Abstract
A leucine zipper motif is conserved in the cytoplasmic domain of glycoprotein gp41 (gp41c) of all HIV-1 subtypes, but is not present in HIV-2 or SIV. The second leucine residue of the leucine zipper was mutated (L95R) to determine the role of this motif in HIV-1 replication and pathogenesis. The L95R mutant replicated to wild-type levels in activated peripheral blood mononuclear cells and CEMx174 cells. However, L95R replication was impaired in SupT1 cells and in the SCID-hu Thy/Liv mouse. Although the infectivity of wild-type virions and that of L95R mutant virions were equally sensitive to heat treatment, we found that L95R produced more defective virions, due to reduced surface expression and virion incorporation of the env glycoprotein. These results suggest that the L95 residue in the leucine zipper of gp41c of HIV-1 plays an important role in the env expression and virion incorporation that is required for viral replication and pathogenesis in the SCID-hu Thy/Liv mouse. The leucine zipper motif in gp41c may provide a novel anti-HIV-1 target.
INTRODUCTION
Like other lentivirus transmembrane (TM) glycoproteins, the TM glycoprotein (gp41) of HIV-1 has a long cytoplasmic domain of 150 amino acids (residues 704–854, gp41c). Deletions of gp41c impair HIV-1 replication and infectivity (Dubay et al., 1992; Gabuzda et al., 1992; Lee et al., 1989; Yu et al., 1993). The gp41c domains appear to be required in a cell type-dependent (Akari et al., 2000; Gabuzda et al., 1992; Murakami and Freed, 2000) and species-dependent fashion (Johnston et al., 1993; Kodama et al., 1989), suggesting that a host cell factor is involved. For instance, simian immunodeficiency virus (SIV) mutants with truncation mutations in gp41c arise from selection in human cells. However, infection of rhesus macaque peripheral blood lymphocytes in vitro or in vivo with the SIV mutant results in rapid reversion to full-length gp41 (Kodama et al., 1989).
The C-terminal domain of gp41 encodes a Tyr-based motif that mediates internalization of HIV-1 glycoproteins via interaction with the AP2 clathrin adaptor protein (Boge et al., 1998) and two lentivirus lytic peptides (LLPs), which are capable of binding and disturbing lipid bilayers (Miller et al., 1991). The ability of LLP-1 (residues 768–788) and LLP-2 (residues 826–854) to interact with lipid bilayers suggests membrane-related functions. LLP-1 also interacts with calmodulin (Miller et al., 1993; Srinivas et al., 1993) and LLP-2 inhibits Ca2+-dependent T-cell activation (Beary et al., 1998), implicating these amphipathic helices in protein–protein interactions and signal transduction. A leucine zipper motif (residues 789–815) in HIV-1 gp41c has also been shown to interact with lipid bilayers (Kliger and Shai, 1997). Mutational analysis has shown that this amphipathic helix region of gp41c is involved in its interaction with p115-RhoGEF, a specific guanine nucleotide exchange factor and activator of the RhoA GTPase (Zhang et al., 1999).
We examined the role of this leucine zipper in HIV-1 replication and pathogenesis by mutating the second leucine residue of this motif (L95, amino acid 95 of gp41c or residue 798 of gp160) to an arginine residue. The L95R mutant replicated to wild-type (NL4-3) levels in peripheral blood mononuclear cells (PBMC) and CEMx174 cells. However, L95R replication was impaired in SupT1 cells and in the SCID-hu Thy/Liv mouse. The L95R mutant env fused efficiently to both SupT1 and CEMx174 cells and L95R mutant virions showed wild-type sensitivity to heat inactivation. We showed that the L95R mutation impaired HIV-1 replication by reducing surface expression and virion incorporation of the env glycoprotein. The findings demonstrate that the L95 residue in the leucine zipper motif of gp41c plays an important role in the surface expression and incorporation of env proteins into infectious HIV-1 virions, which is required for efficient HIV-1 replication and pathogenesis in vivo.
RESULTS
Cell type-dependent phenotypes of the L95R mutant
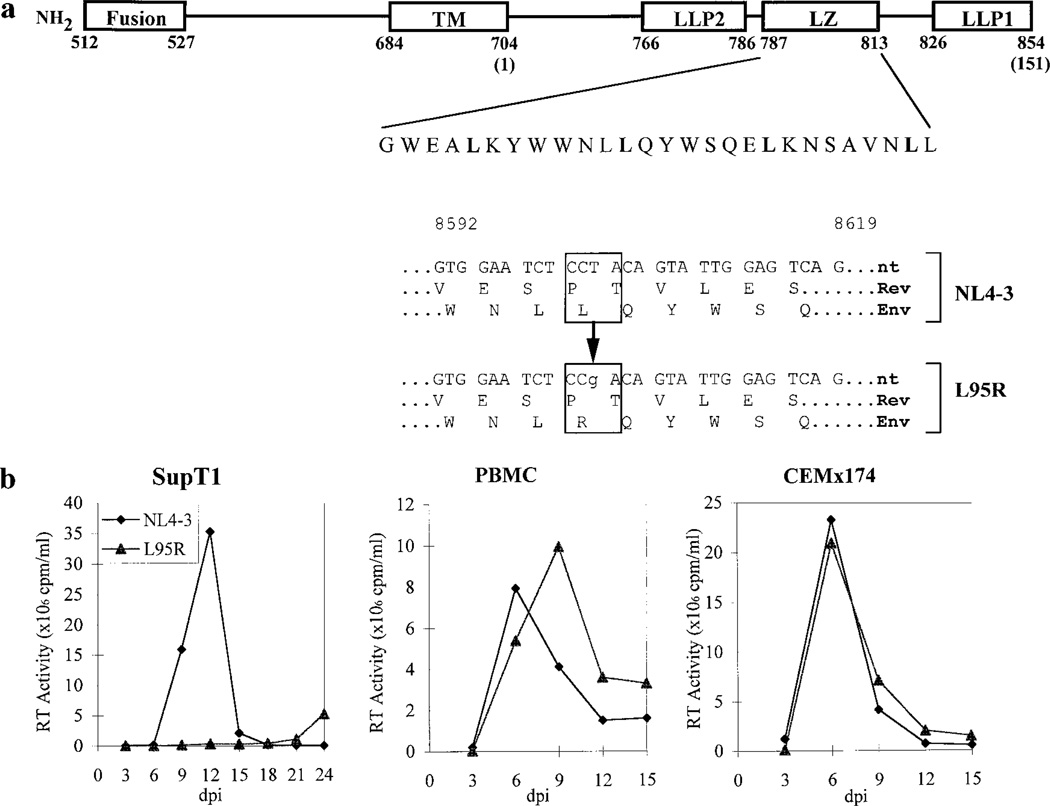
A putative leucine zipper (LZ) motif (LX6LX6LX6L) within the cytoplasmic domain of gp41 (gp41c) of HIV-1 is conserved in all HIV-1 subtypes in the Los Alamos HIV databases. This LZ motif, however, is not present in the HIV-2 or SIV transmembrane proteins (data not shown). To examine the function of the leuzine zipper motif within the cytoplasmic domain of HIV-1 gp41, a point mutation (L95R) was introduced at amino acid position 95 of gp41c (Fig. 1a). L95 is the second leucine within the putative leucine zipper motif. The L95R mutation did not affect the amino acid coded in the overlapping rev open reading frame.
FIG. 1.
Introduction of a mutation in the leucine zipper motif of the cytoplasmic domain of HIV-1 gp41. (a) Functional domains of gp41. Fusion, fusion peptide; TM, transmembrane domain; LLP1 and LLP2, lentivirus lytic peptides 1 and 2. The numbers 1–151 indicate residue positions in the cytoplasmic domain of HIV-1 gp41 (gp41c). The LZ motif is shown in single-letter amino acid code, with the leucines comprising the LZ motif highlighted. The numbers 512–854 indicate residue positions in the HIV-1 gp160 env polypeptide. The Rev open reading frame remains unchanged. The numbers 8592–8619 indicate nucleotide positions in the NL4-3 genome. (b) L95R replication in vitro was cell type-dependent. SupT1, PBMC, or CEMx174 cells were infected with equivalent RT units of NL4-3 or L95R. Virus replication (RT activity) was monitored every 3 days for 15 or 24 days postinfection. The data are representative of three (PBMC) or four (T-cell lines) experiments with similar replication kinetics.
To compare the replication activity of the mutant L95R virus with that of wild-type NL4-3 virus, human T-lymphoid cell lines and phytohemagglutinin (PHA)-stimulated PBMC were infected and virus replication was monitored by measuring reverse transcriptase (RT) activity in the culture supernatants. L95R replication was impaired in SupT1 (Fig. 1b), Jurkat, and H9 cell lines (Zhang et al., 1999). However, L95R showed no observable replication defect in PHA-activated PBMC and CEMx174 cells (Fig. 1b). L95R virus reached similar peak RT activity at a rate equivalent to that of NL4-3. The results suggest that the leucine zipper of gp41c plays a role in cell type-dependent HIV-1 replication in vitro.
Impaired HIV-1 replication and pathogenicity of L95R in the SCID-hu Thy/Liv mouse
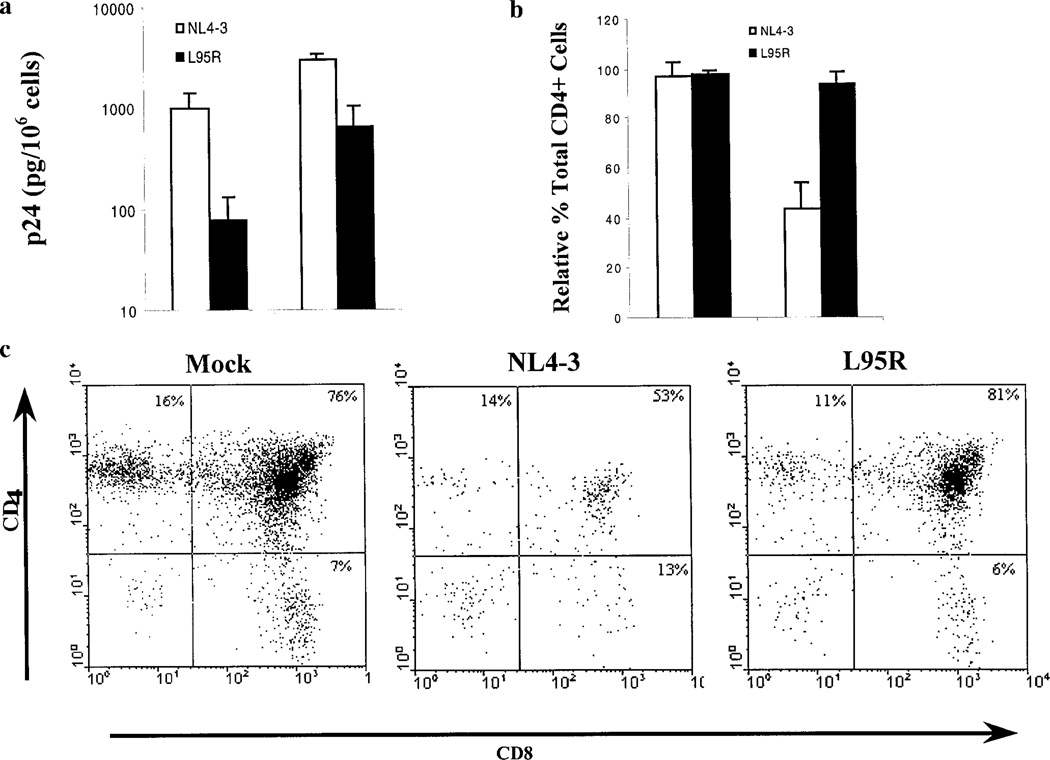
To examine the possible replication defect of the L95R mutant in vivo, SCID-hu Thy/Liv mice were infected with equivalent infectious units of NL4-3 or L95R. NL4-3 virus replication was detected at 2 weeks postinfection (wpi) and peaked at 4 wpi, with an average p24 level of 1037 pg per 106 thymocytes at 2 wpi and 3085 pg per 106 thymocytes at 4 wpi (Fig. 2a). In contrast, L95R virus replication was significantly reduced at 2 and 4 wpi, with an average p24 level of 80 and 660 pg per 106 thymocytes, respectively (Fig. 2a). In accordance with the p24 data, L95R failed to induce MHC I expression at 2 wpi and induced lower levels at 4 wpi (data not shown), which is accompanied by HIV-1 replication in the SCID-hu Thy/Liv mouse (Kovalev et al., 1999).
FIG. 2.
L95R replication/pathogenesis was impaired in the SCID-hu Thy/Liv mouse. (a) HIV-1 replication in SCID-hu Thy/Liv mice. SCID-hu Thy/Liv mice were infected with equivalent infectious units of NL4-3 or L95R. Virus replication (p24, pg/106 thymocytes) was monitored following infection. The data are representative of three independent experiments with 8–10 SCID-hu Thy/Liv mice for each virus. Error bars represent standard errors. (b) HIV-1 pathogenesis in SCID-hu Thy/Liv mice. Pathogenesis was determined by the percentage of total CD4+ (CD4+CD8+ and CD4+CD8−) thymocytes. Values for NL4-3- and L95R-infected Thy/Liv organs are presented relative to the mock-infected Thy/Liv organ (set as 100%). Error bars represent standard errors from 8–10 SCID-hu mice. (c) A representative FACS analysis of CD4/CD8 staining is shown to illustrate the relative thymocyte depletion by NL4-3 and L95R at 4 wpi in comparison to mock infection.
NL4-3 virus replication is accompanied by preferential depletion of CD4+CD8+ and CD4+CD8− thymocytes (Su et al., 1995). In parallel to impaired virus replication, pathogenesis of L95R was impaired (Figs. 2b and 2c). At 4 wpi, the percentage of total CD4+ thymocytes in NL4-3-infected Thy/Liv organs decreased significantly (P < 0.001) compared with that of mock-infected Thy/Liv organs (set at 100%). On the other hand, L95R induced no significant thymocyte depletion by 4 wpi (Figs. 2b and 2c). For example, the total CD4+ thymocytes of NL4-3-infected Thy/Liv organs decreased to about 40% of mockinfected Thy/Liv organs at 4 wpi. In contrast, the L95R-infecteed Thy/Liv organs showed no significant decrease in CD4+ thymocyte populations, compared with mock-infecteed Thy/Liv organs. L95R was also impaired in other measurements of pathogenesis, including a decrease in the total percentage of live cells (based on light scatter profiles) and inversion of the CD4/CD8 ratio (Fig. 2c, and data not shown). Therefore, the L95 residue in the LZ motif of gp41c was required for efficient HIV-1 replication and pathogenesis in vivo.
The L95R mutant env protein did not affect the early stage of the virus life cycle and L95R virions showed similar sensitivity to heat inactivation
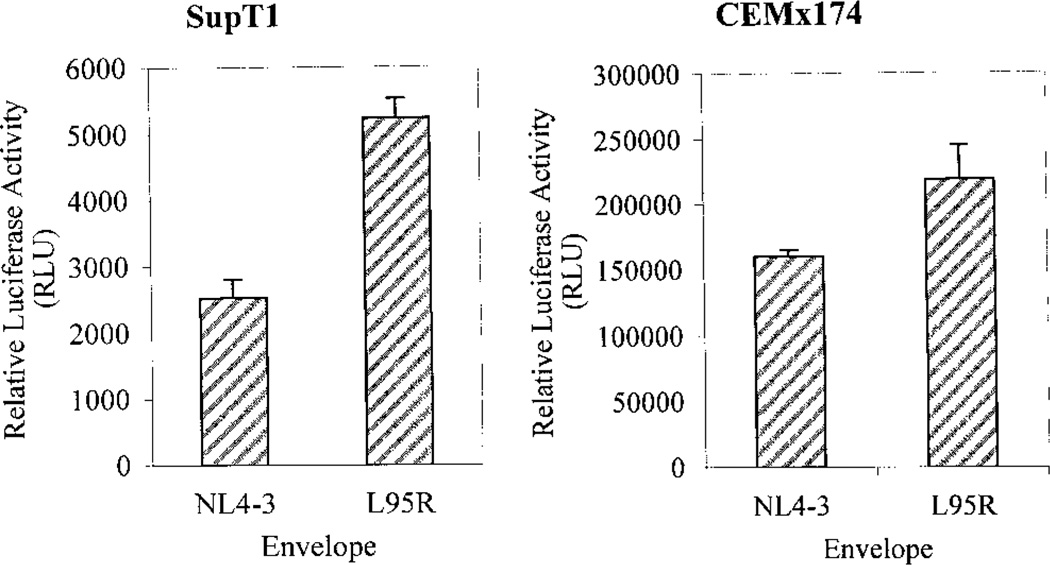
A single-cycle infectivity assay was performed using pseudotyped viruses generated from an env expression vector and an env-deficient HIV-1 provirus containing a luciferase reporter gene, NL4-luc-R−E− (Connor et al., 1995). This infectivity assay assesses the ability of the env protein to mediate binding, fusion, and uncoating of the pseudotyped virion in the context of virus-to-cell transmission. SupT1 or CEMx174 cells were infected with equivalent infectious units of the pseudotyped viruses produced from 293T cells and luciferase expression was assayed at 2 days postinfection (dpi). The L95R-env pseudotyped virus was able to produce levels of luciferase equivalent to that of NL4-3 in SupT1 cells (Fig. 3, left). Similar results were obtained in CEMx174 cells (Fig. 3, right). Virus stocks derived from either defective provirus (NL4-luc-R−E−) or env (pcDNA3-env) alone were not infectious (data not shown). These data suggest that the L95R mutant env functioned normally in the pseudotyped viruses when expressed from a CMV promoter.
FIG. 3.
NL4-3 and L95R env-pseudotyped virions showed similar reporter gene expression in the single-round replication assay. 293T cells were cotransfected with NL4-luc-R-E- and NL4-3- or L95R-env expressing plasmids. SupT1 or CEMx174 cells were infected with equivalent infectious units of viruses pseudotyped with NL4-3 or L95R env glycoproteins. Results are representative of three independent experiments with duplicate samples. Error bars represent standard errors.
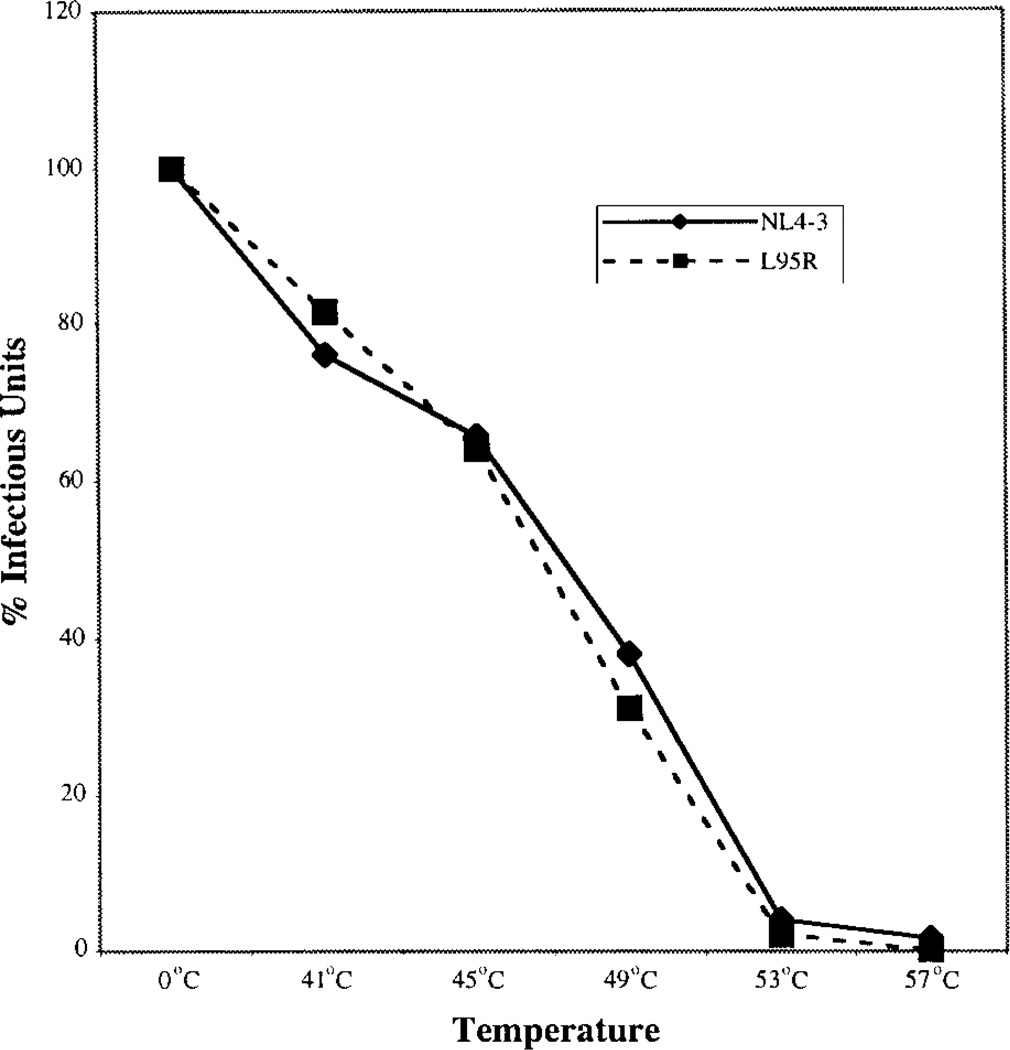
We tested the stability of L95R virions to heat inactivation by incubating NL4-3 and L95R virions at various temperatures (Fig. 4). Both NL4-3 and L95R virions were similarly heat-inactivated at 41 to 49°C and completely inactivated at 53 and 57°C. These results suggest that L95R virions, once assembled and released, were similarly sensitive to heat inactivation in comparison to wildtype virions.
FIG. 4.
NL4-3 and L95R virions showed similar sensitivity to heat inactivation. Heat inactivation of NL4-3 and L95R virions produced from 293T cells was performed with equal number of IUs. Viral stocks incubated on ice (0°C) showed no reduction in IUs and were used as controls (100%). The remaining IU value after heat treatment was determined and is presented as the percentage IU. The experiment was performed three times in duplicate.
The L95R mutation reduced surface expression of the gp120 env glycoprotein
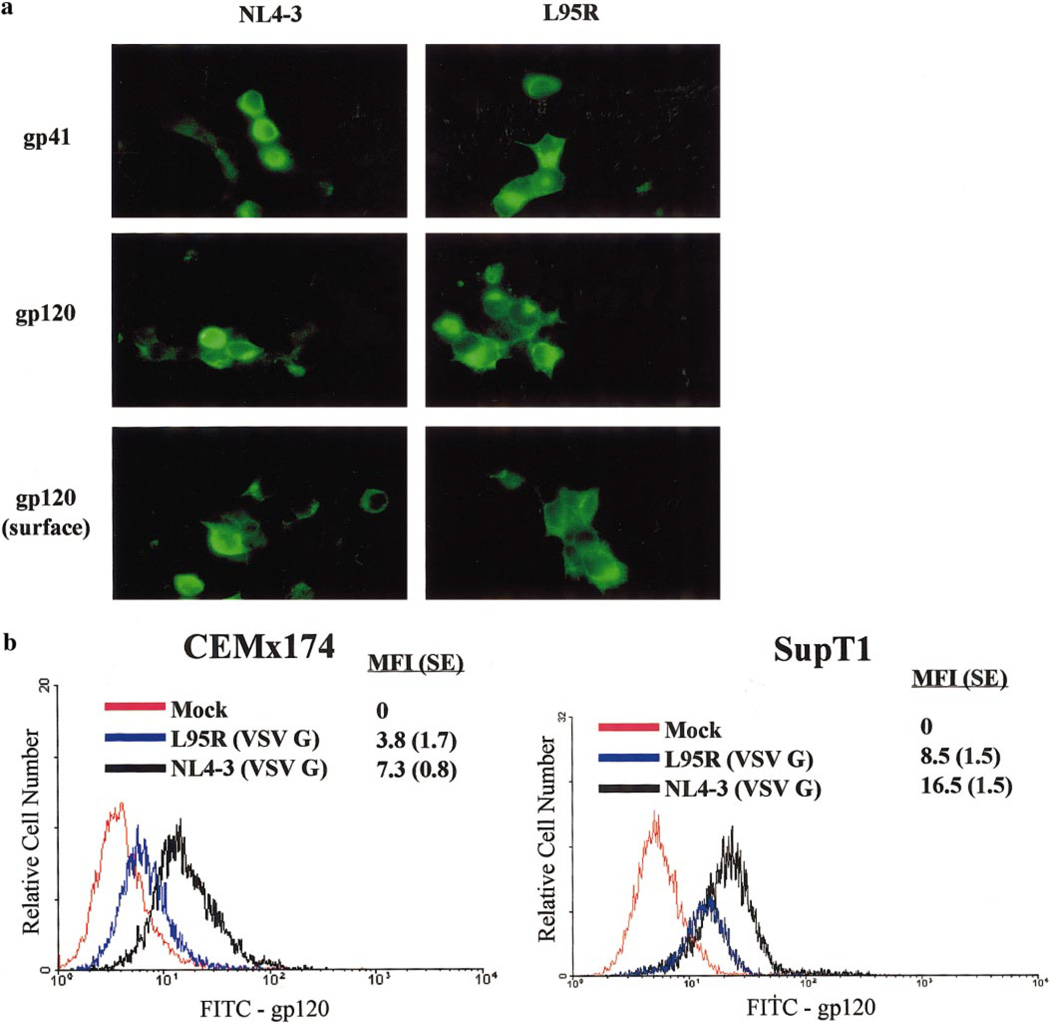
To determine the expression and subcellular localization of L95R mutant glycoproteins, surface expression of gp120 and intracellular expression of gp120 and gp41 were studied by indirect immunofluorescence (Fig. 5a). 293T cells transfected with NL4-3 or L95R proviral DNA showed no discernible differences in subcellular localization of gp120 or gp41.
FIG. 5.
L95R showed reduced cell surface expression of gp120. (a) Subcellular distribution of gp120 and gp41 in 293T cells transfected with NL4-3 or L95R provirus DNA was determined. Intracellular gp41 (top) or gp120 (middle) or surface gp120 (bottom) was visualized by standard fluorescence microscopy. Mock-transfected cells were used as negative controls. (b) FACS analysis of cell surface expression of gp120. CEMx174 or SupT1 cells were infected with VSV G-pseudotyped NL4-3 or L95R viruses. Cell surface gp120 levels were measured by FACS with an anti-gp120 antibody. The MFI is presented and standard errors (SE) are shown. MFI from each cell sample using an isotype control antibody was the same as mock-infected cells (MFI is set to 0). Three independent experiments were performed with similar results.
Cell surface expression of gp120 was measured by FACS analysis of T cells infected with VSV G-pseudotyped NL4-3 and L95R viruses (Fig. 5b). Both CEMx174 and SupT1 cells infected with L95R showed decreased levels of cell surface expression of gp120 compared to NL4-3-infected cells. A twofold reduction of mean fluorescence intensity (MFI) was reproducibly observed in both CEMx174 and SupT1 cells. The data suggest that the L95R mutation reduced the cell surface expression of env glycoproteins.
L95R produced more defective particles
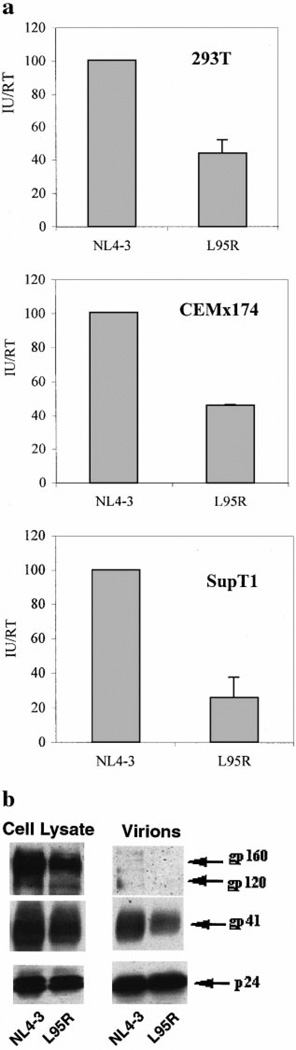
T-cell lines and 293T cells were transfected with proviral DNA and culture supernatants were analyzed by RT (total virions) and infectious unit (IU, infectious virions) assays. Transfection of L95R proviral DNA into 293T cells yielded comparable reverse RT activity (total virions) in culture supernatants to that of wild-type NL4-3 (data not shown). However, L95R viral stocks contained 32% less IU than NL4-3 viral stocks (P < 0.01), indicating that L95R produced more defective viral particles than NL4-3 (Fig. 6a). Transfection of L95R proviral DNA in SupT1 and CEMx174 cells also yielded RT activity equivalent to transfection of NL4-3 provirus, suggesting that the L95R mutant did not affect the expression, assembly, and release of total viral particles (data not shown). However, L95R produced significantly less infectious units in SupT1 (70% reduction) or CEMx174 (50% reduction) cells, as indicated by the ratio of IUs to RT units (IU/RT, Fig. 6a). These data indicate that the L95R mutation affected the assembly or stability of infectious virions.
FIG. 6.
L95R produced more defective virions with reduced incorporation of env glycoproteins. (a) L95R produced more defective particles than NL4-3 in various cell lines. NL4-3 and L95R proviruses were transfected into 293T, CEMx174, and SupT1 T cells. Total virions were measured by RT activity in the supernatants and infectious virions (IU) were titered by MAGI assays. Data shown are the average of four independent experiments. Error bars represent standard errors. The IU/RT ratio of NL4-3 was set to 100%. A significant difference is observed between NL4-3 and L95R using Student’s T test (P < 0.01). (b) L95R virions showed decreased env glycoproteins. Cell lysates and purified virions (pellet) were blotted with antibodies specific for gp41, gp120, or p24. The positions of gp160, gp120, gp41, and p24 are indicated. The experiment was repeated three times with similar results.
The L95R mutation decreased env glycoprotein incorporation into virions
To examine viral protein expression, virion assembly, and virion release, cell lysates or pelleted virions from 293T cells transfected with proviral DNA were analyzed by Western blot using antibodies specific for gp41, gp120, or p24 (Fig. 6b and Table 1). Comparable levels of cell-associated gp41 and gp120 (relative to levels of p24/p25) were synthesized upon transfection of NL4-3 and L95R proviruses, indicating that the L95R mutation did not significantly affect the synthesis and proteolytic cleavage of the env precursor. Virions in culture supernatants were pelleted by centrifugation in a 20% sucrose cushion and also analyzed by Western blot. Relative to p24 levels, L95R virions contained approximately two- to fivefold less gp41 and gp120 than NL4-3 virions, suggesting a possible defect in virion incorporation of gp120/gp41 (Table 1).
TABLE 1.
HIV-1 gp120/p24 and gp41/p24 Ratios
| Proteinsa | C-NL4-3b | C-L95R | V-NL4-3 | V-L95R |
|---|---|---|---|---|
| gp120/p24 | 0.31 (0.13)c | 0.2 (0.08) | 0.34 (0.05) | 0.07 (0.05) |
| gp41/p24 | 0.77 (0.25) | 0.85 (0.31) | 0.57 (0.12) | 0.28 (0.11) |
The level of each protein was calculated by densitometry.
Cell-associated (C−) or virion-associated (V−) proteins were determined by densitometry.
Standard deviations from multiple exposures of two experiments are shown in parentheses.
DISCUSSION
The L95R mutant was constructed to investigate the role of a leucine zipper motif in gp41c in HIV-1 replication and pathogenesis. We report that the L95R mutation in the leucine zipper motif in gp41c impaired HIV-1 replication and pathogenesis in vivo by reducing the surface expression and virion incorporation of env glycoproteins.
L95R replicated to wild-type levels in PHA-stimulated PBMC and CEMx174 cells, but was impaired in SupT1, Jurkat, and H9 cell lines (Fig. 1 and Zhang et al., 1999). In two recent reports, gp41c deletion or truncation mutants replicate in a cell type-dependent manner in vitro probably due to defective env incorporation in virions (Akari et al., 2000; Murakami and Freed, 2000). Using a truncation mutant lacking all but six amino acids in gp41c, Murakami and Freed (2000) have recently reported a similar replication-defective phenotype as L95R in Jurkat and SupT1 cells. This mutant replicates to wild-type levels in MT-4 cells. In contrast to L95R, however, this and other gp41c truncation mutants are also unable to replicate in PBMC (Dubay et al., 1992; Murakami and Freed, 2000; Yu et al., 1993). The discrepancy between gp41c truncation mutants and L95R may be because the truncation mutants also lack potential functional domains in addition to the leucine zipper in gp41c. It is possible that replication in PBMC, unlike that in Jurkat and SupT1 cells, requires functional motifs in gp41c other than the leucine zipper. This difference between the L95R mutant and the gp41c truncation mutants also suggests that the L95R mutation probably disrupted a specific functional domain (LZ) of gp41c, but not the entire gp41c structure.
L95R replication was impaired in the SCID-hu Thy/Liv model (Fig. 2) or in the human fetal thymus organ culture (HF-TOC) model (S. Kao and L. Su, unpublished results). The in vivo models entail virus replication in a repertoire of cell types. The reduced pathogenic effects of L95R were likely a consequence of reduced replication since the L95R replication level was generally below the threshold of thymocyte depletion mediated by NL4-3 (Su et al., 1995). In the SCID-hu Thy/Liv mice with higher levels of L95R replication, reduction in cytopathic effects in L95R-infected SCID-hu Thy/Liv mice was proportional to reduction in HIV-1 replication (Fig. 2; and data not shown).
The block in L95R virus replication may occur after HIV-1 gene expression since L95R-env pseudotyped virions were able to undergo a single round of replication at wild-type levels in both SupT1 and CEMx174 cells (Fig. 3). It is possible the L95R replication defect in SupT1 cells may manifest itself only after multiple rounds of replication. Alternatively, overexpression of L95R mutant env from the strong CMV promoter in the pcDNA3 vector in cotransfected 293T cells may have compensated for the putative defect. Our preliminary results showed that the IU/RT ratio of L95R-env pseudotyped viruses was the same as that for NL4-env pseudotyped viruses (data not shown). This suggests that one defect of L95R replication may be in the level of env expression.
The L95R mutation appeared to interfere with envelope glycoprotein surface expression and incorporation into virions (Figs. 5 and 6). Normal levels of surface env protein are expressed by some gp41c truncation mutants (Dubay et al., 1992; Murakami and Freed, 2000; Yu et al., 1993). However, previous reports have also shown that truncation of gp41c affects virion incorporation of env glycoprotein (Akari et al., 2000; Dubay et al., 1992; Murakami and Freed, 2000). It has been suggested that the cell type-dependent virion incorporation of env glycoproteins of gp41c mutants may result in the cell type-dependent replication phenotype (Akari et al., 2000; Murakami and Freed, 2000). However, other studies have shown that gp41c truncation mutations do not affect env glycoprotein incorporation into virions (Gabuzda et al., 1992; Johnston et al., 1993; Mammano et al., 1995; Salzwedel et al., 1993; Wilk et al., 1992). These discrepancies may be mollified by the specific mutations studied and/or the cell types used to study the effects of the various mutations on virus replication.
In this report, we failed to detect any dramatic differences in permissive (CEMx174) and restrictive (SupT1) cell lines either transfected or infected with L95R and NL4-3. Thus, similar reduction in surface expression of gp120 (Fig. 5b) and production of defective L95R virions (Fig. 6a) were detected in CEMx174 and SupT1 cells. The seemingly less dramatic reduction in IU/RT ratios of L95R in CEMx174 cells (50% of NL4-3) than in SupT1 cells (30% of NL4-3) may contribute to the cell type-dependent replication. The CEMx174 cell line is a fusion product of the human B-cell line 721.174 and human T-cell line CEM (Salter et al., 1985). These cells may contain a host factor (from B cells?) that facilitates virus replication despite decreased surface expression and virion incorporation of env glycoproteins. Transfection of T cells with proviral DNA or infection by VSV-G pseudotyped HIV-1 assesses only a single round of replication. Since L95R replicated to wild-type levels in CEMx174 cells (Fig. 1) over multiple rounds of replication, the defect of L95R mutant in CEMx174 may potentially be compensated for during cell-to-cell transmissions. Interestingly, the L95R mutant replication in SupT1 cells was detectable at 21–24 days postinfection (Fig. 1b). The recovered viruses were not due to reversion of L95R because they showed replication kinetics similar to that of the parental L95R in SupT1 cells (S. Kao and L. Su, unpublished results), suggesting a delayed replication kinetics of L95R in SupT1 cells. In L95R-infected SCID-hu Thy/Liv organs, decreased env surface expression and virion incorporation were likely responsible for the impaired replication and pathogenesis (Fig. 2).
The cell type-dependence phenotype of L95R strongly suggests the involvement of a host factor(s) in mediating the effects of gp41c. It is of interest that the permissive PBMC and CEMx174 cells for L95R replication are also permissive for wild-type SIV replication, whose TM gp41 proteins lack the LZ motif (L. Su, unpublished results). Both the SIV gp41c and the HIV-1 L95R gp41c failed to interact with the host protein p115 RhoGEF that interacts with wild-type HIV-1 gp41c (Zhang et al., 1999). Our data showed that T-cell lines (SupT1, Jurkat, and H9) and SCID-hu Thy/Liv organs expressed high levels of p115, whereas no significant p115 was detected in PHA-activated PBMC (unpublished results). The interaction of gp41c with p115 may be important to overcome the inhibitory effect of p115 and RhoA (Wang et al., 2000).
Other host cell factors may also contribute to the cell type-dependent replication phenotypes of gp41c mutants. For example, a specific tyrosine-based sorting motif (YXXO, where X is any amino acid and O is an amino acid with a bulky hydrophobic side chain) is the principal motif responsible for the basolateral targeting of env glycoprotein and polarized budding of viral particles (Lodge et al., 1997). The interaction of this motif in gp41c with the AP-2 clathrin complex has been reported to mediate rapid glycoprotein internalization (Boge et al., 1998). The gp41c domain has also been shown to interact with α-catenin (Kim et al., 1999), but the biological significance of the interaction is not clear. Further studies need to be conducted to determine the relationship between the host factor(s) and L95R mutant replication. Understanding the role of the LZ motif in gp41c will help to elucidate the structure and function of gp41c and define a novel target for anti-HIV therapy.
MATERIALS AND METHODS
Cells
293T and U-373-MAGI-CXCR4 (Vodicka et al., 1997) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated etal bovine serum (FBS). PBMC and T-cell lines were cultured in RPMI 1640 containing 10% heat-inactivated FBS. The CEMx174 cell line is a fusion product of the human B-cell line 721.174 and human T-cell line CEM (Salter et al., 1985). The SupT1 cell line is a non-Hodgkin’s T-cell lymphoma (Smith et al., 1984).
Construction of the L95R mutant
To construct an infectious NL4-3 proviral clone containing the L95R mutation in gp41, primers with the introduced mutation (nt 8590–8637) were used to perform site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis protocol (Stratagene). The mutated HIV-1 fragment was sequenced and cloned into the proviral DNA clone pNL4-3 (Adachi et al., 1986).
DNA transfection
293T cells were transfected with NL4-3 or L95R proviral DNA using Effectene (Qiagen). To transfect various T-cell lines, cells were electroporated using a Gene Pulser (Bio-Rad) at 0.2 kV and 960 µF. Culture supernatant was harvested 48 h posttransfection (hpt) and total virions were measured by RT activity (Buckheit and Swanstrom, 1991). The infectious virion was titered on U-373-MAGI-CXCR4 indicator cells.
Viral replication in vitro
Relative virus infectivity was assayed in PHA-activated PBMC, CEMx174, and SupT1 cells. A total of 5 × 105 cells were infected with viral supernatants containing 105 RT units. Culture supernatants were monitored for RT activity every 3 days.
HIV-1 replication and pathogenesis in SCID-hu Thy/Liv mice
Infection of SCID-hu Thy/Liv mice was performed as previously described (Kovalev et al., 1999; Su et al., 1995).
Pseudotyped virus production and infection
pcENV-NL4 is a pcDNA3-based vector containing the EcoRI–XhoI fragment of the NL4-3 env gene (H. Zhang and L. Su, UNC-CH). The EcoRI–XhoI fragment of L95R was cloned into pcDNA3 to produce the pcENV-L95R plasmid construct. 293T cells were cotransfected with 0.4 µg each of the env-deficient pNL43-Luc-R−E− reporter genome (Connor et al., 1995) and pcENV-NL4 or pcENV-L95R env expression vector using Effectene Transfection Reagent (Qiagen) as described above. Supernatants containing the pseudotyped viruses were collected 48 hpt and analyzed by luciferase, RT, and MAGI assays.
CEMx174 and SupT1 cells were infected for 2–4 h with supernatants containing equivalent infectious units (MAGI IU, determined as above) of pseudotyped viruses in the presence of 8 µg/ml polybrene. Cells were then washed three times with PBS to remove residual virus particles. Two days postinfection, cells were washed once with PBS and lysed for luciferase activity.
Heat-stability analysis of HIV-1 stocks
Equal numbers of NL4-3 or L95R infectious units determined on MAGI cells were incubated at the indicated temperatures for 10 min and the remaining IUs were determined on U-373-MAGI-CXCR4 indicator cells as above. Triplicate samples were included in each experiment.
Western blot analysis
Transfected 293T cells were lysed 48 hpt in 0.5% NP-40 lysis buffer. To detect viral proteins in virions, supernatants containing viruses were centrifuged for 2 h at 4°C in a standard 20% sucrose cushion (Akari et al., 2000). Viral pellets were lysed in 1% Triton X-100 and analyzed by standard Western blot with anti-gp41 (Abacioglu et al., 1994), anti-gp120 (Chesebro and Wehrly, 1988), and anti-p24 (Chesebro et al., 1992) monoclonal antibodies (provided by the NIH AIDS Research and Reference Reagent Program). The relative level of each protein was determined by densitometry.
Indirect immunofluorescence
293T cells were transfected with NL4-3 or L95R proviral DNA. At 40 hpt, cells were fixed with 2% paraformaldehyde for surface gp120 staining or 2% formaldehyde + 0.05% Triton X-100 for intracellular staining of gp41 or gp120.
Flow cytometric analysis of gp120 cell surface expression
For efficient single-cycle HIV-1 replication assays (Bartz et al., 1996), pci-VSV G DNA was cotransfected with either NL4-3 or L95R proviral DNA in 293T cells using Effectene (Qiagen). Supernatants containing the VSV-G pseudotyped viruses were collected 48 hpt. SupT1 or CEMx174 cell lines were infected with equivalent RT units of VSV G-pseudotyped NL4-3 and L95R viruses in the presence of 100 ng/ml AMD3100 {1,1′-[1,4-phenylenebis(methylene)]-bis-1,4,8,11-tetra-azacyclotetradecane} (kindly provided by J. Moore, Aaron Diamond AIDS Research Center, New York) and 8 µg/ml polybrene. AMD3100 is a potent inhibitor of entry of CXCR4-tropic HIV-1 strains (Donzella et al., 1998; Schols et al., 1997). Cells were then washed three times with PBS and maintained in medium containing 100 ng/ml AMD3100. Two days postinfection, cells were stained with an anti-gp120 antibody (0.5β), which recognizes the V3 loop region of NL4-3 gp120 (Burkly et al., 1995). Cells were then stained with goat anti-mouse IgG-FITC. Mouse IgG2b was used as an isotype control for background.
ACKNOWLEDGMENTS
We dedicate this paper to the memory of Dr. Eric D. Miller. This work was supported by National Institutes of Health (NIH) Grants AI41356 and AI 48407 (L.S.). E.M. was funded in part by the American Foundation for AIDS Research (amfAR 70520-28-RFI). S.K. was partly supported by a Public Health Service predoctoral training grant (AI07273). We thank Drs. R. Swanstrom, J. Ting, A. Baldwin, C. Huang, A. Kaplan, and S. Pettit for critically reading the manuscript and the NIH AIDS Reagent Program for providing the anti-HIV protein hybridomas. We thank K. Duus for advice on immunostaining, S. Pettit for advice on HIV-1 virion precipitation, G. Kovalev and J. Smith for the SCID-hu mouse infection, and L. Wang and M. Townsend for technical assistance. We also thank members of the Su laboratory and the Swanstrom laboratory for discussions and the University of North Carolina at Chapel Hill Center for AIDS Research (CFAR), an NIH funded program (9P30 AI50410), and the UNC DLAM and FACS core facilities for their help with animal care and FACS analysis.
REFERENCES
- Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 2000;74:4891–4893. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz SR, Rogel ME, Emerman M. Human immunodeficiency virus type 1 cell cycle control: vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J. Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beary TP, Tencza SB, Mietzner TA, Montelaro RC. Interruption of T-cell signal transduction by lentivirus lytic peptides from HIV-1 transmembrane protein. J. Peptide Res. 1998;51:75–79. doi: 10.1111/j.1399-3011.1998.tb00419.x. [DOI] [PubMed] [Google Scholar]
- Boge M, Wyss S, Bonifacino JS, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- Buckheit RW, Jr, Swanstrom R. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res. Hum. Retroviruses. 1991;7:295–302. doi: 10.1089/aid.1991.7.295. [DOI] [PubMed] [Google Scholar]
- Burkly L, Mulrey N, Blumenthal R, Dimitrov DS. Synergistic inhibition of human immunodeficiency virus type 1 envelope glycoprotein-mediated cell fusion and infection by an antibody to CD4 domain 2 in combination with anti-gp120 antibodies. J. Virol. 1995;69:4267–4273. doi: 10.1128/jvi.69.7.4267-4273.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: Definition of critical amino acids involved in cell tropism. J. Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Donzella GA, Schols D, Lin SW, Este JA, Nagashima KA, Maddon PJ, Allaway GP, Sakmar TP, Henson G, De Clerq E, Moore JP. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- Dubay JW, Roberts SJ, Hahn BH, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda DH, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PB, Dubay JW, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EM, Lee KH, Kim JW. The cytoplasmic domain of HIV-1 gp41 interacts with the carboxyl-terminal region of α-catenin. Mol. Cells. 1999;9:281–285. [PubMed] [Google Scholar]
- Kliger Y, Shai Y. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers. Biochemistry. 1997;36:5157–5169. doi: 10.1021/bi962935r. [DOI] [PubMed] [Google Scholar]
- Kodama T, Wooley DP, Naidu YM, Kestler HD, Daniel MD, Li Y, Desrosiers RC. Significance of premature stop codons in env of simian immunodeficiency virus. J. Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalev G, Duus K, Wang L, Lee R, Bonyhadi M, Ho D, McCune JM, Kaneshima H, Su L. Induction of MHC class I expression on immature thymocytes in HIV-1-infected SCID-hu Thy/Liv mice: Evidence of indirect mechanisms. J. Immunol. 1999;162:7555–7562. [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hu W, Fisher AG, Looney DJ, Kao VF, Mitsuya H, Ratner L, Wong-Staal F. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res. Hum. Retroviruses. 1989;5:441–449. doi: 10.1089/aid.1989.5.441. [DOI] [PubMed] [Google Scholar]
- Lodge R, Lalonde JP, Lemay G, Cohen EA. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger HG. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Garry RF, Jaynes JM, Montelaro RC. A structural correlation between lentivirus transmembrane proteins and natural cytolytic peptides. AIDS Res. Hum. Retroviruses. 1991;7:511–519. doi: 10.1089/aid.1991.7.511. [DOI] [PubMed] [Google Scholar]
- Miller MA, Mietzner TA, Cloyd MW, Robey WG, Montelaro RC. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retroviruses. 1993;9:1057–1066. doi: 10.1089/aid.1993.9.1057. [DOI] [PubMed] [Google Scholar]
- Murakami T, Freed EO. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- Salzwedel K, Johnston PB, Roberts SJ, Dubay JW, Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 1993;67:5279–5288. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schols D, Este JA, Henson G, De Clerq E. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor Fusin/CXCR-4. Antiviral Res. 1997;35:147–156. doi: 10.1016/s0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- Smith SD, Shatsky M, Cohen PS, Warnke R, Link MP, Glader BE. Monoclonal antibody and enzymatic profiles of 2human malignant T-lymphoid cells and derived cell lines. Cancer Res. 1984;44:5657–5660. [PubMed] [Google Scholar]
- Srinivas SK, Srinivas RV, Anantharamaiah GM, Compans RW, Segrest JP. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J. Biol. Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- Su L, Kaneshima H, Bonyhadi M, Salimi S, Kraft D, Rabin L, McCune JM. HIV-1-induced thymocyte depletion is associated with indirect cytopathogenicity and infection of progenitor cells. in vivo. Immunity. 1995;2:25–36. doi: 10.1016/1074-7613(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Vodicka MA, Goh WC, Wu LI, Rogel ME, Bartz SR, Schweickart VL, Raport CJ, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang H, Solski PA, Hart MJ, Der CJ, Su L. Modulation of HIV-1 replication by a novel RhoA effector activity. J. Immunol. 2000;164:5369–5374. doi: 10.4049/jimmunol.164.10.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- Yu X, Yuan X, McLane MF, Lee TH, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang L, Kao S, Whitehead IP, Hart MJ, Liu B, Duus K, Burridge K, Der CJ, Su L. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr. Biol. 1999;9:1271–1274. doi: 10.1016/s0960-9822(99)80511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]