’s instructions.
Equal amount of each protein sample (20 μg/lane) was subjected to a 12.5 % SDS-PAGE, and the separated proteins were transferred onto a PVDF membrane (Millipore, USA). The membrane was immersed in blocking buffer (5 % skim milk and 1 % BSA) at 4 °C overnight, and then probed with anti-PPARα (1:100 dilution, Santa Cruz®, USA, sc-9000), which is a rabbit polyclonal antibody against a conserved N-terminal epitope of PPARα. After washing with TBST buffer, the blot was reacted with an HRP-conjugated secondary antibody (Santa Cruz®, CA) at room temperature for 2 hours, followed by incubation with ImmobilonTM Western HRP Substrate Reagent (Millipore, USA). A reversible Ponceau S staining of the membrane was carried out to estimate the relative amount of each protein on the membrane [69]. Band intensity on the blot was quantified by the ImageQuantTM TL software (version 7.0, Amersham Biosciences), and the intensity of each band was normalized to the relative value of the corresponding Ponceau-stained protein band.
Bioinformatic Analyses of Target Genes and Determination of Affinity Scores of PPARα
The ChIP-seq data derived from chromatin immunoprecipitation (ChIP) of human hepatoma cells with anti-PPARα antibody were obtained from the GEO database (GSM864671) [36] and mapped to the mouse genome sequence (version mm9) [70]. The number of ChIP-seq peaks located in introns, exons, upstream regions of TSS (transcription start site), downstream regions of TTS, and other regions of all annotated genes in the mouse genome was separately calculated and divided by the total peak number to obtain the peak ratio of a particular location in the genome (see Additional file 3: Fig S7). A PPARα target gene was defined if at least one binding site was located within introns, exons, or 10 kb upstream of TSS of the gene.
The nucleotide sequences of 10 kb upstream of TSS, introns, and exons of all annotated genes of each of the 64 mammalian species (see Additional file 1: Table S6) were retrieved from NCBI, Ensembl, or GigaDB [71]. The position frequency matrix (PFM) of PPARα (f2) (see Additional file 3: Fig S8), which represents DNA sequence patterns recognized by PPARα, was obtained from HOCOMOCO [72]. The matrix scan was first applied to estimate the number of putative binding sites of PPARα in these sequences using parameters ‘-pseudo 1 -decimals 1 -2str -origin end -bg_pseudo 0.01 -return limits -return pval -uth pval 0.0000005’ [73]. The number of PPARα target and non-target genes of each species was calculated based on the results of matrix scan. The ratio of PPARα target gene was derived from the number of target genes divided by that of total genes (see Additional file 4). The downloaded sequences were scanned by f2 PMF using a modified SPeaker algorithm [74] to calculate the affinity score of PPARα binding to each target gene. Only the highest score estimated for a gene was adopted. Results were analyzed by two-tailed Student’s t-test. A P value < 0.05 was considered significant.
To correlate the mammalian hibernation trait with the ratio of PPARα target gene or PPARα affinity to its target genes, 2 different phylogenetic comparison methods, including the quantitative genetic threshold model [52] and the phylogenetic ANOVA [53] were used (see Additional file 1: Table S7, Additional file 2, and Additional file 3: Fig S6). The phylANOVA and threshBayes in the R package phytools were conducted separately to determine the correlations [75].
Availability of supporting data
The data sets supporting the results of this article are available in the Dryad digital repository, [http://dx.doi.org/10.5061/dryad.h6r74] [76] DOI:10.5061/dryad.h6r74%5d%20%5b76. Gene sequences obtained in this study have been deposited in GenBank [http://www.ncbi.nlm.nih.gov/genbank]. Accession numbers are provided in Additional file 1.
Additional files
Branch-site model A test for detection of positively selected sites in ancestral branches of hibernating bats. Table S2. Site model comparisons of 12 bat species. Table S3. Branch model tests of Pparα in 56 mammalian species. Table S4. Site model comparisons of 25 mammals. Table S5. Prediction of transcription factor binding to 3 potential regulatory regions (TSS_Around, TSS_Up, and TSS_Body) of Pparα. Table S6. Affinity scores of PPARα binding to all annotated genes in 64 mammalian species. Table S7. Phylogenetic comparative methods for determining correlations among traits. Table S8. Accession numbers of Pparα sequences of 56 mammals.
Supplementary methods for evolutionary analyses of Ppar α in mammals and analyses of the binding affinity of each of the 205 transcription factors to the 3 potential regulatory regions of Ppar α in various mammalian species.
Geographical locations of bats obtained in China. The degrees of latitude and longitude are shown. Non-hibernating bats in Yinpterochiroptera, hibernating bats in Yinpterochiroptera, and hibernating bats in Yangochiroptera are represented by orange, green, and blue colors, respectively. Figure S2. Red dots A, B, C, and D denote the ancestral branch of Chiroptera; Cetartiodactyla, Perissodactyla, and Carnivora in Laurasiatheria; Rodentia and Lagomorpha; Scandentia and Primates in Euarchontoglires, respectively. The N*d N/S*d S/ω values of A, B, C, and D are shown below the red dots. Abbreviations Af, Au, Am, and M represent Afrotheria, Australidelphia, Ameridelphia, and Marsupialia, respectively. Red asterisks indicate the species that can hibernate. Figure S3. Amino acid sequence alignments of PPARα of bats. Amino acid sequences of non-hibernating and hibernating bat species are denoted with pink and blue colors, respectively. The amino acid positions that are conserved in non-hibernating species but different or diverged among hibernating species are indicated with orange triangles. Amino acid sites that are conserved in hibernating species but different or diverged among non-hibernating species are denoted with red triangles. The green line shows the location of the zinc-finger motif. Figure S4. (A) The ratio of the number of PPARα potential target genes to that of all genes in bats. The small open rectangle indicates the threshold setting for the matrix scan. (B) Mean scores of binding affinity of PPARα to its target genes in bats. Blue and pink dots represent the mean value calculated from hibernating and non-hibernating bats, respectively. Figure S5. The y-axis in (A) represents the ratios of the number of PPARα potential target genes to the number of all genes of different animals. The small open rectangle indicates the threshold setting for the matrix scan. The scores in (B) indicate the binding affinity of PPARα to its target genes in different animals. Blue and pink boxes indicate data calculated from 16 hibernating species (H) and 48 non-hibernating species (N) of mammals. Each dot represents the mean value obtained from each mammalian species. The box plot shows the median, 25/75 percentiles (box), 10/90 percentiles (bars). A P value < 0.05 is considered significant. Figure S6. (A) Phylogenetic tree constructed for 62 mammalian species. Hibernating species are colored in red. (B) Results of phylogenetic ANOVA. The y-axis denotes scores of binding affinity of PPARα to its target genes. The box plot shows the median, 25/75 percentiles (box), and 10/90 percentiles (bars). A P value < 0.05 is considered significant. Figure S7. Mapping of PPARα binding sites to the mouse genome. The y-axis denotes the number of PPARα binding sites located in various regions of all annotated genes, including the sequences upstream of TSS (UpStream), 5’ un-translated sequences (5’UTR), exons (Coding Exon), introns (Intron), 3’ untranslated sequences (3’UTR), the sequences downstream of TSS (DownStream), and the distal sequences (>20 kb) (Distal_Region). The Arabic number shown on top of each bar indicates the ratio of the number of binding sites in the region to the number of all PPARα binding sites. Figure S8. Position frequency matrix (PFM) of sequences recognized by PPARα.
Different threshold settings of P value for the matrix scan of PPARα in mammals.
Acknowledgements
We thank Tengteng Zhu, Guangjian Zhu, and Junpeng Zhang for tissue collections, Tao Fang and Bin Shen for helpful comments on molecular evolutionary analyses, and Yang Liu for the discussions on phylogenetic analyses. We thank Dr. Chao-Hung Lee and Professor John F. Atkins for editing the manuscript and providing valuable advices. This work was supported by grants from the National Science Foundation of China (No. 31100273/C030101) to Yi-Hsuan Pan, Science and Technology Committee of Shanghai (No. 11XD1402000) and National Science Foundation of China (No. 31172077) to Shuyi Zhang.
Abbreviations
- PPARα
Peroxisome proliferator-activated receptor alpha
- RXR
Retinoid X receptor
- TFs
Transcription factors
- FOXL1
Forkhead box protein L1
- NFYA
Nuclear transcription factor Y subunit alpha
- NFYB
Nuclear transcription factor Y subunit beta SP1, Specificity protein 1
- TBP
TATA-box-binding protein 1
- ERG
E-26 related gene
- RELA
v-rel avian reticuloendotheliosis viral oncogene homolog A
- E2F1
E2 Transcription Factor 1
Footnotes
Yijie Han and Guantao Zheng contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YH and GZ carried out evolutionary analyses, performed biochemical experiments, and wrote the manuscript. YP designed the experiments and edited the manuscript. TY and DD performed bioinformatic analyses. YP and SZ provided experimental materials. All authors have read and approved the final manuscript.
Contributor Information
Yijie Han, Email: yijiehan@gmail.com.
Guantao Zheng, Email: guantao.zheng@gmail.com.
Tianxiao Yang, Email: ytxiao1226@hotmail.com.
Shuyi Zhang, Email: syzhang@bio.ecnu.edu.cn.
Dong Dong, Email: ddong.ecnu@gmail.com.
Yi-Hsuan Pan, Email: yihsuanp@gmail.com.
References
- 1.Geiser F. Hibernation. Current Biology. 2013;23(5):R188–R193. doi: 10.1016/j.cub.2013.01.062. [DOI] [PubMed] [Google Scholar]
- 2.Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83(4):1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 3.Storey KB, Storey JM. Metabolic rate depression: the biochemistry of mammalian hibernation. Advances in clinical chemistry. 2010;52:78. [PubMed] [Google Scholar]
- 4.Andrews MT. Advances in molecular biology of hibernation in mammals. Bioessays. 2007;29(5):431–440. doi: 10.1002/bies.20560. [DOI] [PubMed] [Google Scholar]
- 5.Lyman C. Termoregulation and metabolism in bats. In: Wimsatt W, editor. In Biology of bats. New York: Academic; 1970. p. 301–30.
- 6.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307(5709):580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 7.Simmons N. Order Chiroptera. In: DM WDR, editor. Mammal species of the world: a taxonomic and geographic reference. Baltimore, Maryland: Johns Hopkins University Press; 2005. pp. 312–529. [Google Scholar]
- 8.Geiser F, Stawski C. Hibernation and torpor in tropical and subtropical bats in relation to energetics, extinctions, and the evolution of endothermy. Integrative and comparative biology. 2011;51(3):337–348. doi: 10.1093/icb/icr042. [DOI] [PubMed] [Google Scholar]
- 9.Ransome R. The Natural History of Hibernating Bats, In The Natural History of Hibernating Bats. London: Christopher Helm.: Academic Press Inc; 1990.
- 10.Lyman CP. Hibernation and torpor in mammals and birds: Elsevier. 2013. [Google Scholar]
- 11.Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118):456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan L, Zhao X, Lin B, Rossiter SJ, He L, Zuo X, et al. Adaptive evolution of Leptin in heterothermic bats. PloS one. 2011;6(11) doi: 10.1371/journal.pone.0027189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 14.Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Progress in lipid research. 2006;45(2):120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Bernal-Mizrachi C, Weng S, Feng C, Finck BN, Knutsen RH, Leone TC, et al. Dexamethasone induction of hypertension and diabetes is PPAR-α dependent in LDL receptor–null mice. Nature medicine. 2003;9(8):1069–1075. doi: 10.1038/nm898. [DOI] [PubMed] [Google Scholar]
- 16.Tan NS, Michalik L, Desvergne B, Wahli W. Multiple expression control mechanisms of peroxisome proliferator-activated receptors and their target genes. The Journal of steroid biochemistry and molecular biology. 2005;93(2):99–105. doi: 10.1016/j.jsbmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Nelson CJ, Otis JP, Carey HV. A role for nuclear receptors in mammalian hibernation. The Journal of physiology. 2009;587(9):1863–1870. doi: 10.1113/jphysiol.2008.167692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86(2):465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 19.Berger J, Moller DE. The mechanisms of action of PPARs. Annual review of medicine. 2002;53(1):409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell metabolism. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Shao C, Fedorov VB, Goropashnaya AV, Barnes BM, Yan J. Molecular signatures of mammalian hibernation: comparisons with alternative phenotypes. BMC genomics. 2013;14(1):567. doi: 10.1186/1471-2164-14-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin B, Yuan L, Chen J. Selection pressure drives the co-evolution of several lipid metabolism genes in mammals. Chinese Science Bulletin. 2012;57(8):877–885. doi: 10.1007/s11434-011-4862-8. [DOI] [Google Scholar]
- 23.Eddy SF, Morin P, Jr, Storey KB. Cloning and expression of PPARγ and PGC-1α from the hibernating ground squirrel, Spermophilus tridecemlineatus. Molecular and cellular biochemistry. 2005;269(1):175–182. doi: 10.1007/s11010-005-3459-4. [DOI] [PubMed] [Google Scholar]
- 24.Eddy SF, Storey KB. Differential expression of Akt, PPARγ, and PGC-1 during hibernation in bats. Biochemistry and cell biology. 2003;81(4):269–274. doi: 10.1139/o03-056. [DOI] [PubMed] [Google Scholar]
- 25.Kabine M, El Kebbaj Z, Oaxaca-Castillo D, Clémencet M-C, Latruffe N, Cherkaoui-Malki M. Peroxisome proliferator-activated receptors as regulators of lipid metabolism; tissue differential expression in adipose tissues during cold acclimatization and hibernation of jerboa (Jaculus orientalis). Biochimie. 2004;86(11):763–70. [DOI] [PubMed]
- 26.El Kebbaj Z, Andreoletti P, Mountassif D, Kabine M, Schohn H, Dauça M, et al. Differential regulation of peroxisome proliferator-activated receptor (PPAR)-α1 and truncated PPARα2 as an adaptive response to fasting in the control of hepatic peroxisomal fatty acid β-oxidation in the hibernating mamma. Endocrinology. 2009;150(3):1192–1201. doi: 10.1210/en.2008-1394. [DOI] [PubMed] [Google Scholar]
- 27.Pan YH, Zhang Y, Cui J, Liu Y, McAllan BM, Liao CC, et al. Adaptation of phenylalanine and tyrosine catabolic pathway to hibernation in bats. PloS one. 2013;8(4) doi: 10.1371/journal.pone.0062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melvin RG, Andrews MT. Torpor induction in mammals: recent discoveries fueling new ideas. Trends in Endocrinology & Metabolism. 2009;20(10):490–498. doi: 10.1016/j.tem.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaqoob P. Fatty acids as gatekeepers of immune cell regulation. Trends in immunology. 2003;24(12):639–645. doi: 10.1016/j.it.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 31.Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends in Endocrinology & Metabolism. 2012;23(7):351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Meijer AJ, Dubbelhuis PF. Amino acid signalling and the integration of metabolism. Biochemical and biophysical research communications. 2004;313(2):397–403. doi: 10.1016/j.bbrc.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Katz JP, Perreault N, Goldstein BG, Chao H-H, Ferraris RP, Kaestner KH. Foxl1 null mice have abnormal intestinal epithelia, postnatal growth retardation, and defective intestinal glucose uptake. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2004;287(4):G856–G864. doi: 10.1152/ajpgi.00136.2004. [DOI] [PubMed] [Google Scholar]
- 34.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS genetics. 2008;4(7) doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke SD. Polyunsaturated fatty acid regulation of gene transcription: a molecular mechanism to improve the metabolic syndrome. J Nutr. 2001;131(4):1129–1132. doi: 10.1093/jn/131.4.1129. [DOI] [PubMed] [Google Scholar]
- 36.van der Meer DL, Degenhardt T, Väisänen S, de Groot PJ, Heinäniemi M, de Vries SC, Müller M, Carlberg C, Kersten S: Profiling of promoter occupancy by PPARα in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res 2010:gkq012. [DOI] [PMC free article] [PubMed]
- 37.Lim C-A, Yao F, Wong JJ-Y, George J, Xu H, Chiu KP, et al. Genome-wide mapping of RELA (p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Molecular cell. 2007;27(4):622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 38.Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nature Reviews Immunology. 2005;6(1):44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 39.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, et al. Histone deacetylase 1 can repress transcription by binding to Sp1. Molecular and cellular biology. 1999;19(8):5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morin P, Jr, Storey KB. Mammalian hibernation: differential gene expression and novel application of epigenetic controls. International Journal of Developmental Biology. 2009;53(2):433. doi: 10.1387/ijdb.082643pm. [DOI] [PubMed] [Google Scholar]
- 41.Carey H, Frank C, Seifert J. Hibernation induces oxidative stress and activation of NF-κB in ground squirrel intestine. Journal of Comparative Physiology B. 2000;170(7):551–559. doi: 10.1007/s003600000135. [DOI] [PubMed] [Google Scholar]
- 42.Allan ME, Storey KB. Expression of NF-κB and downstream antioxidant genes in skeletal muscle of hibernating ground squirrels, Spermophilus tridecemlineatus. Cell biochemistry and function. 2012;30(2):166–174. doi: 10.1002/cbf.1832. [DOI] [PubMed] [Google Scholar]
- 43.Eddy SF, McNally J, Storey K. Up-regulation of a thioredoxin peroxidase-like protein, proliferation-associated gene, in hibernating bats. Archives of biochemistry and biophysics. 2005;435(1):103–111. doi: 10.1016/j.abb.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 44.Lim WS, Ng DL, Kor SB, Wong HK, Tengku-Muhammad TS, Choo QC, et al. Tumour necrosis factor alpha down-regulates the expression of peroxisome proliferator activated receptor alpha (PPARα) in human hepatocarcinoma HepG2 cells by activation of NF-κB pathway. Cytokine. 2013;61(1):266–274. doi: 10.1016/j.cyto.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Wadosky KM, Willis MS. The story so far: post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation. American Journal of Physiology-Heart and Circulatory Physiology. 2012;302(3):H515–H526. doi: 10.1152/ajpheart.00703.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swoap SJ, Iliff B, Le S. Adenosine, AMP, and Daily Torpor. In: Thomas R, Claudia B, Walter A, Eva M, editors. Living in a Seasonal World. Springer. 2012. p. 337-49.
- 47.Kersten S, Mandard S, Escher P, Gonzalez FJ, Tafuri S, Desvergne B, et al. The peroxisome proliferator-activated receptor α regulates amino acid metabolism. The FASEB Journal. 2001;15(11):1971–1978. doi: 10.1096/fj.01-0147com. [DOI] [PubMed] [Google Scholar]
- 48.Blanquart C, Mansouri R, Fruchart J-C, Staels B, Glineur C. Different ways to regulate the PPARα stability. Biochemical and biophysical research communications. 2004;319(2):663–670. doi: 10.1016/j.bbrc.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 49.Martinelli R, Nardelli C, Pilone V, Buonomo T, Liguori R, Castanò I, et al. P: miR-519d overexpression is associated with human obesity. Obesity. 2010;18(11):2170–2176. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 50.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(D1):D945-50. [DOI] [PMC free article] [PubMed]
- 51.Garland T, Bennett AF, Rezende EL. Phylogenetic approaches in comparative physiology. Journal of experimental Biology. 2005;208(16):3015–3035. doi: 10.1242/jeb.01745. [DOI] [PubMed] [Google Scholar]
- 52.Felsenstein J. A comparative method for both discrete and continuous characters using the threshold model. The American Naturalist. 2012;179(2):145–156. doi: 10.1086/663681. [DOI] [PubMed] [Google Scholar]
- 53.Garland T, Dickerman AW, Janis CM, Jones JA. Phylogenetic analysis of covariance by computer simulation. Systematic Biology. 1993;42(3):265–292. doi: 10.1093/sysbio/42.3.265. [DOI] [Google Scholar]
- 54.Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nature Reviews Genetics. 2012;13(7):505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430(6995):85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 56.Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478(7369):343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 57.Srere HK, Wang L, Martin SL. Central role for differential gene expression in mammalian hibernation. Proceedings of the National Academy of Sciences. 1992;89(15):7119–7123. doi: 10.1073/pnas.89.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villanueva-Cañas JL, Faherty SL, Yoder AD, Albà MM. Comparative Genomics of Mammalian Hibernators Using Gene Networks. Integrative and Comparative Biology. 2014;54(3):452-62. [DOI] [PubMed]
- 59.Murphy WJ, Pevzner PA, O'Brien SJ. Mammalian phylogenomics comes of age. Trends in Genetics. 2004;20(12):631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Thompson JD, Gibson T, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics. 2002;2.3(1–2):3–22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou X, Xu S, Xu J, Chen B, Zhou K, Yang G. Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian mammals. Systematic biology. 2012;61(1):150–164. doi: 10.1093/sysbio/syr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2− ΔΔCT Method. methods. 2001;25((4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 66.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiological genomics. 2005;21(3):389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 67.Soukri A, Valverde F, Hafid N, Elkebbaj M, Serrano A. Occurrence of a differential expression of the glyceraldehyde-3-phosphate dehydrogenase gene in muscle and liver from euthermic and induced hibernating jerboa (Jaculus orientalis). Gene. 1996;181(1):139–45. [DOI] [PubMed]
- 68.68. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology 2002, 3(7):research0034 [DOI] [PMC free article] [PubMed]
- 69.Romero-Calvo I, Ocón B, Martínez-Moya P, Suárez MD, Zarzuelo A, Martínez-Augustin O, et al. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Analytical biochemistry. 2010;401(2):318–320. doi: 10.1016/j.ab.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 70.Chinwalla AT, Cook LL, Delehaunty KD, Fewell GA, Fulton LA, Fulton RS, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 71.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2013. Nucleic Acids Res. 2012;41(D1):D48-55. [DOI] [PMC free article] [PubMed]
- 72.Kulakovskiy IV, Medvedeva YA, Schaefer U, Kasianov AS, Vorontsov IE, Bajic VB, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41(D1):D195–D202. doi: 10.1093/nar/gks1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, et al. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39(suppl 2):W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Megraw M, Pereira F, Jensen ST, Ohler U, Hatzigeorgiou AG. A transcription factor affinity-based code for mammalian transcription initiation. Genome research. 2009;19(4):644–656. doi: 10.1101/gr.085449.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Revell LJ. Phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution. 2012;3(2):217-23.
- 76.Han Y-J, Zheng G-T, Yang T-X, Zhang S-Y, Dong D, Pan Y-H. Supporting data for PPARα and hibernation. In: Dryad Digital Repository. (2015. http://dx.doi.org/10.5061/dryad.h6r74.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Branch-site model A test for detection of positively selected sites in ancestral branches of hibernating bats. Table S2. Site model comparisons of 12 bat species. Table S3. Branch model tests of Pparα in 56 mammalian species. Table S4. Site model comparisons of 25 mammals. Table S5. Prediction of transcription factor binding to 3 potential regulatory regions (TSS_Around, TSS_Up, and TSS_Body) of Pparα. Table S6. Affinity scores of PPARα binding to all annotated genes in 64 mammalian species. Table S7. Phylogenetic comparative methods for determining correlations among traits. Table S8. Accession numbers of Pparα sequences of 56 mammals.
Supplementary methods for evolutionary analyses of Ppar α in mammals and analyses of the binding affinity of each of the 205 transcription factors to the 3 potential regulatory regions of Ppar α in various mammalian species.
Geographical locations of bats obtained in China. The degrees of latitude and longitude are shown. Non-hibernating bats in Yinpterochiroptera, hibernating bats in Yinpterochiroptera, and hibernating bats in Yangochiroptera are represented by orange, green, and blue colors, respectively. Figure S2. Red dots A, B, C, and D denote the ancestral branch of Chiroptera; Cetartiodactyla, Perissodactyla, and Carnivora in Laurasiatheria; Rodentia and Lagomorpha; Scandentia and Primates in Euarchontoglires, respectively. The N*d N/S*d S/ω values of A, B, C, and D are shown below the red dots. Abbreviations Af, Au, Am, and M represent Afrotheria, Australidelphia, Ameridelphia, and Marsupialia, respectively. Red asterisks indicate the species that can hibernate. Figure S3. Amino acid sequence alignments of PPARα of bats. Amino acid sequences of non-hibernating and hibernating bat species are denoted with pink and blue colors, respectively. The amino acid positions that are conserved in non-hibernating species but different or diverged among hibernating species are indicated with orange triangles. Amino acid sites that are conserved in hibernating species but different or diverged among non-hibernating species are denoted with red triangles. The green line shows the location of the zinc-finger motif. Figure S4. (A) The ratio of the number of PPARα potential target genes to that of all genes in bats. The small open rectangle indicates the threshold setting for the matrix scan. (B) Mean scores of binding affinity of PPARα to its target genes in bats. Blue and pink dots represent the mean value calculated from hibernating and non-hibernating bats, respectively. Figure S5. The y-axis in (A) represents the ratios of the number of PPARα potential target genes to the number of all genes of different animals. The small open rectangle indicates the threshold setting for the matrix scan. The scores in (B) indicate the binding affinity of PPARα to its target genes in different animals. Blue and pink boxes indicate data calculated from 16 hibernating species (H) and 48 non-hibernating species (N) of mammals. Each dot represents the mean value obtained from each mammalian species. The box plot shows the median, 25/75 percentiles (box), 10/90 percentiles (bars). A P value < 0.05 is considered significant. Figure S6. (A) Phylogenetic tree constructed for 62 mammalian species. Hibernating species are colored in red. (B) Results of phylogenetic ANOVA. The y-axis denotes scores of binding affinity of PPARα to its target genes. The box plot shows the median, 25/75 percentiles (box), and 10/90 percentiles (bars). A P value < 0.05 is considered significant. Figure S7. Mapping of PPARα binding sites to the mouse genome. The y-axis denotes the number of PPARα binding sites located in various regions of all annotated genes, including the sequences upstream of TSS (UpStream), 5’ un-translated sequences (5’UTR), exons (Coding Exon), introns (Intron), 3’ untranslated sequences (3’UTR), the sequences downstream of TSS (DownStream), and the distal sequences (>20 kb) (Distal_Region). The Arabic number shown on top of each bar indicates the ratio of the number of binding sites in the region to the number of all PPARα binding sites. Figure S8. Position frequency matrix (PFM) of sequences recognized by PPARα.
Different threshold settings of P value for the matrix scan of PPARα in mammals.


