Abstract
In situ visualization has become a popular method for avoiding the slowest component of many visualization pipelines: reading data from disk. Most previous in situ work has focused on achieving visualization scalability on par with simulation codes, or on the data movement concerns that become prevalent at extreme scales. In this work, we consider in situ analysis with respect to ease of use and programmability. We describe an abstraction that opens up new applications for in situ visualization, and demonstrate that this abstraction and an expanded set of use cases can be realized without a performance cost.
1. Introduction and related work
The growing size of simulation data and the problems this poses for subsequent analysis pipelines has driven simulation authors to integrate visualization and analysis tasks into the simulation itself [CGS*13]. The primary advantage of this approach is to perform operations on data while they are still in memory, rather than forcing them through disk, thereby eliminating the most expensive component of the majority of visualization and analysis pipelines.
Scientists and engineers have developed many different approaches to in situ. DART uses RDMA to stage data from supercomputer to potentially separate analysis-focused resources [DPK10], and a system performs computations on the data as they are in transit from one resource to another [MOM*11]. The dominant approach is to use the same supercomputer that is running the simulation for visualization, though potentially on just a subset of cores, in the manner of Damaris/Viz [DSP*13]. Damaris/Viz can provide a wealth of visualization and analysis opportunities due to its ability to act as a front end to both VisIt's [CBW*12] libsim [WFM11] as well as ParaView's Catalyst [FMT*11, BGS13]. Biddiscombe et al. proposed an HDF5-based driver that forwards the data from HDF5 calls to ParaView [BSO*11]; we give an example of our system implementing similar functionality in § 3.2. Abbasi et al. introduce DataStager, a system for streaming data to staging nodes and demonstrate a performance benefit by asynchronously streaming multiple buffers at one time [AWE*09]. In situ libraries can also be used to improve the performance of simulation code [VHP11].
Most work focuses on extreme-scale performance with less regard for the effort required in integrating simulation and visualization software, whereas we focus on the latter concern. Notably, however, Abbasi et al. extend their previous work with a JIT compiler that allows users to customize data coming through ADIOS [LKS*08] using snippets of code written in a subset of C [AEW*11]. Zheng et al. modify OpenMP runtimes, an approach that shares our mentality of working within the constraints of existing infrastructure [ZYH*13]. Others have tightly integrated simulation with visualization to allow steering, but these generally come at high integration costs [LR12, AFS*11].
Existing solutions leave a potentially large segment of the user community behind. Most previous work has integrated or presupposed integration with particular libraries for performing I/O operations, and no such library has achieved universal adoption. Yu et al. note the tight collaboration required for a fruitful integration [YWG*10]. Reasons for not adopting I/O middleware are varied: the difficulty in integrating the library with local tools, perceived lack of benefit, lack of support for existing infrastructure with home-grown formats, or issues conforming to required interfaces, such as synchronous ‘open’ calls.
Moreover, the focus of modern I/O middleware specifically on simulations at the extreme scale leaves a long tail of potential in situ uses behind. The set of simulation authors focused on creating exascale-capable simulations is a small subset of all simulation authors. A large set does not even dream of petascale; and even larger are those who would barely know how to exploit a terascale-capable solver for their science. The distribution gets larger and more diverse as one moves out to lower scalability levels.
At the opposite end of ‘extreme scalability’ uses for in situ, one may find a number of heretofore ignored applications. There is no reason to limit the in situ idea to parallel code running on a supercomputer, for example. Analysis routines embedded into the fabric of network transfer operations would be a boon to distributed research groups (and the success of tools such as Globus [FBC*11] speaks to the multitudes of domains faced with this problem). Those writing simulations in MATLAB® might also benefit from precanned visualization tasks that occur concurrently with their simulation, yet the closed source nature of the product makes the prospect of integrating I/O middleware improbable at best.
The currently-dominant middleware approach to in situ requires significant effort. It is reasonable for simulation authors to spend a week integrating and retooling their code to achieve thousand-way concurrent in situ visualization, but this level of investment is unreasonable to users who simply wants to compute a data range on their files as they move across the country. The cliff between ‘nothing’ and a ‘100%’ solution for in situ visualization with existing middleware solutions is too high to appease such diverse use cases. Worse, the model is unworkable in some situations; it is doubtful that the OpenSSH maintainers would accept patches incorporating ParaView's Catalyst into sftp, for example.
Freeprocessing is an abstraction of previous work. Using it, one can implement classical in situ visualization and analysis, computation or data reduction via staging nodes, unique instrumentation such as gathering power consumption information dynamically [GRP*13], or a number of novel ‘processing while moving data’ ideas. This processing can be synchronous or asynchronous depending on the needs and desires of the user. Developers of a freeprocessor can connect it to existing visualization tools such as VisIt's libsim or ParaView's Catalyst, implement their own analysis routines, and even push data into another language such as Python, all without data copying—or with data copying, should those semantics be preferable. The general nature of Freeprocessing not only allows one to implement the diverse domains of previous work, but also allows novel use cases. Specifically, we contribute:
a new method for inserting data processing code into I/O operations;
the generalization of in situ ideas to heretofore unexplored domains, such as visualization during network transfer;
greatly increased programmability for in situ ideas, making them applicable with considerably less effort;
a sample implementation that demonstrates all of these ideas in real-world cases.
The rest of this paper is organized as follows. First, we explain the technical underpinnings of how the program works. In § 3 we demonstrate Freeprocessing in some classical environments and show that there is almost no overhead. We demonstrate some novel uses before we conclude and note limitations as well as future work in § 5.
2. Instrumentation
Previous in situ solutions have relied on the simulation author explicitly invoking the visualization tool, or the simulation using a custom library for I/O, which is then repurposed for analysis. In this work we demonstrate that there is little need for either; every simulation produces output already, an in situ tool just needs to tap into that output.
Our symbiont uses binary instrumentation to realize that tap. We take unmodified simulation binaries and imbue them with the ability to perform visualization and analysis tasks. In doing so, we remove a potentially complicated component of in situ: modifying the program to work with the visualization or analysis tool. Notably, this approach enables simulation software to produce in situ visualizations even when the source code of the simulation is unavailable. Furthermore, as the symbiont interposes these functions during load time, a user need only change the invocation of the program to enable or disable these features.
The method we use is to redefine some of the standard I/O functions, in a similar manner to the way the GLuRay or Chromium systems operate [BFH12, HHN*02]. These methods rely on features available in runtime dynamic linkers to replace any function implemented within a library at load time. The overridden entry points form what we call the ‘symbiont’, the core of Freeprocessing. The symbiont's purpose is to conditionally forward data to a freeprocessor—a loadable module that implements the desired in situ computation—in addition to fulfilling the function's original duties. Separating the instrumentation itself and the freeprocessor allows users to develop processing elements without knowledge of binary instrumentation.
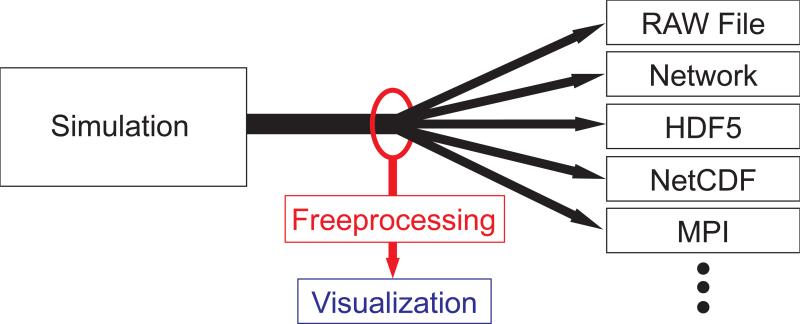
The set of intercepted functions is different depending on the I/O interface that the simulation uses, as shown in Figure 1. For the C language, these functions are those of the POSIX IO layer, such as open(2) and write(2). In Fortran these calls are implementation-specific, and C++ implements I/O differently, but on POSIX-compliant systems all such implementations are ultimately layered on top of the POSIX I/O interface. We also introduce interposition for higher-level functions, such as those that comprise MPI File I/O, and a subset of calls from the HDF5 family. Using this interposition, what the simulation believes is a standard ‘write’ operation actually calls in to our symbiont.
Figure 1.
Freeprocessing works like a vampire tap on the data coming out of a simulation. Without changes to a program's source code, we can intercept the data as it goes to the IO library and inject visualization and analysis tasks.
2.1. Data semantics
Function interposition for higher-level functions from libraries such as HDF5 and NetCDF provide an important benefit: data semantics. As these formats are self-describing, there is enough information in just the stream of function calls to identify data properties—in contrast to raw POSIX I/O functions, which provide little more than an abstract buffer. The symbiont forwards any available data semantics from the interposed library functions to the freeprocessor.
However, in contrast to previous work, Freeprocessing will also willingly forward data without knowledge of any underlying semantics. A freeprocessor can also ignore meta-data simply by not implementing the methods that interpret those messages. This distinction is important, as it both enables Freeprocessing to function in a larger set of scenarios, as well as increases the flexibility of the system. Presumably a freeprocessor would then obtain this information from some external source. We view allowing semantic-less data transfer similar to using ‘dangerous’ constructs in a programming language, such as casts in C. While these constructs are generally frowned upon, with restrained application they can be a powerful and thereby useful tool.
2.2. Data semantics
Meta-information concerning data semantics are required, and are only available through Freeprocessing in limited cases. While we consider such concerns beyond the scope of this work, they need to be provided for the demonstration of the technique. The general nature of Freeprocessing allows any number of solutions: the problem is no different than understanding arbitrary binary data read from a file. One of the solutions we have found works well is a simple text file in the style of Damaris/Viz or ADIOS [DSP*13, LKS*08]. An example of one such configuration is given in Listing 1. However, it is important to note that this configuration is external to Freeprocessing itself. The symbiont does not contain this parsing and metadata acquisition code; the ‘user code’— freeprocessors—implements this only if they desire.
Listing 1: JSON configuration file used for a Silo conversion freeprocessor. Variants that do not require the repeated “i”s are possible, but lack the desirable property of strict adherence to the JSON specification.

Freeprocessing itself does not endorse any specific method for obtaining data semantics, in the same way that the C file I/O routines do not endorse a specific encoding for metadata on binary streams.
2.3. Defining free processors
The module interface for a freeprocessor is simple. The system exposes a stream processing model. Data are input to the processor, utilized (or ignored), and thereafter unavailable. This interface is in principle the same model as GLSL, OpenCL, and CUDA expose, though we do not currently impose the same restrictions. A freeprocessor is free to implement a cache and process data in a more traditional manner, for example.
Listing 2 shows the freeprocessor interface. The symbiont calls Init when a file is first accessed; some of our freeprocessors initialize internal resources here. The filename parameter allows the processor to provide different behavior should the simulation output multiple file formats. The buffer and n parameters are the data and its size in bytes. If the required information is available, the symbiont will call Metadata immediately before a write, communicating the characteristics for the impending data. Likewise, finish cleans up any per-file resources. Finally, the create function implements a ‘virtual constructor’ to create the processor. All functions sans create are optional; if a freeprocessor has no need for metadata, for example, it simply does not implement the corresponding function.
Listing 2: Base class for a freeprocessor.

2.3.1. Configuration
The symbiont reads a configuration file that describes which freeprocessor to execute. Any library that satisfies the interface given in Table 2 is a valid freeprocessor. It is important to note that the operations share the semantics of the simulation code. For example, if a parallel simulation performs only collective writes for a given file, then it is appropriate to perform collective operations in the freeprocessor's Stream call.
It is common for a simulation to produce a large set of output files. Furthermore, MPI runtimes frequently open a number of files to configure their environment, and all these files are ‘seen’ by the symbiont. It is therefore necessary to provide a number of filtering options. Some of these are built in, such as ignoring files that are opened for read-only access. Others the user specifies in the configuration file for the symbiont. The specification uses a match expression for the filenames, so the user can further limit where instrumentation will occur. These match expressions provide a more convenient mechanism to uniquely connect processing elements to streams, but the assignment could also be done by the freeprocessor implementation.
2.3.2. Python
Developers may also implement freeprocessors in Python. We provide a simple freeprocessor that embeds the Python interpreter and exports data and needed metadata. Most notably, it creates the ‘stream’ variable: a NumPy array for the data currently being written. Exposing the array to Python does not require a copy; the simulation data shares the memory with the Python runtime. Should the Python script attempt any write operation on the data, a copy is transparently made inside the Python runtime, which is then managed via Python's garbage collector. We allow only one of the simulation or the Python tool to run at any given time.
The Python script is otherwise indistinguishable from standard Python code; the symbiont imposes no restrictions beyond the unique source of data. Communication via, e.g., MPI4Py is even possible, provided the simulation utilizes synchronous writes. In § 3.2 we demonstrate this method by connecting Freeprocessing with the yt visualization tool [TSO*11].
3. Classical in situ
Freeprocessing can implement a number of in situ ideas, including the traditional use case of in situ: visualization and analysis during a simulation run. In this section, we detail how the corresponding freeprocessors for a few simulation codes operate, and demonstrate that the overhead of the method is negligible.
3.1. PsiPhi
PsiPhi is a Fortan95/2003-based CFD-solver that focuses on Large Eddy Simulation (LES) of flows that include combustion and other types of chemical reactions. The simulation discretizes the governing equations of mass, momentum, and species concentration on a cartesian grid via the finite volume method. Second-order schemes discretize the domain, and an explicit third-order low storage Runge-Kutta scheme advances the solution. The immersed boundary (IB) technique handles diverse geometries in a computationally efficient manner. Besides the solution of the mentioned transport equations in an Eulerian formulation, the code is able to solve the equations of motion for Lagrangian particles. A combination of Lagrangian particles and immersed boundaries describes moving objects. The code is modular, easy to extend and maintain, and highly portable to different machines. PsiPhi parallelizes via the distributed-memory paradigm, using MPI.
PsiPhi simulates highly-resolved simulations of reactive flows, e.g., premixed, non-premixed and stratified combustion, coal and biomass combustion, liquid spray combustion, and nanoparticle synthesis [PCGK11, MSCK13, MMK13]. The software has scaled to thousands of cores on Top500 machines such as SuperMUC and JUQUEEN. Recent tests with the program have shown that the output of the computational results becomes a performance bottleneck when moving up to an even higher number of cores.
There are three types of intermediate outputs in the PsiPhi simulation. The first are actually custom-developed in situ visualizations: slice outputs and volume renderings. The simulation writes out these visualizations in custom ASCII-based formats every n time steps, with typical values of n in between 100 and 1000 [PK13]; Figure 2 shows example visualizations. The second type of output is a simulation-specific binary format used for restart files, which is organized in a ‘one file per process’ manner. Synchronous Fortran ‘unformatted’ WRITE operations create these outputs. The third kind of output is an ASCII-based metadata file that describes the layout of the binary restart files.
Figure 2.
Sample in situ visualizations of the Cambridge stratified flame produced by the PsiPhi code.
The PsiPhi authors are interested in extracting arbitrary 2D slices as well as 3D visualizations with more flexibility than their custom-developed routines allow. Therefore, we developed a custom freeprocessor for the PsiPhi simulation. PsiPhi periodically dumps its state to disk in the form of restart files, at approximately the same cadence as ‘normal’ output files. We utilized the aforementioned restart files as the basis for our freeprocessor, in addition to parsing the ASCII-based metadata to interpret these restart files.
The simulation authors were enthusiastic about the freeprocessor. All the outputs the simulation previously created were redundant with the restart files. Furthermore, PsiPhi users hardcoded postprocessing parameters such as slice numbers into the simulation source, necessitating a recompile to modify the parameters. In light of the visualization options presented by the freeprocessor, the PsiPhi authors elected to remove all custom-developed in situ outputs and create only the restart files.
We therefore reimplemented their outputs in a freeprocessor and measured the performance of the system under both the old and new configurations. As shown in Figure 3, not only was the overhead miniscule, but the simulation actually ran faster with the freeprocessor. The performance difference arose from the difference in how PsiPhi and the freeprocessor organize their writes. In the freeprocessor, we calculate the appropriate file offsets on each rank and output to a shared file directly; the original PsiPhi approach was to gather the data on the root processor and then do all writing from there.
Figure 3.
Scalability of the PsiPhi simulation. ‘existing’ and ‘Freeprocessing’ produce the same outputs via different mechanisms, while ‘raw’ produces only restart files. Freeprocessing's overhead is negligible; new output methodologies can even increase performance.
3.2. Enzo
Enzo is a simulation code designed for rich, multi-physics hydrodynamic astrophysical calculations [TBN*13]. It is of special interest in the visualization community due to its use of adaptively-refined (i.e., AMR) grids. Enzo runs in parallel via MPI and CUDA on some of the world's Top 500 supercomputers, with OpenMP hybrid parallelism under investigation. For I/O, Enzo relies on the HDF5 library.
As Enzo is HDF5-based and HDF5 provides all the data semantics required, the selection of which fields are of interest is the only required work. For HDF5 outputs, the symbiont configuration file specifies the ‘Datasets’ (in the HDF5 sense) of interest as opposed to a filename; the symbiont assumes that all HDF5 files opened for write access are a simulation output.
When Enzo was first investigated, HDF5 support was not available in our symbiont. Generic HDF5 support in the symbiont required only a day of effort. Configuring it to work with Enzo takes seconds. Users must edit a text file to indicate which field[s] they wish to see. To work with Enzo's yt tool, we utilize the aforementioned freeprocessor that exposes data into Python and runs a script (§ 2.3.2); the script we utilized is a standard yt script, except that it pulls its data from the special ‘freeprocessing’ import, instead of a file. Figure 4 demonstrates this. The 100-line freeprocessor is applicable for any in situ application; the 20-line Python script is specific to yt.
Figure 4.
‘Density’ field generated in situ by the Python visualization tool ‘yt’ applied to an Enzo hydrodynamics simulation. A freeprocessor exposed the data into Python and a standard yt script created the visualization.
3.3. N-Body simulation coursework
We taught a course in High-Performance Computing during the preparation of this manuscript. Among the work given in the course was an MPI+OpenMP hybrid-parallel N-Body simulation. We provided our symbiont to the students along with a simple ParaView script, which would produce a visualization given one of their timestep outputs. A sample visualization is shown in Figure 5.
Figure 5.
Sample frame from an animation produced from a student's simulation using our tool. The ease of use allowed the student to quickly get the tool running, allowing fast and simple visual debugging.
The flexibility of the system was a boon in this environment. Visualizing the data in-memory would be difficult. The data were distributed, and the writes were in ASCII; parsing the data from the given stream was daunting for undergraduates. Therefore they elected to delay launching ParaView until after a timestep completed. The system must write and then read particle information from disk, but visualization was still concurrent with simulation and faster than serializing the two tasks. Most importantly, the simplicity allowed application of the technique in tens of minutes.
4. Alternative use cases
The ability to hook into any data movement operation of a process enables Freeprocessing to create novel applications of in situ ideas. In this section, we highlight a couple uses which makes Freeprocessing unique among in situ tools.
4.1. Transfer-based visualization
A heretofore lost opportunity has been in applying visualization methods to data during transport from site to site. This use case shares the primary motivation behind prior in situ visualization work: that we should do operations on data while they are already in memory, instead of writing the data to disk and then reading them back. While most if not all HPC experts agree that—at the largest scale—moving data will no longer be viable for large data, a large userbase still exists for which simulation on a powerful remote supercomputer and analysis on local resources is the norm.
To downplay this drawback, we propose preprocessing during this transit time. As an example of Freeprocessing for this novel case, we use it to instrument the transfer of a dataset using the popular secure copy (scp) tool. The system works by intercepting data as it goes out to or comes in from a socket. The source of the secure shell program itself needs no modification; the system could work with any network service, such as an FTP client or a web browser.
One use case is the computation of an isosurface; Figure 6 shows an example. A freeprocessor computed this iso-surface of a Richtmyer-Meshkov instability during network transfer. This example demonstrates one of the issues with our system: we needed to modify a marching cubes implementation to work in a slice-by-slice manner, as opposed to assuming all data were in-core. Additionally, our marching cubes implementation required at least two slices to operate, which necessitated a cache in the freeprocessor to make up for the small writes utilized by scp. This buffering and our unoptimized marching cubes implementation slows down a gigabit-link transfer by 4x. Although this still proved faster than transferring the dataset and computing the isosurface in series, it highlights the pain associated with the need to rewrite code in a stream processing fashion. On the other hand, with the rise of data parallel architectures and the decreasing memory per core ratio, one might argue that a transition to a stream processing model is inevitable.
Figure 6.
Richtmyer-Meshkov instability isosurface computed by a freeprocessor. Whereas the freeprocessor could be applied to any process that moves data, this particular isosurface was computed during network transfer via scp.
4.2. MATLAB
Users often request methods to read outputs of binary-only commercial software in tools like VisIt.†. We implemented a freeprocessor that accepts raw data, reads a metadata description from a configuration file for semantics, and exports these data into a Silo file that VisIt can easily import. Applying this freeprocessor incurs an additional overhead of 3–10% on a simple Julia set calculation in MATLAB, due to the additional data that it writes.
The alternative of an ‘export to Silo’ MATLAB extension has notable drawbacks. First, one must compile using the ‘mex’ compiler frontend, and every major MATLAB update will require a recompilation or even rewrite. Second, divorcing the code from MATLAB and its interface may require significant effort. In contrast, our freeprocessor is indenden-dent of the MATLAB version it instruments, with neither source changes nor a recompilation required. Furthermore, the same freeprocessor is applicable in other manners, such as creating Silo files during a network transfer.
5. Conclusions
In this paper we have introduced Freeprocessing: an in situ visualization and analysis tool based on binary instrumentation. The method imbues an existing simulation with in situ powers, with little or—in some cases—no effort on the part of the simulation author. The method's generality enables novel applications, such as visualization during network transfer or instrumenting software for which source is unavailable.
The system is, however, not without its drawbacks. The symbiont is stable, but customizing the system via new freeprocessors can require per-simulation effort. Furthermore, the unidirectional communication model precludes simulation steering applications. The ability of Freeprocessing to insert small, ad hoc bits of code in myriad new places uncovers perhaps its greatest limitation: increased programmability requires increased programming.
The work presented here lowers the barrier of entry for a simulation to indulge in in situ processing. Previous work on in situ has largely focused on achieving highly scalable results, with less regard to the amount of integration effort required. The most significant contribution of this work may be that fruitful capabilities can arise from a modicum of effort.
Acknowledgements
Some computations described in this work were performed using the Enzo code, which is the product of a collaborative effort of scientists at many universities and national laboratories. We especially thank Matthew Turk and Sam Skillman for their help interfacing with yt. We thank Burlen Loring for help with ParaView scripting, and Hank Childs for discussions of related work.
This research was made possible in part by the Intel Visual Computing Institute; the NIH/NCRR Center for Integrative Biomedical Computing, P41-RR12553-10; and by Award Number R01EB007688 from the National Institute of Biomedical Imaging and Bioengineering. The content is the sole responsibility of the authors.
Footnotes
c.f. “Using MATLAB to write Silo files to bring data into VisIt”, visit-users mailing list, February 2014.
References
- Abbasi H, Eisenhauer G, Wolf M, Schwan K, Klasky S. Just in time: Adding value to the IO pipelines of high performance applications with JITStaging.. Proceedings of the 20th International Symposium on High Performance Distributed Computing; New York, NY, USA. 2011; HPDC '11, ACM; pp. 27–36. [AEW*11] doi:10.1145/1996130.1996137. [Google Scholar]
- Ament M, Frey S, Sadlo F, Ertl T, Weiskopf D. GPU-based two-dimensional flow simulation steering using coherent structures.. In: Iványi P, Topping BHV, editors. Proceedings of the Second International Conference on Parallel, Distributed, Grid and Cloud Computing for Engineering; Stirlingshire, United Kingdom. 2011; Civil-Comp Press; [AFS*11] doi: http://dx.doi.org/10.4203/ccp.95.18. [Google Scholar]
- Abbasi H, Wolf M, Eisenhauer G, Klasky S, Schwan K, Zheng F. DataStager: Scalable data staging services for petascale applications.. Proceedings of the ; 18th ACM International Symposium on High Performance Distributed Computing; New York, NY, USA. 2009; HPDC '09, ACM; pp. 39–48. [AWE*09] doi:10.1145/1551609.1551618. [Google Scholar]
- Brownlee C, Fogal T, Hansen CD. Eurographics Symposium on Parallel Graphics and Visualization. The Eurographics Association; 2012. GLuRay: Enhanced ray tracing in existing scientific visualization applications using OpenGL interception. pp. 41–50. [BFH12] URL: http://dx.doi.org/10.2312/EGPGV/EGPGV12/041-050. [Google Scholar]
- Bauer AC, Geveci B, Schroeder W. The ParaView Catalyst User's Guide. Kitware; 2013. [BGS13] [Google Scholar]
- Biddiscombe J, Soumagne J, Oger G, Guibert D, Piccinali J-G. Parallel Computational Steering and Analysis for HPC Applications using a ParaView Interface and the HDF5 DSM Virtual File Driver.. In: Kuhlen T, Pajarola R, Zhou K, editors. Eurographics Symposium on Parallel Graphics and Visualization; Llandudno, Wales. 2011; Eurographics Association; pp. 91–100. [BSO*11] [Google Scholar]
- Childs H, Brugger E, Whitlock B, Meredith JS, Ahern S, Bonnell K, Miller M, Weber G, Harrison C, Pugmire D, Fogal T, Garth C, Sander Son A, Bethel EW, Durant M, Camp D, Favre JM, Ruebel O, Navratil P, Wheeler M, Selby P, Vivodtzev F. VisIt: An End-User Tool for Visualizing AND Analyzing Very Large Data. CRC Press; Oct, 2012. pp. 357–372. [CBW*12] [Google Scholar]
- Childs H, Geveci B, Schroeder W, Meredith J, Moreland K, Sewell C, Kuhlen T, Bethel E. Research challenges for visualization software. Computer. 2013 May;46(5):34–42. [CGS*13] doi:10.1109/MC.2013.179. [Google Scholar]
- Docan C, Parashar M, Klasky S. Enabling high-speed asynchronous data extraction and transfer using DART. Concurr. Comput. : Pract. Exper. 2010 Jun;22(9):1181–1204. [DPK10] doi:10.1002/cpe.v22:9. [Google Scholar]
- Dorier M, Sisneros RR, Peterka T, Antoniu G, Semeraro DB. Damaris/Viz: a nonintrusive, adapt able and user-friendly in situ visualization framework. Large Data Analysis and Visualization. 2013 Oct; [DSP*13] [Google Scholar]
- Foster IT, Boverhof J, Chervenak AL, Childers L, Deschoen A, Garzoglio G, Gunter D, Holzman B, Kandaswamy G, Kettimuthu R, Kordas J, Livny M, Martin S, Mhashilkar P, Miller Z, Samak T, Su M-H, Tuecke S, Venkataswamy V, Ward C, Weiss C. Reliable high-performance data transfer via Globus Online. [FBC*11] [Google Scholar]
- Fabian N, Moreland K, Thompson D, Bauer AC, Marion P, Geveci B, Rasquin M, Jansen KE. Large Data Analysis and Visualization (LDAV), 2011 IEEE Symposium on. IEEE; 2011. The ParaView Coprocessing library: A scalable, general purpose In Situ visualization library. pp. 89–96. [FMT*11] [Google Scholar]
- Gamell M, Rodero I, Parashar M, Bennett JC, Kolla H, Chen J, Bremer P-T, Landge AG, Gyulassy A, Mccormick P, Pakin S, Pascucci V, Klasky S. Exploring power behaviors and trade-offs of in-situ data analytics.. Proceedings of SC13: International Conference for High Performance Computing, Networking, Storage and Analysis; New York, NY, USA. 2013; SC '13, ACM; pp. 77:1–77.pp. 12 [GRP*13] doi:10.1145/2503210.2503303. [Google Scholar]
- Humphreys G, Houston M, Ng R, Frank R, Ahern S, Kirchner PD, Klosowski JT. Chromium: A stream-processing framework for interactive rendering on clusters. ACM Trans. Graph. 2002 Jul;21(3):693–702. [HHN*02] [Google Scholar]
- Lofstead JF, Klasky S, Schwan K, Podhorszki N, Jin C. Flexible IO and integration for scientific codes through the adaptable IO system (ADIOS).. Proceedings of the 6th International Workshop on Challenges of Large Applications in Distributed Environments; New York, NY, USA. 2008; CLADE '08, ACM; pp. 15–24. [LKS*08] [Google Scholar]
- Lesage J-D, Raffin B. A hierarchical component model for large parallel interactive applications. J. Supercomput. 2012 Jun;60(3):389–409. [LR12] [Google Scholar]
- Marincola FC, Ma T, Kempf AM. Large eddy simulations of the Darmstadt turbulent stratified flame series. Proceedings of the Combustion Institute. 2013;34(1):1307–1315. [MMK13] [Google Scholar]
- Moreland K, Oldfield R, Marion P, Jourdain S, Podhorszki N, Vishwanath V, Fabian N, Docan C, Parashar M, Hereld M, Papka ME, Klasky S. Examples of In Transit visualization.. Proceedings of the 2nd International Workshop on Petascale Data Analytics: Challenges and Opportunities; New York, NY, USA. 2011; PDAC '11, ACM; pp. 1–6. [MOM*11] doi:10.1145/2110205.2110207. [Google Scholar]
- Ma T, Stein O, Chakraborty N, Kempf AM. A-posteriori testing of algebraic flame surface density models for LES. Combustion Theory and Modelling. 2013 [MSCK13] [Google Scholar]
- Pettit M, Coriton B, Gomez A, Kempf AM. Large-eddy simulation and experiments on non-premixed highly turbulent Opposed Jet flows. Proc. Combust.Inst. 2011;33:1391–1399. [PCGK11] [Google Scholar]
- Proch F, Kempf AM. Numerical analysis of the Cambridge stratified flame series using artificial thickened flame LES with tabulated premixed chemistry. 2013 [PK13] submitted to Combustion and Flame. [Google Scholar]
- The Enzo Collaboration. Bryan GL, Norman ML, O'shea BW, Abel T, Wise JH, Turk MJ, Reynolds DR, Collins DC, Wang P, Skillman SW, Smith B, Harkness RP, Bordner J, Kim J-H, Kuhlen M, Xu H, Goldbaum N, Hummels C, Kritsuk AG, Tasker E, Skory S, Simpson CM, Hahn O, Oishi JS, So GC, Zhao F, Cen R, Li Y. Enzo: An Adaptive Mesh Refinement Code for Astrophysics. Astrophysical Journal Supplement Series. 2013 Jul; [TBN*13] [Google Scholar]
- Turk MJ, Smith BD, Oishi JS, Skory S, Skillman SW, Abel T, Norman ML. yt: A Multi-code Analysis Toolkit for Astrophysical Simulation Data. The Astrophysical Journal Supplement. 2011 Jan;192:9. [TSO*11] [Google Scholar]
- Vishwanath V, Hereld M, Papka M. Toward simulation-time data analysis and I/O acceleration on leadership-class systems. Large Data Analysis and Visualization (LDAV) 2011 Oct;:9–14. [VHP11] [Google Scholar]
- Whitlock B, Favre JM, Meredith JS. Proceedings of the 11th Eurographics conference on Parallel Graphics and Visualization. Eurographics Association; 2011. Parallel in situ coupling of simulation with a fully featured visualization system. pp. 101–109. [WFM11] [Google Scholar]
- Yu H, Wang C, Grout RW, Chen JH, Ma K-L. In situ visualization for large-scale combustion simulations. IEEE Comput. Graph. Appl. 2010 May;30(3):45–57. doi: 10.1109/MCG.2010.55. [YWG*10] [DOI] [PubMed] [Google Scholar]
- Zheng F, Yu H, Hantas C, Wolf M, Eisenhauer G, Schwan K, Abbasi H, Klasky S. Proceedings of SC13: International Conference for High Performance Computing, Networking, Storage and Analysis. ACM; 2013. GoldRush: Resource efficient in situ scientific data analytics using fine-grained interference aware execution. p. 78. [ZYH*13] URL: http://doi.acm.org/10.1145/2503210.2503279. [Google Scholar]