Abstract
MicroRNAs (miRNAs) are a class of highly conserved small noncoding RNAs that negatively regulate gene expression by imperfectly base pairing to the 3’-untranslated region of their target mRNAs, leading to mRNA degradation or translational inhibition. The emerging field of miRNA biology has begun to unravel roles for these regulatory molecules in a variety of biological processes. This review concentrates on the roles of miRNAs in skeletogenesis as well as in skeleton-related disease processes. Before describing these data, we present a brief review of the biogenesis and action of miRNAs, the approaches to miRNAs study, and miRNAs as global regulators of development. We finish by emphasizing that the study of the biological functions of miRNAs in skeletogenesis and dysplasia represents an entirely new avenue in the exploration of bone and cartilage biology, and large gaps remain in our knowledge of miRNAs in skeletogenesis in vivo and in our knowledge of the molecular events underlying miRNA-mediated musculoskeletal disorders.
Keywords: MicroRNAs, Skeletogenesis, Bone, Cartilage, Arthritis, Implants, Review
2. INTRODUCTION
MicroRNAs (miRNAs) are a class of short (~20-24nt) non-coding single-stranded RNA molecules that play an important role in regulating cellular gene expression. They function at the post-transcriptional level, by binding to and inhibiting mRNA. The first miRNA, lin-4 and its target, lin-14 were identified in 1993 as regulators of early larval development in C. elegans. Since then, the number of miRNAs to be identified or predicted has grown to >600, and it is thought that the human genome may code for 800-1000 miRNAs (1-4). miRNAs are currently thought to target the expression of approximately one-third of all mammalian genes (5). Of particular interest, miRNAs have been found to play important roles in mediating the fundamental biological processes of cell proliferation and differentiation in a variety of tissues types. In mammals, miRNAs have been found to play an important role in the development of a broad range of tissues and organ systems including the heart, skin, skeletal muscle, brain, neurons, kidneys, lung, adipocytes, and hematopoiesis (6-13). Recent evidence has suggested that miRNAs may also play a significant role in bone and cartilage development. In this review, we summarize what is currently known about the roles of miRNAs in the skeletogenesis and musculoskeletal disorders, including arthritis.
3. MICRORNAS—HOW DO THEY WORK?
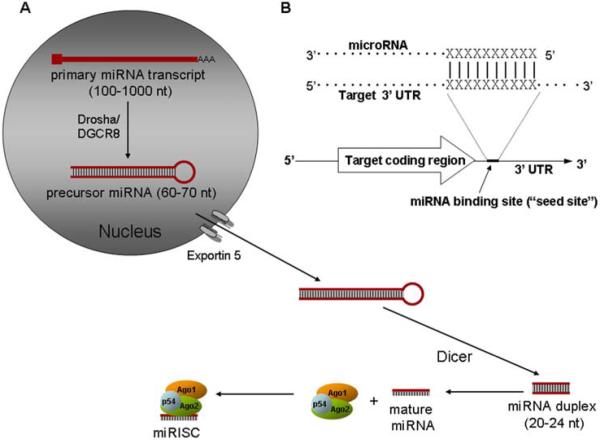
The expression of mature miRNA from a primary genomic template has been well described (Figure 1A). Long primary genomic transcripts (pri-miRNAs) are processed in the nucleus by the enzymes Drosha and DGCR8, resulting in the excision of a stem loop structure. The 60-80-nucleotide stem-loop product, also known as precursor miRNA (pre-miRNA) is exported from the nucleus by Exportin 5. In the cytoplasm, precursor miRNA undergoes processing by the RNase III enzyme Dicer, which generates a short RNA duplex. This is then processed into the single-stranded mature miRNA, which is incorporated into an RNA-induced silencing complex (RISC), an RNA-protein complex that includes members of the Argonaute protein family (Figure 1A).
Figure 1.
Schematic of microRNA biogenesis. Panel A. Biogenesis of a miRNA-induced silencing complex (RISC). Panel B. Binding of microRNA to the 3’-UTR of mRNA. See text for details.
MicroRNAs suppress target gene expression by initially binding to the 3’-untranslated region (3’-UTR) of mRNA before directing the repression of translation and/or mRNA degradation (Figure 1B). This requires that the 3’-UTR contains at least one specific ~7nt sequence that displays at least partial complementarity to a “seed site,” located within the 5’ region of the miRNA. Binding with full complementarity is required for mRNA cleavage while partial complementarity results in repression of translation. However, binding with either full or partial complementarity to the seed site is not always sufficient for gene repression; indeed a multitude of other factors, such as the relative position of the seed site binding sequence within the 3’ UTR, are likely involved (14). Following the binding of a particular mRNA by RISC, the whole complex is thought to be relocated to a cytoplasmic focus known as a processing body (P-body), while translation initiation is inhibited (15,16).
4. APPROACHES TO MICRORNA STUDY
The discovery of novel miRNAs and insights into their function generally follow a logical sequence of study with different experimental approaches making contributions at various stages. The initial prediction of miRNA sequences within the genome has been performed bioinformatically by screening for hairpin stem-loop structures characteristic of pre-miRNAs (2,17). However, predicted miRNAs must then be further supported with cloning or expression evidence before they can be included in miRBase, a database that catalogues all known miRNA (1).
An investigation often begins by screening for miRNAs which exhibit a tissue-specific pattern of expression or which are differentially expressed during different stages of various biological processes. Microarray technology has been implemented to analyze the expression of hundreds of miRNA simultaneously (18,19). The results of this analysis can then be confirmed via various techniques including real-time RT-PCR with miRNA-specific primers, in situ hybridization using locked-nucleic acid (LNA) probes, and the repression of reporter transgenes (20-22). The biological function of miRNA can also be confirmed through the cellular overexpression of miRNA, either through direct transfection of antisense miRNA or via the construction of a miRNA expression vector (23-29). Similarly, the repression of miRNAs can be accomplished through the transfection of 2’-O-methyl antisense oligoribonucleotides (30), or through a vector expressing siRNA that targets the hairpin portion of the miRNA precursor.
miRNA target prediction is critical to explaining the biological function of miRNAs. Multiple resources are available for algorithms that compare miRNA and 3’-UTR mRNA sequences, thus identifying a group of potential target genes (31). Although different algorithms can often result in divergent results, it has been suggested that at least a certain class of conserved miRNA targets can be confidently predicted. Once a list of predicted targets is acquired, individual targets can be confirmed by observing the repression of reporter transgenes in a miRNA overexpression model.
5. MICRORNAS AS GLOBAL REGULATORS OF DEVELOPMENT
miRNAs play an important role in a variety of biological processes, including development and differentiation in a variety of organisms and tissue types. Indeed, one of the first insights into the significance of miRNA function was reached through the disruption of the Dicer gene. Dicer is encoded at a single locus and is required for miRNA processing; Dicer ablation results in a significant reduction in the levels of nearly all mature miRNAs (32-34). Dicer mutants were found to have profound developmental defects across a whole range of animal and plant species, including Arabidopsis, Drosophila, C. elegans, and zebrafish (32,35-39). In mice, Dicer ablation resulted in embryonic lethality by day 7.5 (32).
More recently, individual miRNAs have been found to play important roles in the development of various tissues and organs. For example miR-1, which is widely conserved during evolution, is essential for proper muscle development in Drosophila (40,41). miR-1, which targets the genes Hand2 and HDAC4, is expressed downstream of muscle differentiation regulators SRF, MyoD and Mef2 (13). miR-1 has also been implicated in muscle-related disease processes, such as cardiac hypertrophy (42-44).
miR-143 has been found to stimulate adipogenesis; inhibition of miR-143 resulted in the lowered expression of adipocyte-specific genes (8). miR-221 and miR-222 inhibit angiogenesis by targeting the stem cell factor receptor c-Kit, and have been implicated in melanoma progression (45,46). miR-124a and miR-124b stimulate neuronal differentiation by targeting lin-28 (47,48).
6. MICRORNAS IN SKELETOGENESIS
A recent study demonstrated that functional miRNAs are essential for normal skeletal growth in vivo (49). Mice whose Dicer was differentially disrupted in chondrocytes (by site-specific Cre recombination under the control of a Col2a1 promoter) displayed significant skeletal growth defects. Analysis of the growth plates revealed decreased chondrocyte proliferation and accelerated hypertrophic differentiation, resulting in a reduced number of proliferating chondrocytes, an expanded hypertrophic region, and significant defects in skeletal development. This suggests that at least several miRNAs may be involved in inhibition of the chondrogenic differentiation pathway.
Despite mounting evidence that miRNAs play a significant role in embryonic development and other biological processes, the function of only a handful of miRNAs has been determined thus far. And of these miRNAs, only a small subset has been implicated in cartilage and/or bone development (Table 1). Using in situ hybridization of locked nucleic acid probes, it has been demonstrated that a high degree of tissue specificity exists for certain miRNAs, suggesting that these miRNAs may play a key role in tissue differentiation and development (22). In particular, miR-140 was found to be expressed exclusively in the cartilage tissue of embryonic zebrafish. Other miRNAs were also found to be highly (although not exclusively) expressed in the developing tissues of the zebrafish skeletal system, including miRNAs 146, 140*, 199a, 199a*, 145, as well as others that were ubiquitously expressed (22). For miRNAs 140, 199a, and 199a*, this pattern of expression is conserved in medaka and chicken embryos (50). Although one study (51) did not detect high expression levels of these three miRNAs in mouse embryos, in situ staining of later-stage mouse embryos confirmed that miR-140 is expressed specifically in mouse embryonic cartilage tissue (52). Although the expression of miR-199a in mouse connective tissue has been established via small RNA library sequencing (53), the expression patterns of miR-199a and miR-199a* in mouse embryos has yet to be confirmed through in situ hybridization.
Table 1.
MicroRNAs in skeletogenesis
| miRNA | Role in Differentiation | Expression in Embryonic Bone/Cartilage | Verified Targets | Citations |
|---|---|---|---|---|
| miR-140 | Unknown | Exclusively expressed in zebrafish, medaka, chicken, and mouse | HDAC4 | (22, 50, 52) |
| miR-140s | Unknown | Highly expressed in zebrafish | (22) | |
| miR-199a | Inhibits chondrocyte1 | Highly expressed in zebrafish, medaka, chicken, and mouse | (22, 50, 53) | |
| miR-199as | Inhibits chondrocyte1 | Highly expressed in zebrafish, medaka, chicken, and mouse | SMAD11 | (22, 50) |
| miR-26a | Inhibits osteoblast | SMAD1 | (57, 58) | |
| miR-126 | Unknown | VCAM1, HOXA9 | (61, 62) | |
| miR-125b | Inhibits osteoblast | ERBB2 | (64) | |
| miR-145 | Unknown | Highly expressed in zebrafish | (22) | |
| miR-146 | Unknown | Highly expressed in zebrafish | (22) |
VCAM1, vascular cell adhesion molecule 1; HOXA9, homeobox A9; ERBB2, v-erb-b2 erythroblastic leukemia viral oncogene homolog 2
Lin and Liu, 2008, unpublished data
Other studies involving differentiation of human mesenchymal stem cells into osteocytes and chondrocytes implicated a different subset of miRNAs. miR-638 and miR-663 were found to be upregulated in chondrocytes, while miR-24, let-7a, let-7b, let-7c, miR-138, and miR-320 were associated with osteocyte maturation (54). The transcription of many upregulated miRNAs was found to be under the control of platelet-derived growth factor (PDGF)-responsive transcription factors, suggesting that PDGF may mediate the differential regulation of miRNAs during osteogenesis (54). Interestingly, miR-140 has recently been found to target PDGF receptor alpha, thus acting as a negative regulator of PDGF during craniofacial development (55). However, it is unknown whether this is also significant for osteogenic differentiation.
Although miR-140 is unique as the only identified miRNA that is specifically expressed in cartilage, it remains unknown whether miR-140 plays any significant role in chondrogenic or osteoblastic differentiation. miR-140 has been shown to target HDAC4, a known corepressor of Runx2, which is an essential transcription factor for chondrocyte hypertrophy and osteoblast differentiation (52). However, HDAC4 is also targeted by various other miRNAs, including miR-1, which when overexpressed, has been shown to promote muscle differentiation without affecting osteoblastic differentiation (7,56). Thus, a single gene can be differentially regulated by various tissue-specific miRNAs, thus playing separate roles in different developmental pathways.
Recent findings from our lab have implicated miR-199a and miR-199a* as having a significant role in the course of chondrogenic differentiation. Experiments involving real-time PCR have shown miR-199a* to be dramatically downregulated in early chondrogenesis, suggesting that miR-199a* may play an inhibitory role in the early stage of chondrocyte differentiation. Overexpression of miR-199a* precursor in a chondrogenic differentiation cell model resulted in the repression of early chondrogenesis markers (Lin et al, 2008, unpublished data). Interestingly, both miR-199a and miR-199a* are predicted to target SMAD1, a downstream transcription factor of the bone morphogenic protein (BMP) signal transduction pathway. Thus, by the inhibition of SMAD1, miR-199a and miR-199a* may act as important regulators of chondrocyte and/or osteoblast differentiation.
A greater number of microRNAs have been implicated in the regulation of osteoblast differentiation than chondrocyte differentiation. miR-26a, which promotes myogenesis by targeting a histone methyltransferase (57), has been identified as an inhibitor of late osteoblast differentiation (58). This suggests that miR-26a may act as a facilitator to maintain cells in one differentiation fate over another—silencing osteoblast differentiation genes while promoting the expression of myogenic factors. There is evidence to show that miR-26a inhibits osteoblast differentiation by targeting the important cell-signaling molecule, SMAD1 (58). Interestingly, SMAD1 interacts with the transcriptional repressor HOXA9, which binds to the OPN promoter and represses OPN gene expression (59,60). OPN is an intermediate marker of osteoblastic differentiation. Therefore, downregulation of SMAD1 by miR-26a would relieve the inhibition on HOXA9, thus increasing the inhibition of OPN.
More recent evidence suggests that HOXA9 is itself under the regulation of miRNAs, particularly miR-126 (61). Although miR-126 has yet to be formally implicated in the osteoblast differentiation process, miR-126 and miR-26a have counteracting effects on HOXA9 expression and the summation of their activities may serve to fine tune HOXA9 expression levels. Thus the combined activity of multiple miRNAs may provide various stages of regulation along the same signal transduction pathway. Of note, miR-126 has been shown to downregulate VCAM-1 expression in TNF-α-stimulated endothelial cells (62). Since VCAM-1 is an important mediator of osteoblast adhesion to osteoclasts, the potential that miR-126 plays a role in bone formation and remodeling is significant (63).
Recent evidence has shown that miR-125b is also involved in osteoblastic differentiation. When compared to the control, miR-125b is inhibited in cells undergoing BMP-4 induced osteoblastic differentiation, suggesting an osteoblast inhibitory effect. Indeed, overexpression of miR-125b resulted in the repression, while inhibition of miR-125b resulted in the promotion of osteoblastic differentiation (64). miR-125b was also found to inhibit cell proliferation—confirming previous findings that this miRNA modulates proliferation in a variety of cell types, including human carcinoma cells through its target gene ErbB2 (65-67). Interestingly, suppression of ErbB2 via siRNA transfection also results in reduced osteoblastic differentiation. This suggests that by regulating a single target gene, such as one that regulates cell proliferation, a particular miRNA may have wide-ranging effects on many biological processes in multiple tissue types.
7. MICRORNAS AND IMPLANTS
A number of studies have examined the effect of various biocompatible materials on bone formation. Because miRNAs have been implicated in growth and differentiation, several labs have sought to determine the effect of biocompatible materials on microRNA expression profiles. Many of these studies have focused on bone grafting or artificial implants in the context of dental procedures. For example, the synthetic bone graft material calcium sulfate (CaS) has been found to upregulate miR-377, while downregulating miRNAs-22, 93, 27a, 31, and 16 (68). Many of these miRNAs target genes that are involved in regulating osteoblast activity, such as BMP1, BMP7, FGFR1, PTH, CALCA and GHRHR. Interestingly, homeobox regulatory genes such as HOXA13, DLX5, and SHOX were also found as potential targets. P-15, a collagen analog that is used for bone grafting, has been found to upregulate miR-26a, an inhibitor of late osteoblast differentiation (58,69). These studies suggest that miRNAs play an important role in bone healing and remodeling following surgical procedures involving implants.
8. MICRORNAS IN ARTHRITIS
The possible role of miRNAs in the disease processes of rheumatoid arthritis (RA) and osteoarthritis (OA) has only been recently explored. One study found a significant upregulation of miR-155 and miR-146a in synovial fibroblasts (RASFs) and synovial fliud derived from patients with RA (70). Furthermore, stimulation of cultured RASFs with proinflammatory mediators, such as TNF-α, IL-1β and LPS, markedly induced miR-155 and miR-146 expression levels (70,71). Similarly, proinflammatory ligands stimulated miR-155 expression in synovial fibroblasts derived from patients with OA. Additionally, both miR-155 and miR-146 expression were significantly higher in RA synovial tissues when compared to OA, which is consistent with previous work outlining the role of both miRNAs in immune system function (70-74).
The exact mechanism behind these findings is still unclear. Overexpression of miR-155 in RASFs resulted in the inhibition of MMP-3, a known contributor to cartilage destruction in RA. This suggests that miR-155 may serve to protect joint tissues from excessive inflammatory damage. However, since miR-155 is not predicted to directly target MMP-3, it is likely that the mechanism of action is indirect, perhaps through the regulation of LPS signaling (75). The mechanism for the role of miR-146 is better understood; miR-146 is known to target TRAF6 and IRAK1, which are key downstream molecules of TNF-α and IL-1β signaling (76). Thus, it is likely that both miR-155 and miR-146 serve to temper immune responses in rheumatoid arthritis by providing negative feedback through the targeting of key immune signaling pathway genes.
9. SUMMARY
Based on the literature to date, it is clear that miRNAs play a key role in bone and cartilage development. Although several miRNAs have been implicated in regulating chondro- and osteogenesis, much work remains to elucidate the complex interplay of different miRNAs and their gene targets, and the molecular events underlying miRNA-mediated skeletogenesis and musculoskeletal disorders, particularly with in vivo models. Nevertheless, the study of miRNAs represents a novel and exciting avenue in the exploration of bone and cartilage biology.
ACKNOWLEDGMENTS
This work was aided by NIH research grants AR050620, AR052022, AR053210, AG029388 and a grant from Arthritis National Research Foundation.
REFERENCES
- 1.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 3.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 7.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 9.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 14.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol. 2006;3:97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 17.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 18.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 20.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 21.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 22.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda Y, Kawasaki H, Taira K. Construction of microRNA-containing vectors for expression in mammalian cells. Methods Mol Biol. 2006;338:167–173. doi: 10.1385/1-59745-097-9:167. [DOI] [PubMed] [Google Scholar]
- 24.Zeng Y, Cai X, Cullen BR. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol. 2005;392:371–380. doi: 10.1016/S0076-6879(04)92022-8. [DOI] [PubMed] [Google Scholar]
- 25.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 27.Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 29.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S. Double-stranded regions are essential design components of potent inhibitors of RISC function. Rna. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 33.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 34.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 36.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 37.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 40.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 43.Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Downregulation of MIRNA-1/MIRNA-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem. 2008 May 5; doi: 10.1074/jbc.M801035200. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 44.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Carè A. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 46.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 47.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105:1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 52.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 53.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lakshmipathy U, Love B, Goff LA, Jornsten R, Graichen R, Hart RP, Chesnut JD. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakajima N, Takahashi T, Kitamura R, Isodono K, Asada S, Ueyama T, Matsubara H, Oh H. MicroRNA-1 facilitates skeletal myogenic differentiation without affecting osteoblastic and adipogenic differentiation. Biochem Biophys Res Commun. 2006;350:1006–1012. doi: 10.1016/j.bbrc.2006.09.153. [DOI] [PubMed] [Google Scholar]
- 57.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 58.Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 59.Li X, Nie S, Chang C, Qiu T, Cao X. Smads oppose Hox transcriptional activities. Exp Cell Res. 2006;312:854–864. doi: 10.1016/j.yexcr.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 60.Shi X, Bai S, Li L, Cao X. Hoxa-9 represses transforming growth factor-beta-induced osteopontin gene transcription. J Biol Chem. 2001;276:850–855. doi: 10.1074/jbc.M005955200. [DOI] [PubMed] [Google Scholar]
- 61.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 Regulates HOXA9 by Binding to the Homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka Y, Morimoto I, Nakano Y, Okada Y, Hirota S, Nomura S, Nakamura T, Eto S. Osteoblasts are regulated by the cellular adhesion through ICAM-1 and VCAM-1. J Bone Miner Res. 1995;10:1462–1469. doi: 10.1002/jbmr.5650101006. [DOI] [PubMed] [Google Scholar]
- 64.Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda A, Amemiya T, Kondoh Y, Tashiro H, Okazaki Y. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun. 2008;368:267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 65.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 66.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 67.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, Liu CG, Petrocca F, Troncone G, Calin GA, Scarpa A, Colato C, Tallini G, Santoro M, Croce CM, Fusco A. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 68.Palmieri A, Pezzetti F, Brunelli G, Scapoli L, Lo Muzio L, Scarano A, Martinelli M, Carinci F. Calcium sulfate acts on the miRNA of MG63E osteoblast-like cells. J Biomed Mater Res B Appl Biomater. 2008;84:369–374. doi: 10.1002/jbm.b.30880. [DOI] [PubMed] [Google Scholar]
- 69.Palmieri A, Pezzetti F, Brunelli G, Zollino I, Scapoli L, Martinelli M, Arlotti M, Carinci F. Differences in osteoblast miRNA induced by cell binding domain of collagen and silicate-based synthetic bone. J Biomed Sci. 2007;14:777–782. doi: 10.1007/s11373-007-9193-z. [DOI] [PubMed] [Google Scholar]
- 70.Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S, Kyburz D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 71.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Georgantas RW, 3rd, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 75.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 76.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]