Abstract
BACKGROUND
A novel murine mitochondria-associated neutral sphingomyelinase (MA-nSMase) has been recently cloned and partially characterized. The subcellular localization of the enzyme was found to be predominantly in mitochondria. In this work, the determinants of mitochondrial localization and its topology were investigated.
METHODS
MA-nSMase mutants lacking consecutive regions and fusion proteins of GFP with truncated MA-nSMase regions were constructed and expressed in MCF-7 cells. Its localization was analyzed using confocal microscopy and sub-cellular fractionation methods. The sub-mitochondrial localization of MA-nSMase was determined using protease protection assay on isolated mitochondria.
RESULTS
The results initially showed that a putative mitochondrial localization signal (MLS), homologous to an MLS in the zebra-fish mitochondrial SMase is not necessary for the mitochondrial localization of the murine MA-nSMase. Evidence is provided to the presence of two regions in MA-nSMase that are sufficient for mitochondrial localization: a signal sequence (amino acids 24–6) that is responsible for the mitochondrial localization and an additional 'signal-anchor' sequence (amino acids 77–99) that anchors the protein to the mitochondrial membrane. This protein is topologically located in the outer mitochondrial membrane where both the C and N-termini remain exposed to the cytosol.
CONCLUSIONS
MA-nSMase is a membrane anchored protein with a MLS and a signal-anchor sequence at its N-terminal to localize it to the outer mitochondrial membrane.
GENERAL SIGNIFICANCE
Mitochondrial sphingolipids have been reported to play a critical role in cellular viability. This study opens a new window to investigate their cellular functions, and to define novel therapeutic targets.
1. Introduction
The mitochondrion is emerging as a novel compartment of ceramide metabolism and function. Mitochondria have been shown to contain many sphingolipids including sphingomyelin (SM) and ceramide [1, 2]. Many ceramide generating enzymes have been suggested to reside in this organelle, including ceramide synthases (CerS1, CerS2, CerS4 and CerS6) [3–5], zebrafish and mouse neutral sphingomyelinases [6, 7], and neutral ceramidases [8].
Besides the occurrence of these enzymes, various studies have also suggested the biological significance of ceramide generation in this compartment. Birbes et al. [9] showed that the selective targeting of bacterial sphingomyelinase to mitochondria and not to other compartments resulted in apoptosis, and over-expression of Bcl-2 prevented these effects [9]. Dai et al. showed that UV-induced apoptosis is marked by an increase in SM in all sub-cellular locations particularly in mitochondria in HeLa cells, and ceramide level was found to be elevated in mitochondria at 2–6hrs, consistent with cell death time course. D609, an inhibitor of sphingomyelin synthase, rescued the cells from the spike in SM and ceramide and consequently cell death [10] suggesting the involvement of the SM hydrolysis in the cell death triggered by UV irradiation.
In another study in Caenorhabditis elegans, subsequent to inactivation of ceramide synthase, somatic apoptosis was unaffected but ionizing radiation-induced apoptosis of germ cells was obliterated and this phenotype was reversed by microinjection of long-chain natural ceramide. Radiation induced ceramide accumulation in mitochondria and consequent activation of CED-3 caspase and apoptosis [11].
In the studies on isolated mitochondria, exogenous synthetic N-acetylsphingosine (C2-ceramide) elicited inhibition of state 4 respiration (respiration upon exhaustion of ADP) and inhibition of electron transport complex I [12]. In another study, respiratory chain complex III function was reduced by C2-ceramide whereas N-acetylsphinganine (C2-dihydroceramide), which lacks the functional double bond, did not alter mitochondrial respiration or complex III activity [13].
Sphingomyelinases (SMases) catalyze the hydrolysis of SM to ceramide. There are three major classes of SMases depending on their optimum pH of activity, acid, neutral, and alkaline SMases. Acid SMase (SMPD1) exists in both the lysosomal and secretory forms; both of them operate at a pH optima of ~5; deficiency of this enzyme causes Niemann-Pick disease. Alkaline SMase is found in the mucosa of the gastrointestinal tract and bile in humans; it has a pH optima of 9; it plays a role in dietary SM digestion but also displays broad substrate specificity [14].
Neutral sphingomyelinases operate at pH optima of ~7. Mouse and human nSMase 1 (SMPD2), first cloned in 1998, [15] were found to be ubiquitously expressed in various tissues and co-localized with ER and Golgi [16]. Human nSMase 2 (SMPD 3) was found to be localized in the Golgi and plasma membrane [17]. Human nSMase 3 (SMPD4) biochemically ‘behaved’ like neutral sphingomyelinase; it was found to be a C-tail anchored transmembrane protein [18], and enriched in the ER [19]. The family also includes Isc1p from S. cerevisiae [20] and its homolog css1 in S. pombe [21].
Among the sphingomyelinases identified so far, the Zebra-fish mitochondrial SMase was the first SMase found in the mitochondria to be cloned and characterized. Over-expression of this protein in HEK293 cell lines localized it to mitochondria whereas mutants lacking the first N-terminal 35 residues did not localize to mitochondria. Topologically, it was found to be in the mitochondrial inter membrane space and/or inner membrane of zebrafish embryonic cells [6]. Murine mitochondria associated sphingomyelinase (MA-nSMase) was the second mitochondrial member of this family [7]. In our previous work, it was found that MA-nSMase retained a significant number of conserved amino acids that are necessary for cation binding; a P-loop like domain, and two critical residues, D470 and H471 required for its catalytic activity. The sequence homology with zebrafish reveals a putative mitochondrial localization signal (MLS) extending from 24–56aa, and a putative transmembrane domain (TMD) extending from 77–99aa. Biochemical characterization of MA-nSMase using lysates from transiently transfected HEK293 cells disclosed that the murine MA-nSMase belongs to the neutral sphingomyelinase group [7].
The majority of mitochondrial proteins are synthesized in the cytosol and imported into mitochondria. These precursor proteins are thought to be stabilized by chaperones, especially Hsp70 and Hsp90 [22, 23]. Normally, these precursors contain a presequence, a signal at the most N-terminal region of the protein (or matrix-targeting sequences or MTS) to target the protein to mitochondria. In most cases, the presequence is cleaved after reaching the matrix by mitochondrial-processing peptidase (MPP) [24, 25]. However, there are many mitochondrial proteins that do not have an N-terminal signal but rather possess an internal targeting signal. Precursors of mitochondrial outer membrane proteins have such a signal. Outer membrane proteins might be N-terminally anchored as in Tom 20 and Tom 70 or C-terminally anchored as in Tom 5 and Bcl2 [26]. A third type spans the outer membrane twice with a small loop in the intermembrane space as in Fzo 1 [27]. Porins and Tom 40 traverse the outer-membrane many times forming β barrel structure [28]. N-terminal anchored proteins are called ‘signal anchored’ proteins because the transmembrane domain TMD and the residues flanking it serve for both its intracellular sorting and anchoring functions [29]. Signal anchored proteins do not share sequence similarity but they may be characterized by certain features such as moderate hydrophobicity of its transmembrane domain and positively charged C terminal residues flanking the TMD as in Tom 20 [30]. Also, at least three basic amino acids should be flanking the TMD for membrane anchorage as in Tom 70 [31]. The salient features of C-terminally anchored proteins are relatively short TMD with moderate hydrophobicity and positive charged residues at its flanking regions. The relative contribution of each of these structural features seems to vary from protein to protein. For instance, a net positive charge flanking the TMD is extremely important for Bcl-2 and Bcl-XL. The TMD of both proteins has similar hydrophobicity and length. However, two positively charged amino acids at either side of TMD in Bcl-xL make it go exclusively to mitochondria [32] whereas in Bcl2- one positive charge on either side of the TMD makes it indiscriminately targeted to mitochondria, ER and nuclear envelopes. The targeting signals of many of these outer membrane proteins have previously been reviewed [33].
Since MA-nSMase appeared to localize to the mitochondrion which is a critical organelle involved in the cell survival and growth, this study was undertaken to define the critical domain in the protein that targets it to the mitochondria and define its topology.
2. Materials and Methods
2.1 Site directed mutagenesis and generation of fusion constructs
For making all deletion mutant constructs except Δ24–99, Quickchange site-directed mutagenesis kit (Stratagene, Catalogue # 200518) was employed as per the manufacturer’s instruction. For generating Δ24–99 construct and bacterial SMase fusion constructs, the overlap extension method was followed as described previously [34]. This was a two-step cloning technique. It was based on performing two independent PCR reactions with partially matching overhangs. In the subsequent fusion reaction, the PCR products of the previous two reactions were used as templates in which the overlapping ends anneal allowing the 3’ end of each strand acting as primer for 3' extension of the complementary strand. The most upstream forward primer and the most downstream reverse primer (primers are listed in supplemental section Table 2) serve as primers to amplify the full length fused product. Taq polymerase was used to perform the third PCR to leave an ‘A’ overhang which was gel purified and subcloned into pEF-6/V5-His TOPO TA expression kit from Invitrogen (Catalogue #K9610-20). For making the GFP constructs, forward and reverse primers (primers are listed in supplemental section Table 3) with restriction sites (Kpn I/Hind III respectively) were used to generate the desired products and subcloned into pEGFP-N1 plasmid from Clontech (Catalogue #6085-1).
For bacterial SMase (bSMase) fusion constructs, the catalytic domain of bSMase was used. Bioinformatics prediction using the Signal P 4.1 software suggested that there is a signal peptide within the first 1–27 residues of the protein followed by a putative cleavage site between residues 27 and 28. Thus, to make the fusion constructs and to prevent protein mistargeting, we deleted the first 27 amino acids of the bacterial SMase protein.
For generation of fusion proteins with tags at different locations, the overlap extension method as described previously was used with primers carrying sequences for the V5 tags (primers are listed in supplemental section Table 4) and restriction enzyme sites corresponding to Bam H1/Pme I. The resultant PCR products were subjected to restriction enzyme digestion, gel purified, and then subcloned into pEF-6/V5-His vector. For generating the truncation fusion constructs 1–27, 1–66 and 1–198, PCR was done to amplify the desired length product and the resultant construct was subcloned into pEF-6/V5-His TOPO TA expression kit as mentioned previously. For making of 1–45 and 1–56, PCR was done using 1–66 fusion construct as a template with primers having restriction site for BamH I/ Not I (primers are listed in supplemental section Table 1). The resultant PCR products were subjected to restriction enzyme digestion, gel purification, and subcloning into pEF-6/V5-His vector. The authenticity of these constructs was verified by DNA sequencing using multiple primers designed to cover the whole gene sequence in these constructs.
2.2 Cell culture and plasmid DNA transfection
MCF-7 (ATCC HTB-22) cells were cultured in RPMI 1640 media (Invitrogen) supplemented with 10% of fetal bovine serum. DNA transfection was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
2.3 Crude fractionation
MCF-7 cells were incubated in isotonic buffer containing sucrose, disrupted by passage through 25 gauge needle and centrifuged at 1,000 × g for 10 min and 10,000 × g for 10 min for collection of the nuclear fraction and the mitochondria-enriched heavy membrane fraction respectively. The supernatant obtained after the 10,000 × g was used for light membrane and cytosolic fractions. Fractions were analyzed by western blot.
2.4 Western blot analysis
The cells were collected in lysis buffer (25 mM Tris; pH 7.4, 1 mM EDTA, 1×protease inhibitor cocktail (Roche)); homogenized by sonication and protein concentration was determined using BCA assay (Bio-Rad Laboratories). Ten µg of total proteins from each lysate were loaded onto a 4–20% gradient SDS polyacrylamide gel, subjected to electrophoresis, and then transferred to nitrocellulose membranes. The blots were probed using 1:1000 dilution of primary antibody followed by horseradish peroxidase labeled secondary antibody (1:5000 dilution) (Santa Cruz Biotechnology, Inc.). The signals were detected using ECL chemiluminescence reagents (Pierce). V5- antibodies are from Invitrogen and GAPDH antibodies are from Santa-Cruz Biotechnology.
2.5 Immunostaining and confocal microscopy
MCF-7 cells were grown on polylysine coated 35-mm glass-bottom culture dishes (MatTek). Transient transfection of plasmids (1µg plasmid DNA/ dish) was performed using lipofectamine reagent (Invitrogen) according to the manufacturer's recommendations. At 24h after transfection, cells were washed with PBS, fixed with 3.7% formaldehyde for 10 min, permeabilized with methanol for 5 min, and incubated with 2% human serum in PBS for 1h at room temperature. Anti-V5 (Invitrogen) primary antibody was co-incubated with anti-Tom 20 (Santa Cruz) or anti-calreticulin (Sigma) at 4°C overnight or 2h at room temperature, respectively. Following the primary antibody treatment, cells were washed with PBS and probed with fluorescent secondary antibody (Cell probes) at 1:500 dilution at room temperature for 1h and washed three times with PBS. Cells were stored at 4°C under PBS prior to be viewed on a Leica LAFF confocal microscope. Images were quantified with Pearson correlation coefficient software that accompanied the microscope.
2.6 Protease protection assay
Cells were incubated in isotonic buffer containing 20 mM HEPES, 250 mM Sucrose and 150 mM NaCl, disrupted by passage through 25 gauge needle and centrifuged at 1,000 × g for 10 min, 10,000 × g for 10 min for collection of the nuclear fraction and mitochondria-enriched heavy membrane fraction, respectively. The 10,000 ×g pellet was suspended in the isotonic buffer and the protein concentration was measured using BCA assay (Bio-Rad Laboratories). The crude mitochondrial preparation was used as an input for the assay. Proteinase K was added from freshly prepared stocks in water. Unless otherwise indicated, the final proteinase K concentration was 4 µg/ml and the detergent Triton X-100 was added at a final concentration of 0.2% (w/v). Digestion reactions were performed for 20 min on ice and quenched by addition of PMSF to a final concentration of 2 mM. Finally, the samples were analyzed by western blotting.
2.7 Statistical analysis
Results are expressed as the mean ±SD. Differences between groups were identified using one-way analysis of variance (ANOVA) followed by Fisher’s post-hoc tests with Bonferroni correction. A P value of 0.05 or less was considered as statistically significant.
3. Results
3.1 Prediction of a putative MLS, a TMD and catalytic domain in MA-nSMase
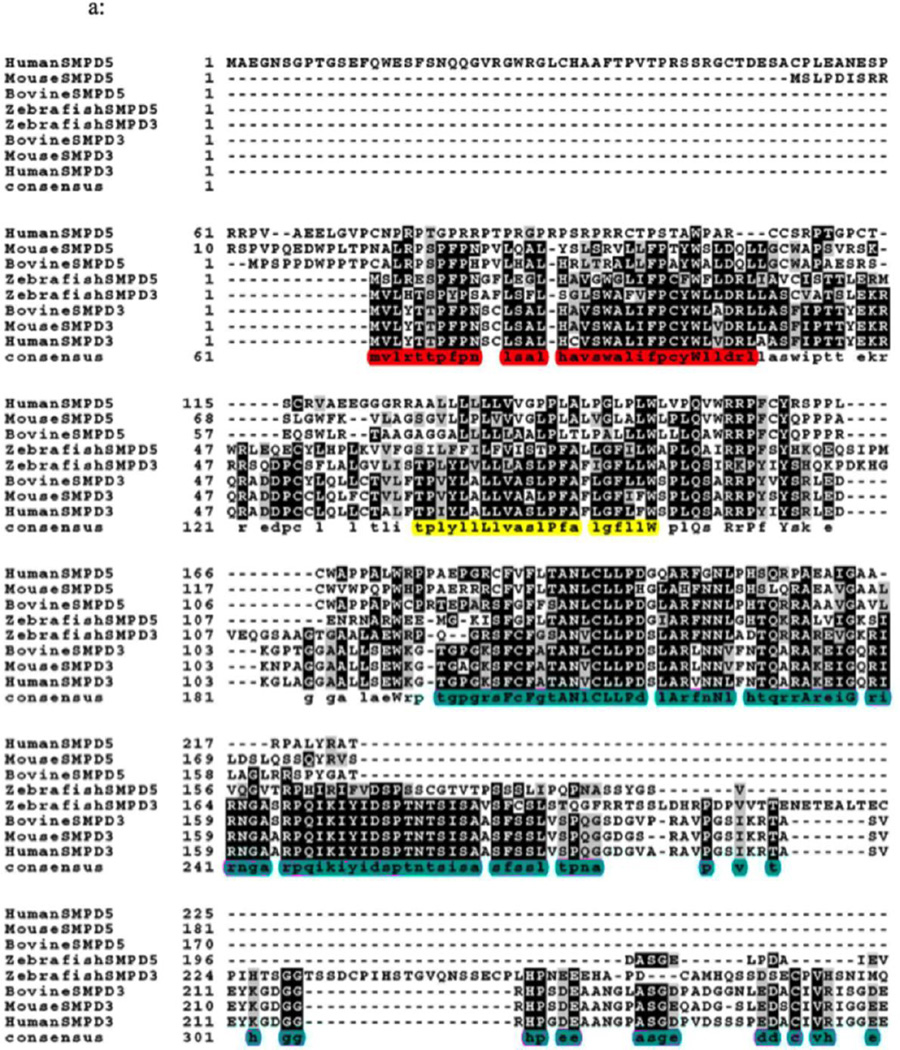
Neutral sphingomyelinase type activity had been found in mammalian mitochondrial fractions; however, it had not been shown clearly what specific form of the enzyme associated to the mitochondria. In a previous work, our group showed murine MA-nSMase to be associated to mitochondria in MCF-7 [7]. However, the signal that targets MA-nSMase to mitochondria in mammals and its intra-mitochondrial localization is still unknown. The MLS of MA-nSMase zebrafish homologue has been defined experimentally, in that a deletion construct spanning the residues 1–35 prevented its mitochondrial localization [6]. In order to ascertain the putative MLS, possible TMD, and the catalytic core of MA-nSMase, we aligned the protein sequences of sphingomyelin phosphodiesterase 5 (SMPD5, the gene for MA-nSMase) and of sphingomyelin phosphodiesterase 3 (SMPD3, which is not localized in the mitochondria) from various species (Figure 1A). The accession numbers of these protein sequences were: human (NP001182466.1), mouse (XP002692632.1), bovine (XP003120137.2) and zebrafish (NP001071083.1) for SMPD5; and human (NP001116222.1), mouse (XP005256088.1), bovine (NP001179292.1), and zebrafish (AAH43077.1) for SMPD3. Interestingly, most of the SMPD5 shared a conserved sequence coinciding with zebrafish MLS (highlighted in red in Figure 1A) whereas the human SMPD5 did not share this consequence. The MLS of the zebrafish mitochondrial SMase (1–35) is homologous to residues 24–56aa of MA-nSMase. This putative MLS in mouse MA-nSMase is conserved in all species except the predicted human sequence which had the least homology; the TMD of the zebrafish mitochondrial SMase, that spans 64–85aa and is homologous to residues 77–99aa of MA-nSMase, is approximately 50% conserved in all species (highlighted in yellow in Figure 1A); the catalytic domain, predicted based on nSMase 2 sequence (highlighted in pink in Figure 1A), starts from residue 129 until 483 in the mouse MA-nSMase and is fairly conserved (Figure 1B).
Fig. 1.
Fig. 1A. Sequence alignment of SMPD 5 (MA-nSMase) with SMPD 3 (nSMase 2). Alignment of the deduced amino acid sequences of human (NP001182466.1), mouse (XP002692632.1), bovine (XP003120137.2) and zebrafish (NP001071083.1) SMPD5; and human (NP001116222.1), mouse (XP005256088.1), bovine (NP001179292.1), and zebrafish (AAH43077.1) SMPD3. The sequences were aligned by Clustal omega program. Identical residues in all the three sequences are indicated by bold characters. The mitochondrial localization signal (MLS) of the zebrafish mitochondrial SMase that spans 1–35aa which is homologous to residues 24–56aa of MA-nSMase highlighted in red, predicted transmembrane domain (TMD) sequence spanning between 77–99aa in MA-nSMase is highlighted in yellow and the catalytic domain that spans 129–483 in the mouse MA-nSMase, predicted based on nSMase 2 sequence is highlighted in pink.
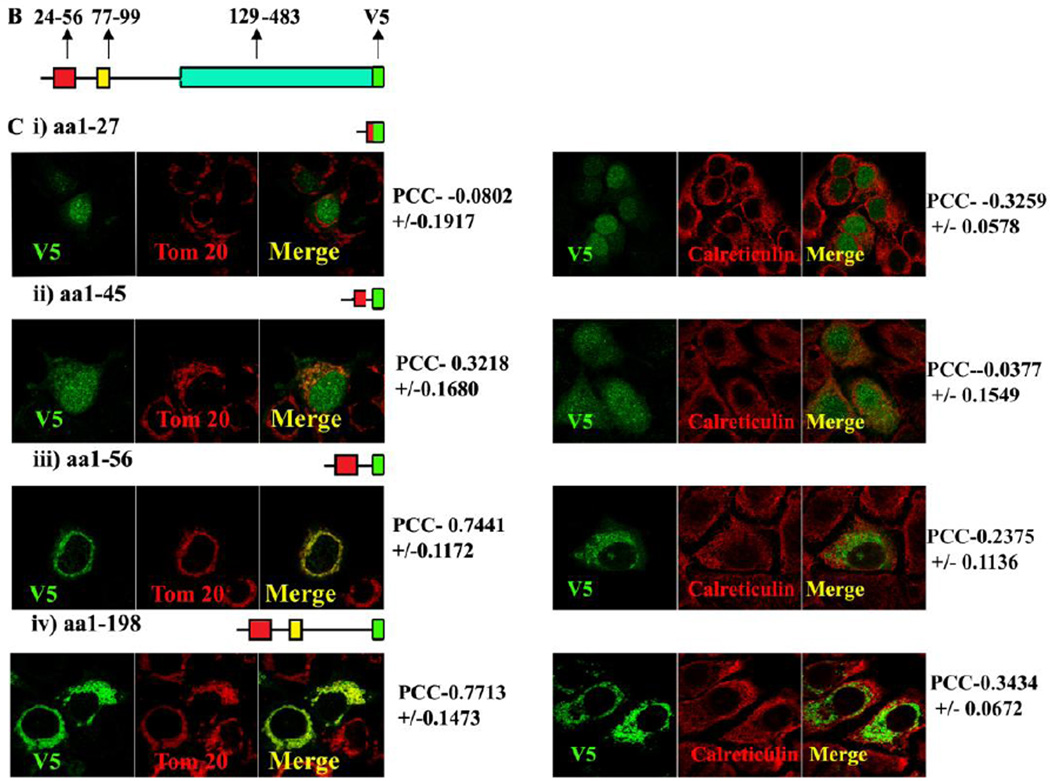
Fig. 1B, Schematic representation of the MLS, TMD and catalytic domain of MA-nSMase. C, Truncated mutant constructs and its co-localization with mitochondrial marker (Tom 20). Table 1 showing the summary of the results of truncation mutant constructs.
3.2 A Mitochondria Localization Signal (MLS) resides in the first 56 residues of the MAnSMase
Human SMPD5 did not possess the conserved MLS motif. A putative MLS is present in the mouse SMPD 3 but this protein does not localize to mitochondria. These incongruence prompted us to investigate whether the predicted MLS in mouse functions to localize the protein to mitochondria. In order to accomplish this goal, a series of fusion proteins were constructed with the following residues: aa1–27, aa1–45, aa1–56 and aa1–198 of mouse MA-nSMase fused to V5. The fusion constructs were transiently expressed in MCF-7 cells, and their subcellular localization was analyzed for co-localization with known organelle markers using confocal microscopy. In order to quantify the data, the Pearson correlation coefficient (PCC) which measures the pixel-by-pixel covariance in the signal levels of two images was employed. In order to define the range of PCC, well established markers for mitochondria (Tom 20, cytochrome C), ER (calreticulin) and Cytosol (GAPDH) were tested (Supplemental Figure 1), and compared with the PCC values measured for each of the mutant constructs. The constructs aa1–27, aa1–45 did not co-localize with mitochondrial marker Tom 20 as evidenced by poor PCC (−0.08020+/−0.1917 and 0.3218+/−0.1680, respectively) whereas constructs 1–56 and 1–198 highly co-localized with Tom 20, as evidenced by high PCC (0.7461+/−0.1171 and 0.7713+/−0.1473 respectively) (Figure 1C and Table 1). When these constructs were analyzed for co-localization with the ER marker calreticulin, constructs aa1–27, aa1–45 did not co-localize with ER, as evidenced by poor PCC (−0.3259+/−0.0578 and − 0.0377+/−0.1549, respectively) whereas constructs 1–56 and 1–198 mildly co-localized with ER, as evidenced by slightly higher PCC (0.2375+/−0.1136 and 0.3434+/−0.0672 respectively). The result of this experiment show that residues 1–56 and 1–198 are sufficient for mitochondrial localization, and they suggest that the MLS resides in the first 1–56 amino acids of the protein.
Table 1.
Truncated MA-inSMase contructs and their localization
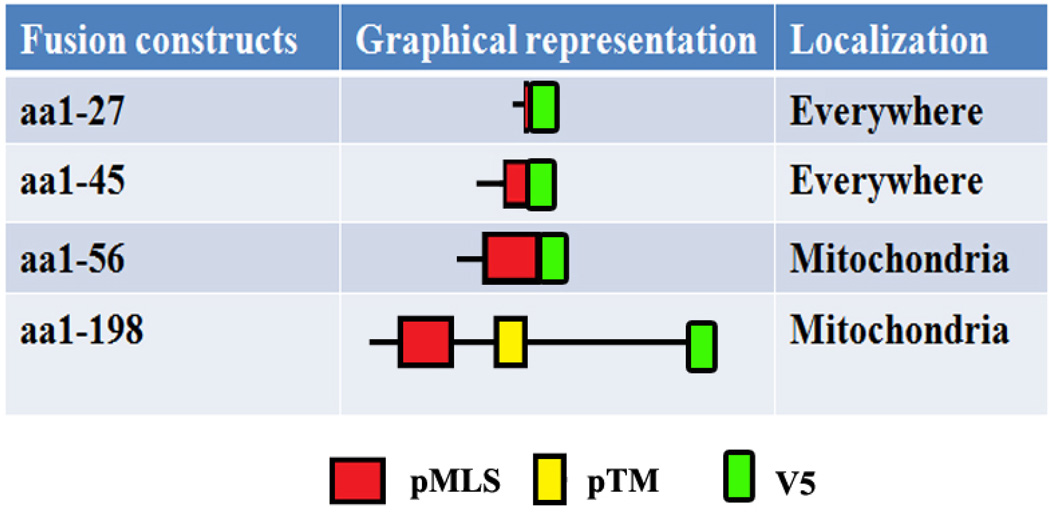 |
3.3 The putative MLS is not necessary for mitochondrial localization of MA-nSMase
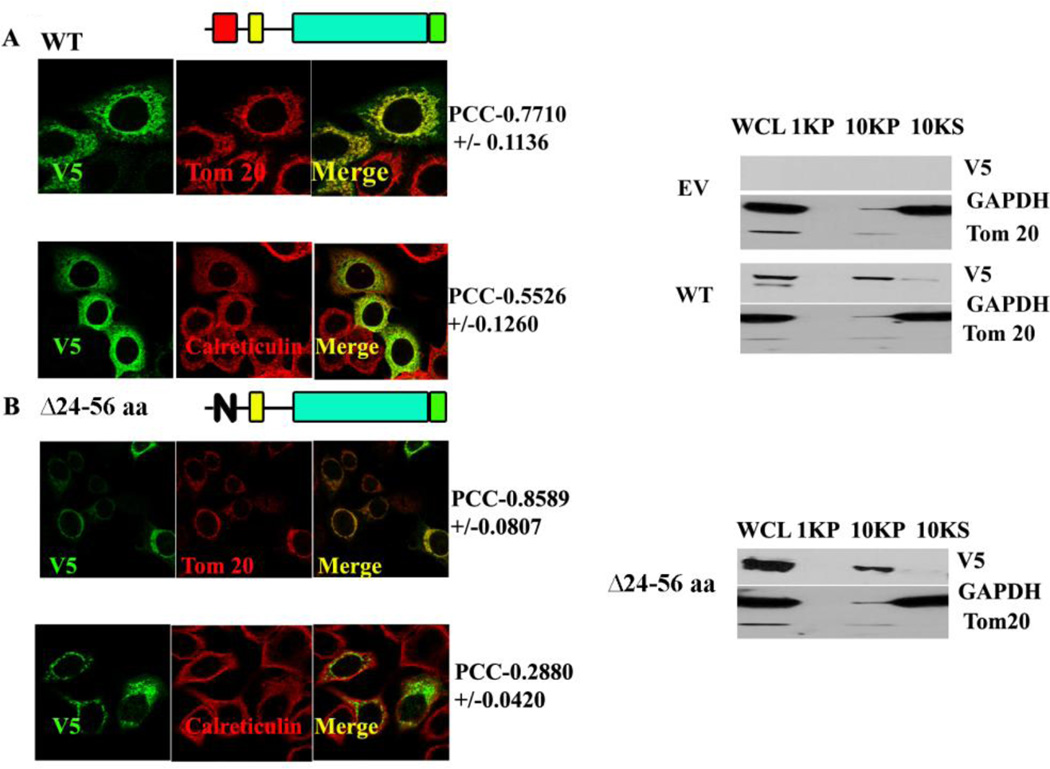
To determine if the minimally sufficient MLS was also necessary for the mitochondria localization of the SMPD5 protein and to test if other regions preceding the catalytic domain, play any regulatory role in mitochondrial localization, several deletion mutants spanning the entire 1–198aa were constructed. First, the WT (full length) MA-nSMase protein was tested for its localization. Surprisingly, it was found to localize not only to mitochondria but also to ER, as indicated by the high PCC with Tom 20 (0.7710 +/−0.1136) and calreticulin markers (0.5526+/−0.1260) in MCF-7 cells (Figure 2A and Table 2). Next, a deletion construct spanning the 24–56 region was constructed and tested. Unexpectedly, deletion of the putative mitochondrial localization signal (Δ24–56aa) did not prevent the protein to reach the mitochondria (PCC with Tom 20- 0.8589+/−0.0807) (Fig. 2B and Table 2) while preventing its co-localization with ER (PCC with calreticulin- 0.2880+/−0.0420). In order to validate the above findings, differential centrifugation was resorted to in order to separate the heavy membranes, rich in mitochondria (indicated as 10KP in Figure 2 western blot), from the light membrane and soluble components of the cell (marked as 10KS in Figure 2 western blot). As with the confocal visualization, the wild type and Δ24–56 were highly enriched in the 10,000 × g pellet, demonstrating that the 24–56 region is not necessary for the mitochondrial localization (Figure 2B).
Fig. 2. Co-localization and cellular fractionation of Wild type and deletion of MLS (ΔMLS) MA-nSMase constructs.
A and B, MCF-7 transfected with WT (panel A) and MCF-7 transfected with ΔMLS (Δ24–56) (panel B) cells were transfected with MA-nSMase expression vector. After 24 h, the cells were fixed and co-stained with an antibody against V5 (green) for MA-nSMase signal and antibodies against various subcellular markers (red), including Tom20 (mitochondrial markers), calreticulin (ER marker) and then subjected to confocal microscopic observation. The colocalization signals were observed as yellow or orange and quantified using Pearson correlation coefficient (PCC) which measures the pixel-by-pixel covariance in the signal levels of two images. On to the right of WT (panel A), ΔMLS (Δ24–56) (panel B), cellular lysates from corresponding constructs were incubated in isotonic buffer containing 250mM sucrose, disrupted by passage through 25 gauge needle and centrifuged at 1,000 × g for 10 min (indicated as 1KP in the western blot figure) and 10,000 × g for 10 min (indicated as 10 KP in the western blot figure) for collection of the nuclear fraction and the mitochondria-enriched heavy membrane fraction respectively. The supernatant obtained after the 10,000 × g was used for light membrane and cytosolic fraction. The whole cell lysate was used to show the input. Fractions were analyzed by western blotting.
WCL: whole cell lysate; 1KP: 1,000g pellet; 10KP: 10,000g pellet; 10KS: 10,000g supernatant.
Table 2.
Deletion mutant constructs and their localization
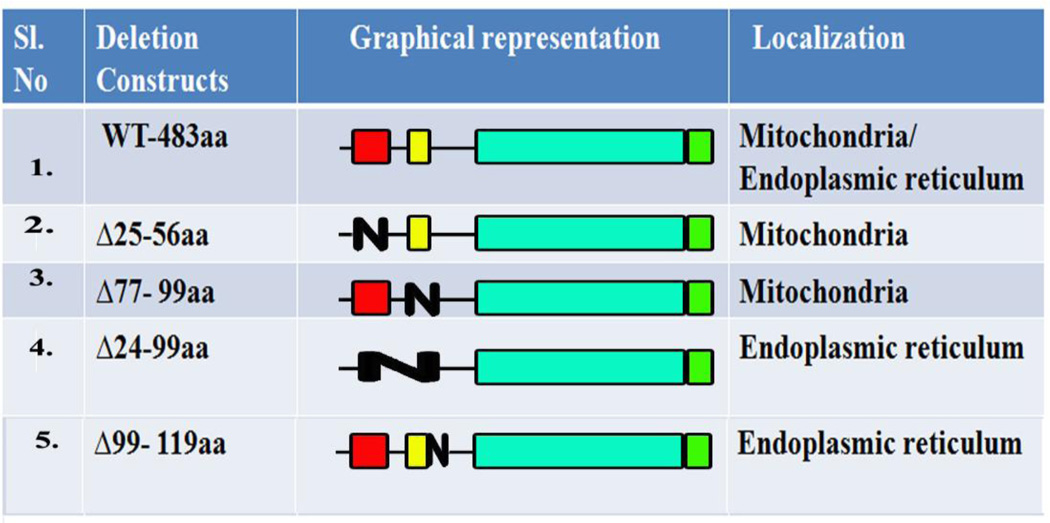 |
3.4 The TMD is not necessary for mitochondrial localization of the protein
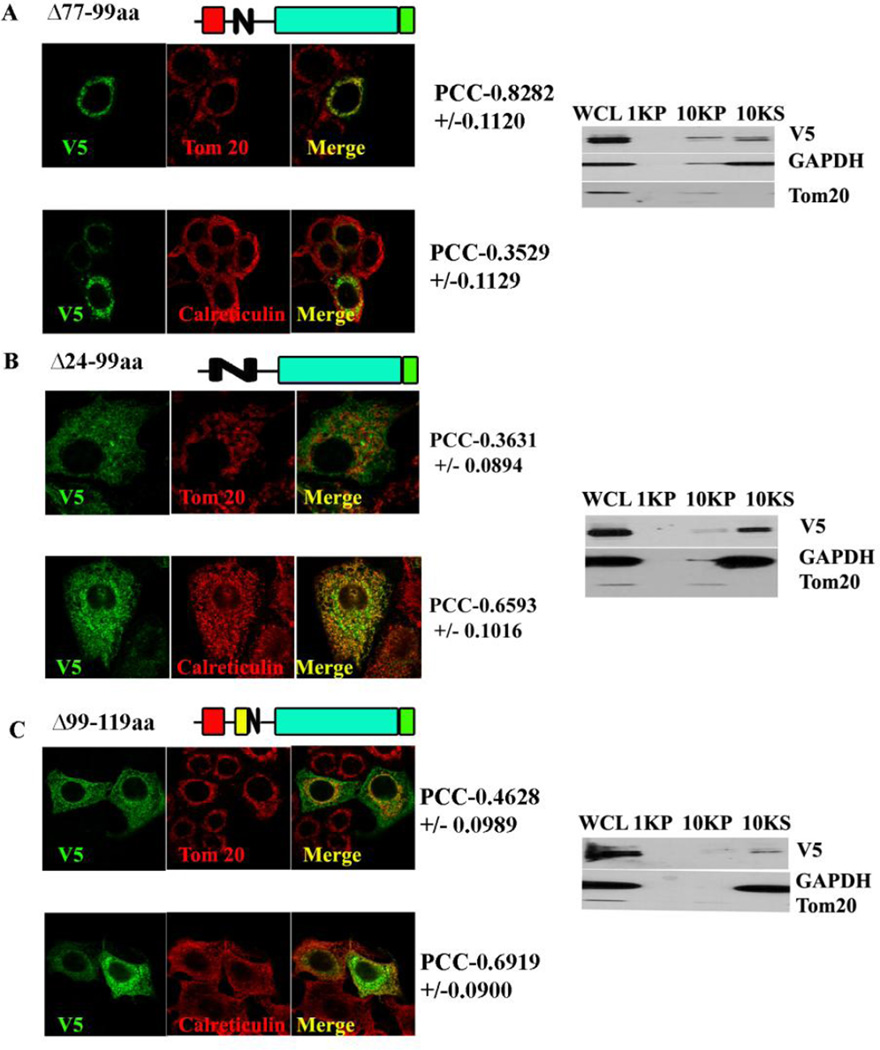
Since the predicted MLS is not necessary for mitochondrial localization, it was investigated if the TMD functions as signal-anchor; sorting this protein to mitochondria and incorporating it to the mitochondrial membrane, as has been demonstrated in the case of Tom 20/Tom70 [30, 31]. In order to test this hypothesis, the TMD spanning between 77–99aa was deleted, and this construct was subjected to confocal visualization and differential centrifugation. The Δ77–99 deletion, did not prevent the mitochondrial localization of the protein as concluded from the high co-localization with Tom 20 (PCC of 0.8282+/− 0.1120) and mild co-localization with the ER marker (PCC with calreticulin- 0.3529+/−0.1129). Despite this high PCC found using confocal microscopy, the Δ77–99 partitioned equally in the 10,000 × g supernatant and pellet, suggesting it might be only loosely associated with mitochondria (Figure 3A and Table 2) due to the lack of hydrophobic interactions that may tether it more tightly. Therefore, the TMD is not necessary for the mitochondrial localization but may be important for membrane incorporation.
Fig. 3. Co-localization and cellular fractionation of deletion of TMD (Δ77–99).
A. MCF-7 cells were transfected with an expression construct of deletion of TMD MA-nSMase; co-localization and fractionation were done as described previously. B. Co-localization and cellular fractionation of double deletion of MLS and TMD (Δ24–99): MCF-7 cells were transfected with an expression construct of double deletion of MLS and TMD MA-nSMase; co-localization and fractionation were done as described previously. C. Co-localization and cellular fractionation of deletion of residues 99–119aa flanking the transmembrane domain(Δ99–119): MCF-7 cells were transfected with an expression construct of deletion of residues 99–119aa flanking the transmembrane domain of MA-nSMase; co-localization and fractionation were done as described previously.
3.5 Double deletion of MLS and TMD abrogated mitochondrial localization of MAnSMase
The above results show that single deletion of MLS (24–56aa) or TMD (77–99aa) do not abrogate mitochondrial localization, indicating that neither is necessary. Since the putative MLS is sufficient, but not necessary for mitochondrial localization, it was therefore hypothesized that there are two mitochondrial signals, MLS serving as one and TMD serving as the other. In order to test this hypothesis, these two regions were deleted simultaneously. As shown in the Figure 3B and Table 2, the deletion of both of these regions (del 24–99) prevented the targeting of the protein to mitochondria as evidenced by its low co-localization with Tom 20 (PCC- 0.3631+/−0.0894) and its presence in the 10,000 ×g supernatant. On the other hand it did not prevent its co-localization with the ER (PCC with calreticulin- 0.6593+/−0.1016). All together these results indicate that there are two MLS (24–56aa and 77–99aa) and deletion of both prevents the mitochondrial association of MA-nSMase.
3.6 Residues 99–119aa flanking the transmembrane domain might be necessary for mitochondrial protein import
Features such as moderate hydrophobicity of TMD and at least 3 basic amino acids close to the C-terminal of the TMD (at residues 130,131 and 132) present in MA-nSMase but not in mouse neutral SMase2 suggested that MA-nSMase might be a signal anchored protein with the region flanking TMD functioning as a signaling component. Therefore, a deletion of this flanking region (99–119aa) was performed, and this construct was subjected to confocal visualization and differential centrifugation. Δ99–119aa showed a poor co-localization with Tom 20 (PCC−0.4627+/−0.0958) and high co-localization with the ER marker (PCC with calreticulin- 0.6919+/−0.0900). In agreement with the confocal results, it was also enriched in the 10,000×g supernatant (Figure 3C and Table 2). Thus, in addition to the putative MLS and the TMD, this TMD-flanking region seems to play an important role in the mitochondrial localization.
3.7 The TMD region (residues 77–99) is sufficient to target a heterologous protein to mitochondria
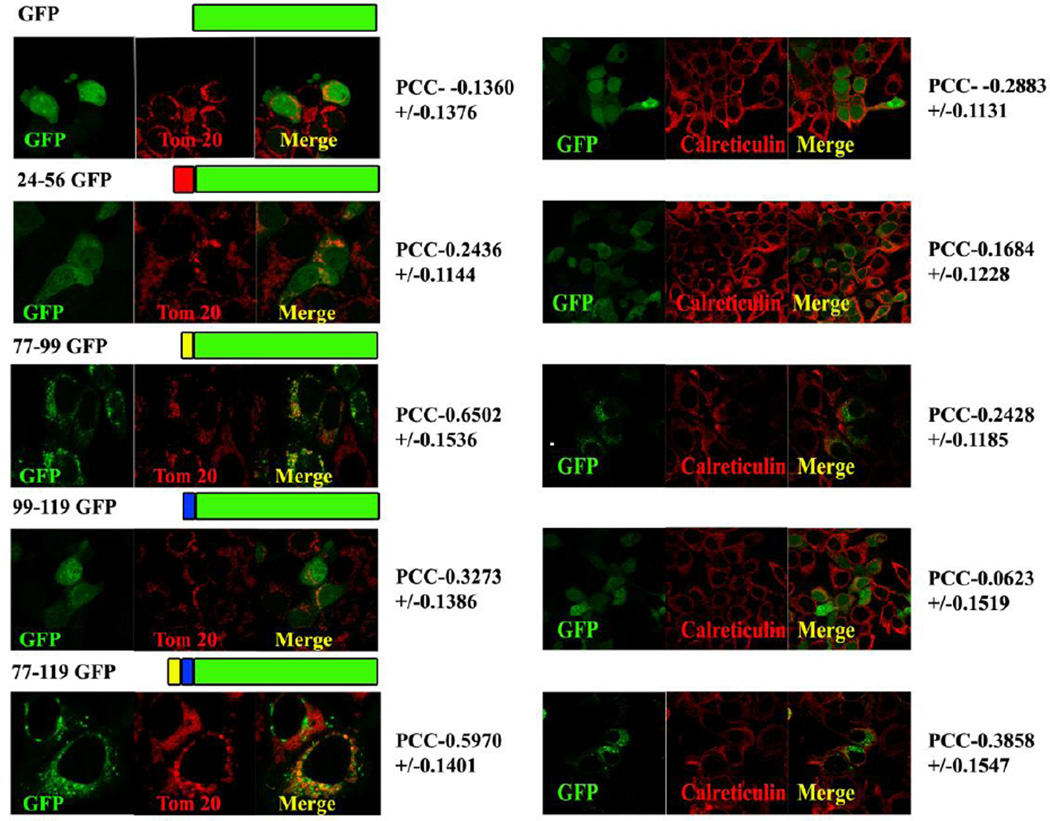
Once the regions which were necessary for mitochondrial localization of MA-nSMase were identified, we evaluated if they were sufficient for its localization. Therefore, a series of GFP fusion constructs were made with aforementioned critical regions such as the putative MLS 24–56-GFP, the TMD 77–99-GFP, the TMD flanking region 99–119-GFP, and the TMD with its flanking region 77–119-GFP and they were subjected to confocal visualization (Figure 4 and Table 3). Only the constructs 77–99 GFP and 77–119 GFP showed high co-localization with the mitochondrial marker (PCC- 0.6502 +/− 0.1536 and PCC- 0.5970+/− 0.1401 respectively) and mild co-localization with the ER marker (PCC- 0.2428+/−0.1185 and PCC- 0.3858 +/− 0.1546 respectively), suggesting that this region has a signal that is sufficient to target the GFP to mitochondria. All other GFP constructs co-localized poorly with the Tom 20 marker suggesting poor co-localization with mitochondria. The TMD-flanking region (99–119 GFP) although being necessary for mitochondrial localization, was not sufficient for its localization.
Fig. 4. Co-localization of various GFP fusion constructs.
MCF-7 cells were transfected with a series of GFP fusion constructs made with aforementioned critical regions such as putative MLS 24–56-GFP, TMD 77–99-GFP, the TMD flanking region 99–119-GFP, and the TMD with its flanking region 77–119-GFP and subjected to confocal visualization as described previously.
Table 3.
GFP constructs and their localization
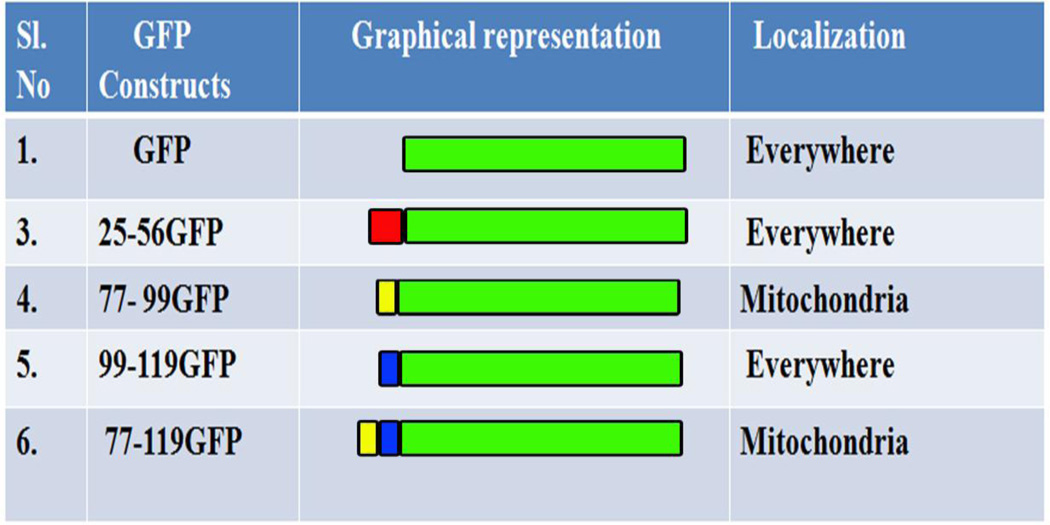 |
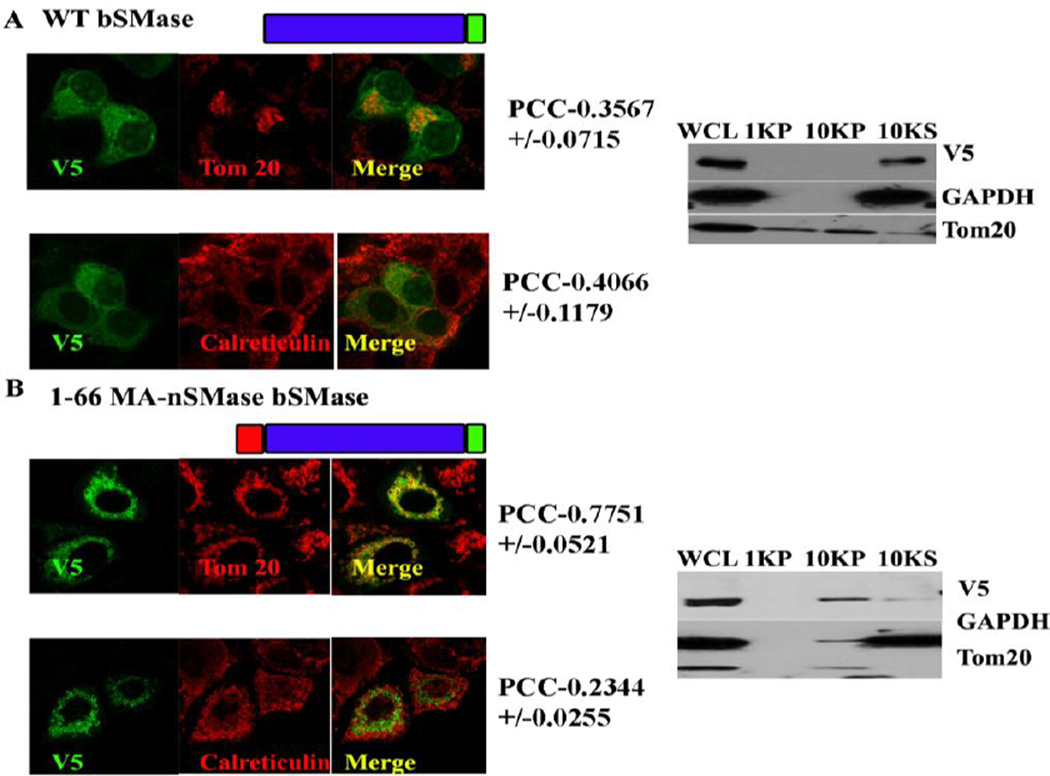
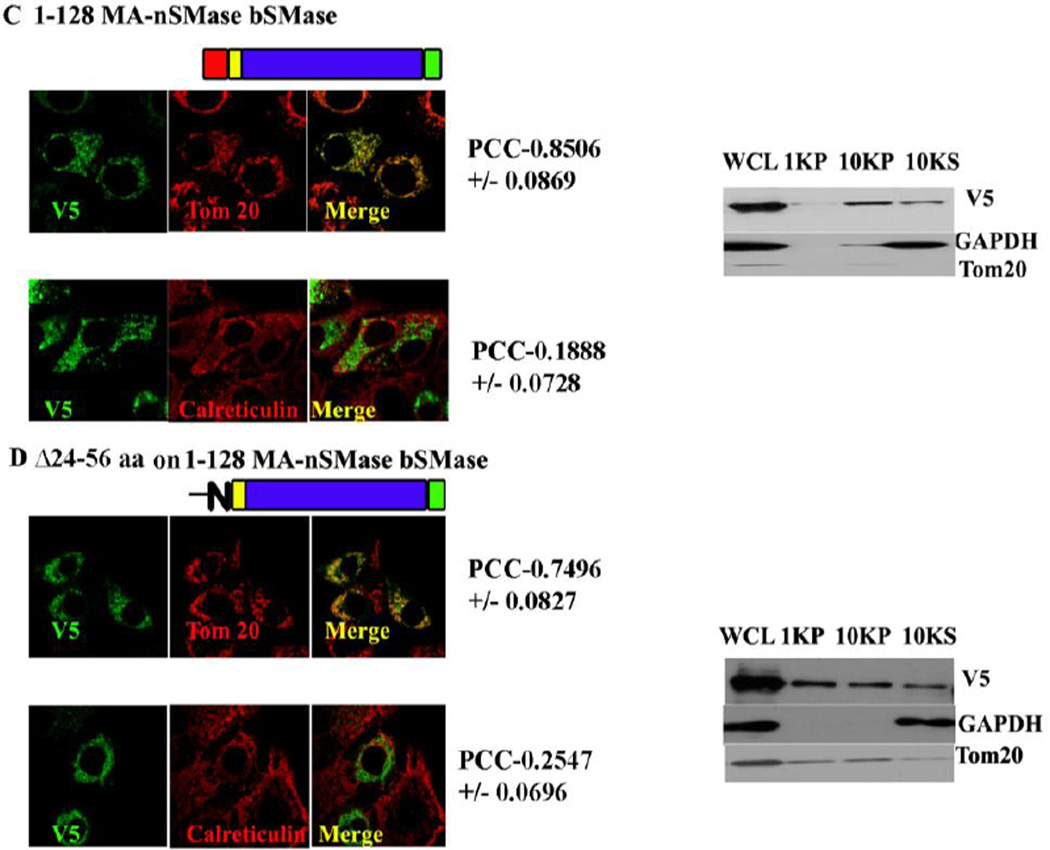
3.8 Putative MLS and TMD are sufficient to target bacterial SMase constructs to mitochondria
The mitochondrial localization signal deduced with the GFP constructs was validated by using the MLS to target a non-mitochondrial protein (bacterial SMase) to mitochondria. Three construct were made: 1–66, 1–128 of MA-nSMase fused to bacterial SMase and Δ24–56 on 1–128 bSMase construct. These constructs were subjected to confocal visualization and differential centrifugation (Figure 5 and Table 4). The result suggested that WT bSMase did not localize with mitochondria as obvious from its low PCC with Tom 20 (PCC-0.3567+/− 0.0706) and mildly co-localizated with the ER marker (PCC with calreticulin- 0.4065+/− 0.1198); whereas 1–66 MA-nSMase bSMase construct clearly co-localized with mitochondria with high co-localization coefficient with Tom 20 (PCC- 0.7751+/− 0.0512) and mildly colocalizated with the ER marker (PCC with calreticulin- 0.2343+/−0.0258). The construct 1–128 MA-nSMase bSMase showed increased PCC with Tom 20 (PCC-0.8506 +/− 0.0869) while the PCC with calreticulin went down (PCC- 0.1858+/− 0.0728). Finally the construct Δ24–56 on 1–128 bSMase co-localized with mitochondria (PCC-0.7496+/−0.0827) and mild co-localization with the ER marker (PCC with calreticulin- 0.2547+/−0.0696) which was lower compared to 1–66 MA-nSMase bSMase fusion (24–56) but significantly higher than the WT bSMase. There was a clear enrichment of all of these constructs in the 10,000 × g pellet with the differential centrifugation except that the WT bSMase which partitioned in the 10,000 ×g supernatant as expected further confirming the above findings that there are two sufficiency regions (24–56 and 77–99) targeting MA-nSMase to mitochondria.
Fig. 5. Co-localization of bacterial SMase fusion constructs.
MCF-7 cells were transfected with a series of bacterial SMase fusion constructs such as 1–66, 1–128 of MA-nSMase fused to bacterial SMase and Δ24–56 on 1–128 bSMase construct. These constructs were subjected to confocal visualization and differential centrifugation as described previously.
Table 4.
Bacterial SMase fusion constructs and their localization
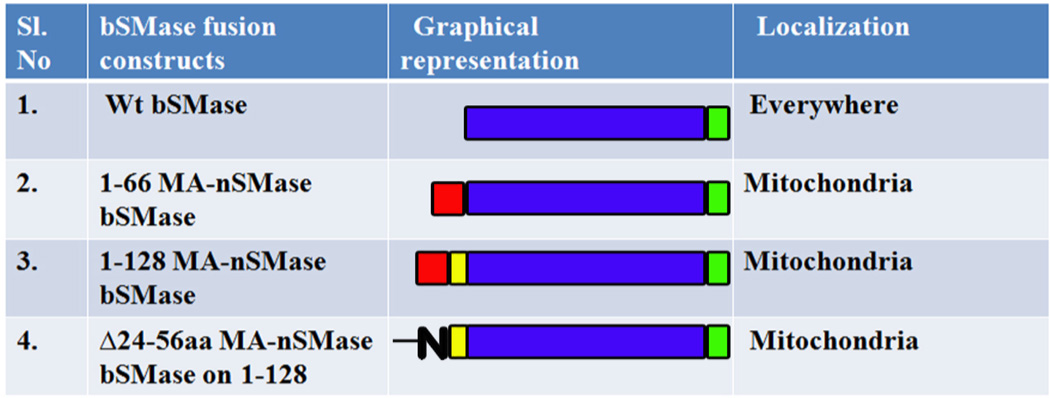 |
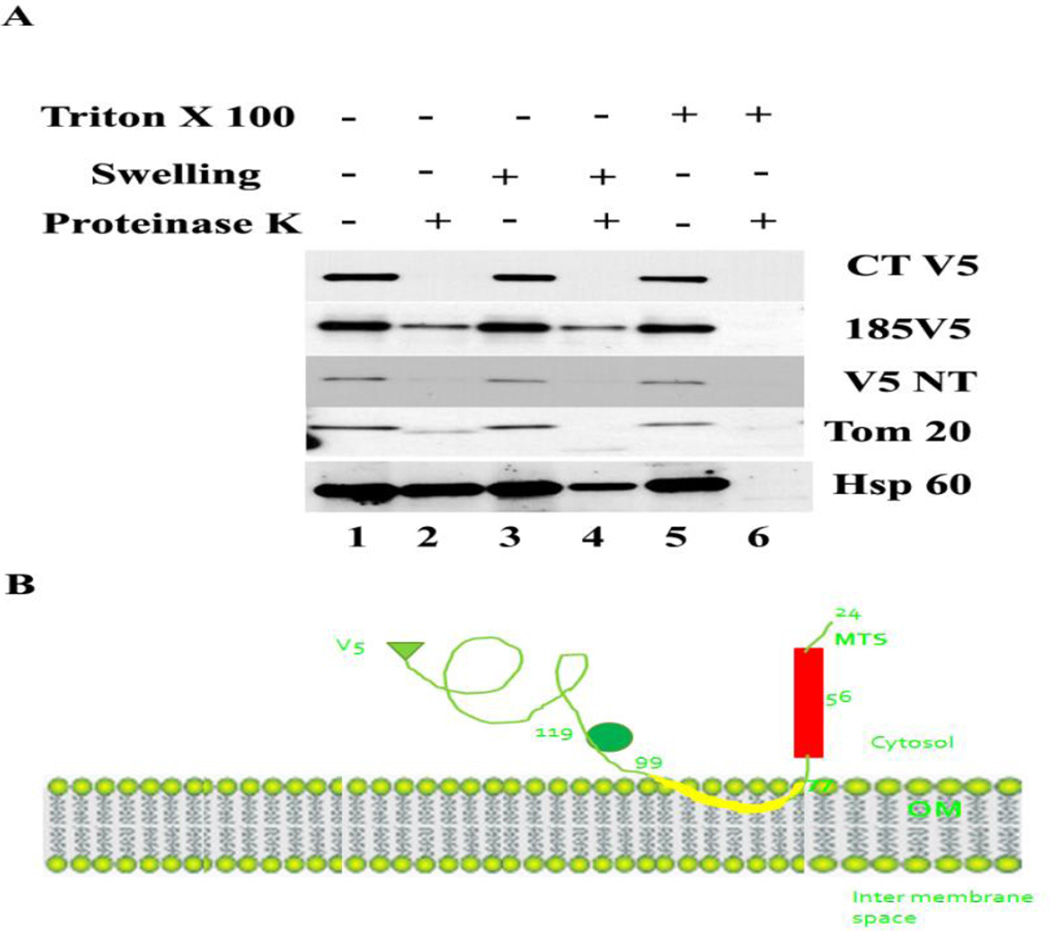
3.9 MA-nSMase is topologically localized in the outer mitochondrial membrane with the C-terminal facing the cytosol
The results, thus far, suggested that MA-nSMase might have a MLS at its N terminal and a ‘signal -anchor’ sequence that anchored the protein in mitochondrial membranes. Since the mitochondrial outer membrane is enriched with such proteins, e.g. Tom 20 and Tom 70, it was hypothesized that MA-nSMase might be an outer mitochondrial membrane anchored protein. In order to determine the sub-mitochondrial topology of MA-nSMase, the mitochondria enriched 10,000 × g pellet was subjected to a protease protection assay with Tom 20 as an outer membrane marker and HSP-60 as a matrix marker (Figure 6 C-terminal V5). The results showed that MA-nSMase was digested completely in the presence of protease even when the mitochondrial outer membrane is intact; suggesting MA-nSMase is an outer membrane anchored protein with its C-terminal carrying the tag oriented towards the cytosolic side. In order to further probe the orientation, the V5 tag was inserted between residues 185 and 186 and separately at the N terminus, and protease protection assay was performed. The results showed that both these constructs behaved similarly to the C-terminal tag suggesting the entire protein faces the cytosolic side of the mitochondria with the putative transmembrane domain partially buried in the OM or tethered to mitochondria. A schematic model of this prediction of the protein is presented (Figure 6B).
Fig. 6. Topology of MA-nSMase as demonstrated by the protease protection assay.
V5 tags were introduced at C terminal, between residues 184 to 185 and at N terminal. Crude heavy membrane fraction (10,000 ×g pellet) was collected as described previously from MCF-7 cells expressing these constructs. The pellet was suspended in the isotonic buffer and the protein concentration measured using BCA assay (Bio-Rad Laboratories). 5mg/mL of protein were used as an input for the protease protection assay. Proteinase K was added from freshly prepared stocks in water. Unless otherwise indicated, the final proteinase K concentration was 4µg/ml and the detergent Triton X-100 was added at 0.2% (w/v) final concentration. Digestion reactions were performed for 20 min on ice and quenched by addition of PMSF to a final concentration of 2 mM. Finally, the samples were analyzed by western blotting.
4. Discussion
In this work, the recently discovered mouse sphingomyelinase SMPD5 was found to be localized primarily in mitochondria and also partially in the ER in mammalian cells. Two different regions preceding the catalytic sequence were found to target MA-nSMase to the mitochondria; each MLS was sufficient in itself for this localization, and neither was necessary. The second MLS was also found to be a signal-anchor region, tethering MA-nSMase to the mitochondrial outer membrane.
Mitochondrial proteins are synthesized in the cytosol and targeted to the mitochondria by recognition of a sequence of amino acids, typically upstream of the catalytic region of the enzyme. In this study, we attempted to delineate the domains responsible for mitochondrial localization of MA-nSMase. Localization predictions using bioinformatics approach (pSORT II) did not confirm the existence of a mitochondrial signal. However, the zebrafish homologue contains a mitochondrial pre-sequence that was found to be highly conserved in MA-nSMase (residues 24–56). The results showed that this homologous conserved sequence in MA-nSMase, also targeted fusion proteins to the mitochondria (1–55V5 and 1–128V5).
However, quite contrary to the findings with the previously characterized zebrafish mitochondrial SMase, the putative MLS of MA-nSMase spanning the regions 24–56, although sufficient, was not necessary for MA-nSMase to localize the protein to mitochondria. This finding suggested that there could be another MLS in MA-nSMase that targets the protein to mitochondria. Some mitochondrial proteins contain what it is called a ‘signal-anchor’ sequence in which the localization signal and membrane anchor are in the same sequence motif. When the predicted transmembrane domain (TMD) of MA-nSMase was deleted, it showed high mitochondrial co-localization. However, upon cell lysis and fractionation, the deletion mutant was enriched in the soluble fraction. This apparent contradiction between the strong co-localization pattern with mitochondrial markers when analyzed with confocal microscopy and the enrichment in the cytosolic fraction with differential centrifugation suggested that the deletion mutant was transported to the mitochondria, but its interaction with the organelle was weak and it was lost during the differential centrifugation procedure. It has been previously demonstrated that, in the case of the bacterial preprotein proOMP-A carrying a synthetic ‘stop-transfer sequence’, that the translocation arrest across the membrane is controlled by the translocase SecYEG while lateral release into the lipid bilayer and subsequent membrane integration is governed by the hydrophobicity of the protein itself [28]. It is therefore possible that deletion of TMD makes the protein loosely tethered to mitochondria due to lack of lateral release of the protein into membranes.
Moreover, residues 99–119aa were found to be necessary for mitochondrial localization. They flank the TMD on one side, but they do not contribute to the hydrophobicity of the domain. This region might be necessary for mitochondrial protein import as deletion of this region showed no mitochondrial co-localization with both the confocal visualization and fractionation methods. However, the GFP fused to 99–119 region failed to localize to mitochondria with both these analysis. These results suggest that it is highly likely that this region might not be a signal (MLS) but may be necessary for processes upstream of mitochondrial protein import such as chaperon binding and correct protein folding which is essential for the correct mitochondrial protein import.
Of note and quite surprisingly, the 24–56 GFP fusion protein, which carries a MLS, did not co-localize with the mitochondrial marker whereas the 77–99 GFP co-localized with mitochondrial marker. On the other hand, the 1–56 V5 fusion protein colocalized with the mitochondria (Figure 1C) suggesting that perhaps the stretch containing aa 1 to 23 may cooperate with the 24–56 MLS for targeting to mitochondria. Nevertheless aa 1–24 were neither necessary (Supplemental Figure 2A) nor sufficient (Supplemental Figure 2B) for mitochondrial localization thus it is also possible that the reason for the discrepancy in localization between the 24–56 GFP fusion protein and the 77–99 GFP might be due to folding of the GFP. For instance, in the 24–56 GFP protein, the MLS could have been masked whereas in the construct 77–99 GFP, it could have been accessible to the import receptors of mitochondria. This phenomenon has been previously documented in the literature. A fusion protein made by fusing GFP and the C terminal of Qcr6p, subunit 6 of the cytochrome bc1 complex [35], has been found to mis-localize to the cytosol (instead of the mitochondria); also fusion proteins of GFP with N or C termini of porin were found to localize in the cytosol [36].
In order to circumvent the potential GFP mis-folding problem, bacterial SMase fused to either the MLS or TMD or both were constructed and tested. The bacterial SMase chimeric proteins clearly show that the putative MLS (24–56) is sufficient to target the chimeric protein to mitochondria whereas the simultaneous presence of the MLS and TMD, the ‘signal anchor’ increases the mitochondrial localization even more. The construct Δ24–56 on 1–128 bSMase also co-localized with mitochondria suggesting that there are two signals that are sufficient to drive the bSMase to mitochondria and the signal-transfer sequence has a signaling component to it.
In addition, the data from protease protection assay and bioinformatics modeling predict that this protein is indeed in the outer mitochondrial membrane with both its N and C-terminal containing the catalytic domain pointing towards the cytosolic side. Sphingomyelinase with such orientation might hydrolyze the SM of the outer-membrane of the mitochondria or the SM associated with ER and mitochondria associated membranes (MAMs) and subsequent metabolites might be exchanged to the mitochondria for its biology. Interestingly, in addition to mitochondria, WT MA-nSMase colocalizes in part with the ER (Figure 2A) and, in absence of both mitochondrial signals (24–56 or 77–99) or absence of presumed regulatory region (99–119aa), the protein localizes exclusively to the ER. This could suggest a couple of possibilities: a) WT MA-nSMase transits through the ER for presumed post-translational modifications before it is transported to the mitochondria, and, if the protein does not have the mitochondrial signals, it is retained in the ER; or b) the WT protein resides in MAMs, as MAMs are also enriched in crude 10KP, and the different targeting signals may be used to dynamically redistribute MA-nSMase from MAMs to mitochondria in conditions that could favor generation of ceramide in mitochondria and apoptosis. These are interesting possibilities that will be investigated in future studies.
The outer mitochondrial membrane localization of MA-nSMase may be brought about exclusively by its transmembrane domain and its flanking region. MA-nSMase has features corresponding to the class of signal-anchored proteins that were previously reported in the literature [30, 31], GFP fusion constructs and bSMase with intact TMD (77–99 GFP and 77–119 GFP, Δ24–56 on 1–128 bSMase) also convincingly proved it is sufficient to target the protein to mitochondria.
In summary, we have identified MA-nSMase as a protein with N –terminal mitochondrial localization signal and a ‘signal anchor’ sequence to anchor it to the outer mitochondrial membrane. In one of the earlier studies by Birbes et al. [9], bacterial SMase GFP was fused with targeting sequence isolated from subunit VIII of human cytochrome c oxidase. This construct presumably might have been translocated to the inner mitochondrial membrane/mitochondrial matrix and caused apoptosis in MCF-7 cells. In our study, MA-nSMase targeted to the outer mitochondrial membrane did not cause apoptosis (data not shown). Taken together, these data suggest that topologically distinct sub- mitochondrial ceramide changes might dictate distinct sphingolipid biology which is presently explored in our lab.
Supplementary Material
Highlights.
Evidences for mammalian Ma-nSMase localization in to the mitochondria
The putative MLS is not necessary but sufficient in murine Ma-nSMase
There is a 'signal-anchor' sequence following the MLS
MA-nSMase is topologically located in the outer mitochondrial
Acknowledgements
This work was supported by NIH Grant GM 04823.
Abbreviations used are
- SMase
sphingomyelinase
- N-SMase
neutral sphingomyelinase
- SMPD
sphingomyelin phosphodiesterase
- SM
sphingomyelin
- MAMS
mitochondria-associated membranes
- CerS
Ceramide synthase
- Hsp
heat shock protein
- MLS
mitochondrial localization signal
- TMD
transmembrane domain
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- bSMase
bacterial sphingomyelinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ardail D, Popa I, Alcantara K, Pons A, Zanetta JP, Louisot P, Thomas L, Portoukalian J. Occurrence of ceramides and neutral glycolipids with unusual long-chain base composition in purified rat liver mitochondria. FEBS Lett. 2001;488:160–164. doi: 10.1016/s0014-5793(00)02332-2. [DOI] [PubMed] [Google Scholar]
- 2.Tserng KY, Griffin R. Quantitation and molecular species determination of diacylglycerols, phosphatidylcholines, ceramides, and sphingomyelins with gas chromatography. Analytical biochemistry. 2003;323:84–93. doi: 10.1016/j.ab.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Novgorodov SA, Chudakova D, Zhu H, Bielawska A, Bielawski J, Obeid LM, Kindy MS, Gudz TI. JNK3 signaling pathway activates ceramide synthase leading to mitochondrial dysfunction. J Biol Chem. 2007;282:25940–25949. doi: 10.1074/jbc.M701812200. [DOI] [PubMed] [Google Scholar]
- 4.Novgorodov SA, Chudakova DA, Wheeler BW, Bielawski J, Kindy MS, Obeid LM, Gudz TI. Developmentally regulated ceramide synthase 6 increases mitochondrial Ca2+ loading capacity and promotes apoptosis. J Biol Chem. 2011;286:4644–4658. doi: 10.1074/jbc.M110.164392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Futerman AH. Intracellular trafficking of sphingolipids: relationship to biosynthesis. Biochim Biophys Acta. 2006;1758:1885–1892. doi: 10.1016/j.bbamem.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Yabu T, Shimuzu A, Yamashita M. A novel mitochondrial sphingomyelinase in zebrafish cells. J Biol Chem. 2009;284:20349–20363. doi: 10.1074/jbc.M109.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu BX, Rajagopalan V, Roddy PL, Clarke CJ, Hannun YA. Identification and characterization of murine mitochondria-associated neutral sphingomyelinase (MA-nSMase), the mammalian sphingomyelin phosphodiesterase 5. J Biol Chem. 2010;285:17993–18002. doi: 10.1074/jbc.M110.102988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novgorodov SA, Wu BX, Gudz TI, Bielawski J, Ovchinnikova TV, Hannun YA, Obeid LM. Novel pathway of ceramide production in mitochondria: thioesterase and neutral ceramidase produce ceramide from sphingosine and acyl-CoA. J Biol Chem. 2011;286:25352–25362. doi: 10.1074/jbc.M110.214866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birbes H, El Bawab S, Hannun YA, Obeid LM. Selective hydrolysis of a mitochondrial pool of sphingomyelin induces apoptosis. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:2669–2679. doi: 10.1096/fj.01-0539com. [DOI] [PubMed] [Google Scholar]
- 10.Dai Q, Liu J, Chen J, Durrant D, McIntyre TM, Lee RM. Mitochondrial ceramide increases in UV-irradiated HeLa cells and is mainly derived from hydrolysis of sphingomyelin. Oncogene. 2004;23:3650–3658. doi: 10.1038/sj.onc.1207430. [DOI] [PubMed] [Google Scholar]
- 11.Deng X, Yin X, Allan R, Lu DD, Maurer CW, Haimovitz-Friedman A, Fuks Z, Shaham S, Kolesnick R. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science. 2008;322:110–115. doi: 10.1126/science.1158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Paola M, Cocco T, Lorusso M. Ceramide interaction with the respiratory chain of heart mitochondria. Biochemistry. 2000;39:6660–6668. doi: 10.1021/bi9924415. [DOI] [PubMed] [Google Scholar]
- 13.Gudz TI, Tserng KY, Hoppel CL. Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J Biol Chem. 1997;272:24154–24158. doi: 10.1074/jbc.272.39.24154. [DOI] [PubMed] [Google Scholar]
- 14.Duan RD, Bergman T, Xu N, Wu J, Cheng Y, Duan J, Nelander S, Palmberg C, Nilsson A. Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J Biol Chem. 2003;278:38528–38536. doi: 10.1074/jbc.M305437200. [DOI] [PubMed] [Google Scholar]
- 15.Tomiuk S, Hofmann K, Nix M, Zumbansen M, Stoffel W. Cloned mammalian neutral sphingomyelinase: functions in sphingolipid signaling? Proc Natl Acad Sci U S A. 1998;95:3638–3643. doi: 10.1073/pnas.95.7.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomiuk S, Zumbansen M, Stoffel W. Characterization and subcellular localization of murine and human magnesium-dependent neutral sphingomyelinase. J Biol Chem. 2000;275:5710–5717. doi: 10.1074/jbc.275.8.5710. [DOI] [PubMed] [Google Scholar]
- 17.Milhas D, Clarke CJ, Idkowiak-Baldys J, Canals D, Hannun YA. Anterograde and retrograde transport of neutral sphingomyelinase-2 between the Golgi and the plasma membrane. Biochim Biophys Acta. 2010;1801:1361–1374. doi: 10.1016/j.bbalip.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krut O, Wiegmann K, Kashkar H, Yazdanpanah B, Kronke M. Novel tumor necrosis factor-responsive mammalian neutral sphingomyelinase-3 is a C-tail-anchored protein. J Biol Chem. 2006;281:13784–13793. doi: 10.1074/jbc.M511306200. [DOI] [PubMed] [Google Scholar]
- 19.Corcoran CA, He Q, Ponnusamy S, Ogretmen B, Huang Y, Sheikh MS. Neutral sphingomyelinase-3 is a DNA damage and nongenotoxic stress-regulated gene that is deregulated in human malignancies. Molecular cancer research : MCR. 2008;6:795–807. doi: 10.1158/1541-7786.MCR-07-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 21.Feoktistova A, Magnelli P, Abeijon C, Perez P, Lester RL, Dickson RC, Gould KL. Coordination between fission yeast glucan formation and growth requires a sphingolipase activity. Genetics. 2001;158:1397–1411. doi: 10.1093/genetics/158.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihara K, Omura T. Cytoplasmic chaperones in precursor targeting to mitochondria: the role of MSF and hsp 70. Trends Cell Biol. 1996;6:104–108. doi: 10.1016/0962-8924(96)81000-2. [DOI] [PubMed] [Google Scholar]
- 23.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 24.Braun HP, Schmitz UK. The mitochondrial processing peptidase. The international journal of biochemistry & cell biology. 1997;29:1043–1045. doi: 10.1016/s1357-2725(97)00032-0. [DOI] [PubMed] [Google Scholar]
- 25.Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 26.Ridder G, VonBargen E, Burgard D, Pickrum H, Williams E. Quantitative analysis and pattern recognition of two-dimensional electrophoresis gels. Clin Chem. 1984;30:1919–1924. [PubMed] [Google Scholar]
- 27.Chu A, Patterson J, Berger C, Vonderheid E, Edelson R. In situ study of T-cell subpopulations in cutaneous T-cell lymphoma. Diagnostic criteria. Cancer. 1984;54:2414–2422. doi: 10.1002/1097-0142(19841201)54:11<2414::aid-cncr2820541118>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Duong F, Wickner W. Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. Embo J. 1998;17:696–705. doi: 10.1093/emboj/17.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Bloois E, Haan GJ, de Gier JW, Oudega B, Luirink J. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J Biol Chem. 2006;281:10002–10009. doi: 10.1074/jbc.M511357200. [DOI] [PubMed] [Google Scholar]
- 30.Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, Maeda M, Mihara K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J Cell Sci. 2002;115:1895–1905. doi: 10.1242/jcs.115.9.1895. [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann T, Schlipf S, Sanz J, Neubert K, Stein R, Borner C. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J Cell Biol. 2003;160:53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapaport D. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 35.Van Loon AP, De Groot RJ, De Haan M, Dekker A, Grivell LA. The DNA sequence of the nuclear gene coding for the 17-kd subunit VI of the yeast ubiquinol-cytochrome c reductase: a protein with an extremely high content of acidic amino acids. Embo J. 1984;3:1039–1043. doi: 10.1002/j.1460-2075.1984.tb01924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto K, Perlman PS, Butow RA. Chapter 16 Targeting of green fluorescent protein to mitochondria. In: Liza EAS, Pon A, editors. Methods in Cell Biology. Vol. 65. Academic Press; 2001. pp. 277–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.