Abstract
The ability of Staphylococcus aureus to infect tissues is dependent on precise control of virulence through gene-regulatory systems. While the SaeR/S two-component system has been shown to be a major regulator of S. aureus virulence, the influence of the host environment on SaeR/S-regulated genes (saeR/S targets) remains incompletely defined. Using QuantiGene 2.0 transcriptional assays, we examined expression of genes with the SaeR binding site in USA300 exposed to human and mouse neutrophils and host-derived peptides and during subcutaneous skin infection. We found that only some of the saeR/S targets, as opposed to the entire SaeR/S virulon, were activated within 5 and 10 min of interacting with human neutrophils as well as α-defensin. Furthermore, mouse neutrophils promoted transcription of saeR/S targets despite lacking α-defensin, and the murine skin environment elicited a distinctive expression profile of saeR/S targets. These findings indicate that saeR/S-mediated transcription is unique to and dependent on specific host stimuli. By using isogenic USA300ΔsaeR/S and USA300Δagr knockout strains, we also determined that SaeR/S is the major regulator of virulence factors, while Agr, a quorum-sensing two-component system, has moderate influence on transcription of the saeR/S targets under the tested physiological conditions.
Key Words: Staphylococcus aureus, USA300, SaeR/S, Two-component systems, Virulence, Gene expression, Neutrophils, α-Defensin
Introduction
Staphylococcus aureus is a pathogen of major medical concern that can cause infections ranging from mild skin abscesses to necrotizing pneumonia and sepsis in immunocompromised and healthy individuals. It contributes to 12 million annual outpatient visits for skin and soft tissue infections and is the leading cause of infective endocarditis worldwide [1, 2]. The pathogen's success in hospital and community settings can be partially attributed to elaborate two-component gene-regulatory systems that tightly control expression of virulence and immunomodulatory factors in response to environmental stimuli. The SaeR/S system is recognized as a major regulator of S. aureus virulence and is required for successful establishment of skin and invasive infections as well as survival following human neutrophil phagocytosis [3, 4, 5, 6]. Upon activation of the histidine kinase SaeS, the cytosolic response regulator SaeR promotes expression of genes with the SaeR binding site which include leukotoxins, hemolysins, adhesins and proteases that are responsible for saeR/S-mediated virulence [3, 4, 5, 7]. While expression of genes with the consensus SaeR binding site was shown to be dependent on the SaeR/S system in vitro, rot and arlR transcription was unaffected by the absence of saeR/S, suggesting that expression of genes with the SaeR binding site might be influenced by other regulatory networks and/or environmental conditions [3]. Furthermore, the SaeR/S system has 2 membrane proteins, SaeP and SaeQ, that most likely serve to negatively control the system's activity [8]. Despite the essential role of the SaeR/S system in evading innate immunity, the conditions under which SaeR/S becomes activated as well as the subsequent transcription of genes with SaeR binding sites have not been completely defined.
To this end, we designed QuantiGene 2.0 assays with S. aureus genes that contain the SaeR recognition sequence in the promoter region (referred to as saeR/S targets) and control targets that do not have the SaeR binding site (agrA, mecA and dltA) [3, 7]. Using this technique, we were able to measure gene expression changes in S. aureus during initial interactions with human and mouse neutrophils and host-derived peptides as well as during subcutaneous skin infection. We also began to elucidate the influence of the quorum-sensing gene regulator Agr on SaeR/S regulon expression, as agr controls the expression of a number of virulence factors, including those with SaeR binding sites, depending on the growth conditions [9]. By measuring transcript levels of saeR/S targets in saeR/S and agr isogenic mutants, we found that saeR/S is the predominant regulator of genes containing the SaeR binding domain while agr has a moderate impact on saeR/S target transcription during initial interactions with human neutrophils and in vivo during murine skin infection. Furthermore, we demonstrate that while saeR/S regulates expression of a number of virulence factors under physiologically relevant conditions, only a subset of these effectors is transcriptionally upregulated in the presence of neutrophils as well as α-defensin. Supporting this finding, we show that the mouse skin environment promotes transcription of specific saeR/S targets, differing from the expression profile elicited following neutrophil interaction or α-defensin, further emphasizing that activation of this two-component system and the subsequent downstream regulation are dependent on the stimuli sensed by S. aureus.
Materials and Methods
Bacterial Strains and Culture
Wild-type USA300 (LAC) was used to generate USA300ΔsaeR/S, USA300ΔsaeR/S+Comp (complemented with a plasmid encoding saeR and saeS) and USA300Δagr (isogenic knockout of agrBDCA), as previously described [3, 4, 10]. Bacteria used in the studies were cultured in tryptic soy broth with 0.5% glucose and harvested at midexponential growth phase (optical density at 600 nm = 1.5) [11].
Neutrophil and Purified Peptide Assays
Human neutrophils (polymorphonuclear leukocytes, PMNs) were isolated from heparinized venous blood of healthy donors in accordance with a protocol approved by the Institutional Review Board for Human Subjects at Montana State University. All donors provided written consent to participate in the study. Erythrocytes were removed by dextran sedimentation, and PMNs were separated from peripheral blood mononuclear cells using Histopaque-1077® (Sigma Aldrich) followed by water lysis of remaining erythrocytes under endotoxin-free conditions (<25.0 pg/ml) as described elsewhere [11]. Bone marrow-derived mouse neutrophils were isolated from BALB/c mice using a Percoll gradient and resuspended in RPMI plus 5 mM HEPES media [12]. PMN phagocytosis was synchronized by centrifuging PMNs and bacteria at 380 g for 8 min at 4°C [11]. For activation assays with host-derived components, USA300 [5 × 107 colony-forming units (CFUs)] was treated with water or a sublethal dose (confirmed by CFU assays) of either human α-defensin (HNP-1, Millipore), β-defensin (hBD-3, AnaSpec), LL-37 (AnaSpec) or dermcidin (DCD, AnaSpec), all resuspended in water and tested at 0.48 µM, apart from H2O2, which was tested at 0.55 mM (AMRESCO). Strains were incubated at 37°C with shaking (250 rpm) for 10 and 30 min (as indicated). The control experiment with lethal doses (determined using CFU assays) was performed by exposing USA300 to α-defensin (4.8 µM) or LL-37 (9.6 µM) for 30 min. At designated time points, samples were lysed with RLT-β-mercaptoethanol buffer and homogenized using FastPrep, and S. aureus RNA was isolated using the RNeasy method (Qiagen) [11, 13].
Mouse Studies
Mouse studies performed for this investigation conformed to the NIH guidelines and were approved by the Institutional Animal Care and Use Committee at Montana State University. Crl:SKH1-hrBR mice (Charles River) were inoculated subcutaneously with 2 × 107 CFUs of USA300, USA300ΔsaeR/S or USA300Δagr in 50 µl of PBS [3, 14]. Eight hours after infection, mice were sacrificed, and skin biopsies were harvested with a 9-mm punch, homogenized and centrifuged. The supernatants were removed, the remaining bacteria pellets were lysed with RLT-β-mercaptoethanolbuffer (Qiagen), and S. aureus RNA was harvested as described above [11, 13].
QuantiGene 2.0 Assays
QuantiGene 2.0 assays (Affymetrix) were performed on 2 biological replicates of the aforementioned experiments [15]. In brief, purified RNA samples were captured by microsphere beads unique to each gene of interest and hybridized overnight to target-specific DNA probes with biotinylated sites, which were recognized by addition of streptavidin-conjugated R-phycoerythrin. The amplified signal was detected together with the capture bead specific to each gene of interest using a Luminex flow cytometer (BioRad). Quality control of QuantiGene 2.0 was assessed in preliminary experiments using varied doses of RNA to determine the optimum amount of RNA for subsequent assays. Each S. aureus RNA sample (50-100 ng) was assayed in duplicate, and transcript abundance of 15 genes with the SaeR recognition sequence and 3 genes (agrA, mecA and dltA) without the SaeR binding site was detected as the mean fluorescent signal normalized to gyrB expression levels. This endogenous control was constitutively expressed in S. aureus under the tested conditions. Changes in S. aureus gene expression in response to host stimuli were determined by comparing transcript levels in S. aureus strain(s) treated with the described stimulus to those in the same strain grown in media (time-matched) (fig. 1, 2). To define the influence of saeR/S and agr on gene expression, transcript levels in the wild-type strain treated with the relevant host component were compared to gene expression in mutant strains exposed to the same stimulus (fig. 3, 5). Samples that consisted of host RNA alone demonstrated that the probes were S. aureus RNA specific (data not shown). QuantiGene 2.0 results were confirmed using TaqMan quantitative RT-PCR (qRT-PCR) as previously described [3, 11], demonstrating that this assay is a suitable alternative to global gene expression assessment methods.
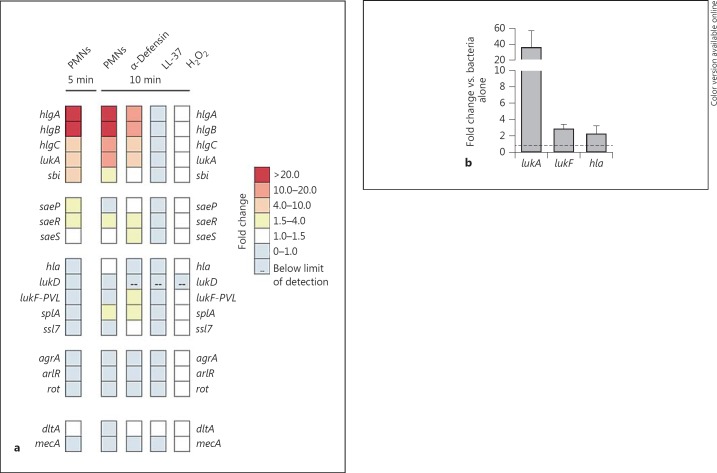
Fig. 1.
Differential transcription of saeR/S targets is dependent on the host stimulus. a QuantiGene 2.0 assays were used to assess transcript levels of the listed saeR/S targets in USA300 exposed to human neutrophils (10:1, bacteria to PMN), α-defensin (0.48 µM), LL-37 (0.48 µM) or H2O2 (0.55 mM). Data are reported as the mean fold change of 2 separate experiments as shown in online supplementary table 1. Transcript levels in S. aureus exposed to the stimulus were normalized to gyrB and calibrated to those from S. aureus strains grown in media only (time-matched). b TaqMan qRT-PCR confirmation of QuantiGene 2.0 data. The fold change is the relative gene expression of lukA, lukF-PVL and hla measured in USA300 calibrated to gyrB levels after 10-min interaction with human neutrophils (10:1, bacteria to PMN) and normalized to baseline gene expression of USA300 grown in media (indicated by the dotted line).
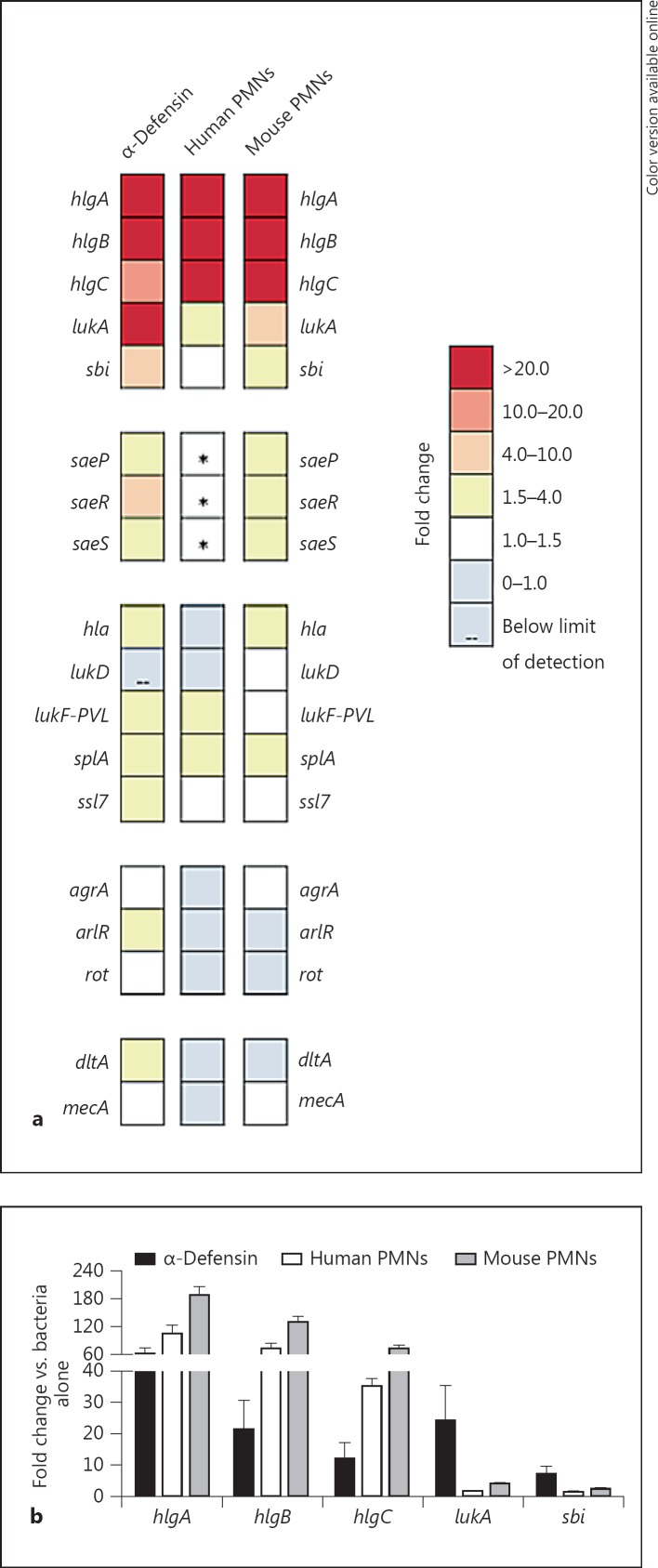
Fig. 2.
α-Defensin is not required for neutrophil-mediated activation of the SaeR/S system. USA300 was exposed to α-defensin (0.48 µM), human neutrophils and mouse neutrophils (10:1, bacteria to PMN) for 30 min, and changes in expression were measured using the QuantiGene 2.0 platform. a Fold change of saeR/S targets. b Quantitative comparison of saeR/S targets (from a) substantially affected by α-defensin, human neutrophils and mouse neutrophils. Data are shown as the mean fold change of 2 separate experiments calculated as stated in figure 1 and listed in online supplementary table 2. Asterisks indicate gene targets with expression above the limit of detection.
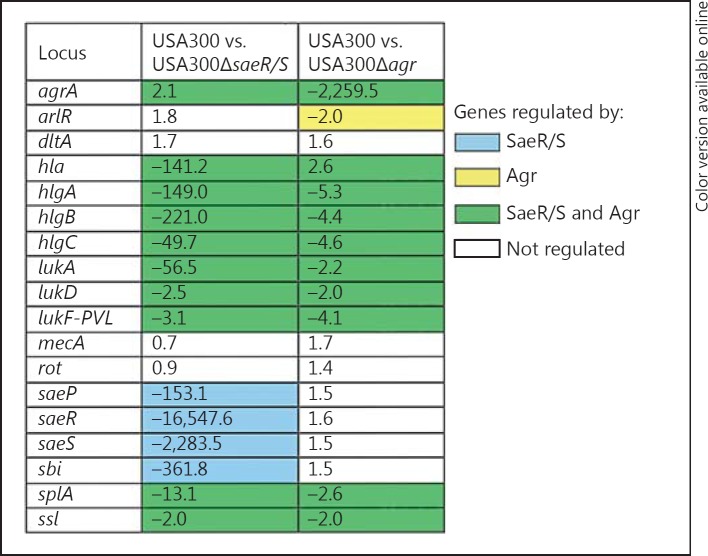
Fig. 3.
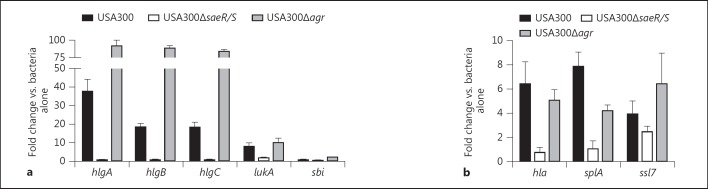
USA300 genes regulated by saeR/S and agr following interaction with human neutrophils. QuantiGene 2.0 assays were used to assess transcript levels of the listed saeR/S-regulated genes in USA300, USA300ΔsaeR/S or USA300Δagr exposed to human neutrophils (10:1, bacteria to PMN). The mean fold change of 2 separate experiments was calculated by comparing gyrB-calibrated transcript levels in wild-type USA300 to those in USA300ΔsaeR/S and USA300Δagr isogenic knockout strains 10 min after neutrophil interaction. Blue and yellow depict genes regulated by saeR/S and agr, respectively, and genes influenced by both systems are shaded green.
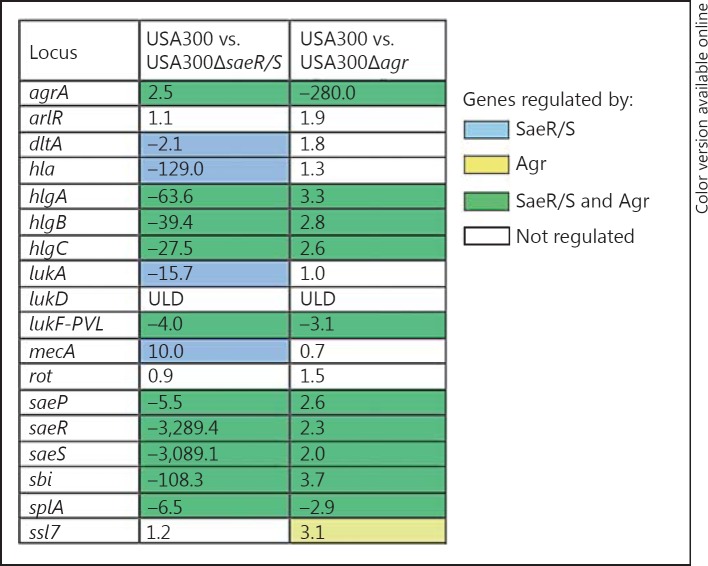
Fig. 5.
USA300 genes regulated by saeR/S and agr during subcutaneous skin infection. The mean fold change of 2 separate experiments, described in figure 4, was calculated by comparing gyrB-calibrated transcript levels in wild-type USA300 to those in USA300ΔsaeR/S and USA300Δagr 8 h after inoculation. ULD = Under the limit of detection. Blue and yellow depict genes regulated by saeR/S and agr, respectively, and genes influenced by both systems are shaded green.
Results
A Subset of saeR/S-Regulated Genes Is Activated during Early Human Neutrophil Interactions
While promoter activity and transcript levels of saeR/S have been shown to be affected by PMN phagocytosis, individual PMN components and pH changes [9, 11, 16, 17], the effect of host-derived components on saeR/S targets has not been completely defined. To this end, we employed QuantiGene 2.0 assays to measure expression changes of 15 genes with the SaeR binding site [3, 7] in a USA300 wild-type strain (LAC) during initial interactions with human neutrophils (fig. 1a). By comparing transcript levels in neutrophil-treated USA300 to those in USA300 grown in media, we identified targets that were immediately activated and thus may be essential to successful initiation of infection by S. aureus. Five and 10 min after interaction, transcription of a specific subset of genes, namely hlgA, hlgB, hlgC (γ-hemolysins), lukA (leukotoxin A/B or G/H) and sbi (immunoglobulin-binding protein), was strongly upregulated in USA300 (fig. 1a; online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000351200). In contrast, lukD (leukotoxin E/D), lukF-PVL (Panton-Valentine leukocidin), hla (α-hemolysin), ssl7 (superantigen-like protein) and splA (serine protease) expression did not increase despite the presence of the SaeR binding region (fig. 1a; online suppl. table 1). Although saeS transcript levels were unaffected by the treatment, the transcriptional activity of saeP and saeR increased slightly after interaction with neutrophils while agrA and arlR expression did not increase (fig. 1a; online suppl. table 1). To confirm the validity of the QuantiGene 2.0 assay, we used TaqMan qRT-PCR to measure changes in the gene expression of USA300 in the presence of neutrophils. In congruence with the QuantiGene 2.0 data, after 10 min, lukA transcript was upregulated while lukF-PVL and hla expression did not change significantly (fig. 1b). Together, these findings suggest that only a subset of saeR/S-regulated genes is transcriptionally activated during initial interactions with human neutrophils.
Specific Host Stimuli Promote Transcription of saeR/S-Regulated Genes
Unlike LL-37, which does not promote activation of the sae promoter, α-defensin and H2O2 have been shown to increase saeR/S transcript levels [16]. However, the effect of neutrophil-derived components on saeR/S-regulated genes has not been completely defined. To address this, we treated USA300 with α-defensin, LL-37 and H2O2 for 10 min and investigated changes in transcript levels of saeR/S targets. Human α-defensin had the most robust effect on saeR/S targets, relatively similar to that of USA300 treated with neutrophils (fig. 1a; online suppl. table 1). Expression of γ-hemolysins (hlgA, hlgB, hlgC), leukotoxin AB/GH (lukA) and genes of the SaeR/S system was increased, while changes in expression of the remaining target genes were minimal. Despite their antimicrobial properties, LL-37 and H2O2 had no effect on transcript abundance of saeR/S targets (fig. 1a; online suppl. table 1).
We next wanted to determine whether peptide-mediated killing could promote transcription of saeR/S targets to test if cell death is a prominent activator of the SaeR/S system. We measured expression of saeR/S targets in S. aureus subjected to a lethal dose of α-defensin or LL-37 for 30 min. Even at a higher dosage, LL-37 did not elicit a transcriptional change in saeR/S targets, while α-defensin triggered a similar response to that of the sublethal dose (data not shown), suggesting that the transcriptional activation of saeR/S targets noted in our studies is due to specific host-derived components as opposed to any antimicrobial insult.
While the expression profiles of saeR/S targets were similar between neutrophil- and α-defensin-treated groups (fig. 1a), the difference in the magnitude of expression suggested that α-defensin might contribute only partially to activating saeR/S targets in USA300 following neutrophil interaction. To address this, we examined gene expression in saeR/S targets after 30-min exposure to purified human α-defensin, human neutrophils and bone marrow-derived mouse neutrophils that do not produce α-defensins [18] (fig. 2a, b; online suppl. table 2). Despite the absence of α-defensins, saeR/S-regulated gene expression increased substantially in response to mouse PMNs (fig. 2a, b; online suppl. table 2). Transcript levels of hlgA, hlgB, hlgC, lukA, sbi, hla, splA, saeP, saeR and saeS were upregulated, suggesting that, in addition to α-defensin, there are other neutrophil-specific signals that promote activation of saeR/S targets (fig 2a). Also, lukF-PVL was only upregulated in the presence of α-defensin and human neutrophils, while α-hemolysin (hla) and immunoglobulin-binding protein (sbi) were increased due to α-defensin and mouse PMNs, further highlighting the dependence of saeR/S target expression on the external environment (fig. 2a). We next compared the magnitude of responses among the 3 treatment groups and found that hlgABC transcript levels were the highest in the mouse neutrophil-treated group, while α-defensin promoted strong transcription of lukA and sbi when compared to transcript levels in USA300 treated with human or mouse neutrophils (fig. 2b). In addition, expression of saeR, saeS and saeP was above the limit of detection in USA300 treated with human neutrophils. To confirm that the regulatory system becomes activated by human neutrophils at 30 min, as reported previously [11], we employed TaqMan qRT-PCR to measure saeR levels under the same conditions (data not shown). Transcript levels of saeR were increased 10-fold, demonstrating that human neutrophils promote SaeR/S activation 30 min after interaction. These findings suggest that the downstream activation of saeR/S targets is independent of saeR transcript levels and specific to the host-derived signal.
The SaeR/S Two-Component System Is the Major Regulator of Virulence Factors during Neutrophil Interaction
Although extensive in vitro studies have identified genes regulated by the SaeR/S system [3, 4, 7], we wanted to determine if expression of the relevant targets was also saeR/S-dependent under physiologically relevant conditions. We addressed this by investigating gene expression in USA300ΔsaeR/S and USA300Δagr, following early interaction with human neutrophils. USA300Δagr was included because previous studies have shown agr to have an influence on expression of saeR/S targets [9]. By comparing transcript levels of USA300 to those in the mutant strains in the presence of neutrophils, we identified that genes triggered in USA300 (hlgA, hlgB, hlgC, lukA and sbi) were abolished in USA300ΔsaeR/S (fig. 3). Although expression of saeP, hla, lukF-PVL and splA was not upregulated in USA300 treated with neutrophils (fig. 1a), their transcript levels were substantially downregulated in USA300ΔsaeR/S (fig. 3). In contrast, expression of rot and arlR was similar in USA300 and USA300ΔsaeR/S, suggesting that these targets are not solely regulated by saeR/S under the conditions tested, an observation supported by in vitro studies (fig. 3) [3]. By assessing the fold change between USA300 and USA300Δagr gene expression in response to neutrophils, we found that transcription of hlgA, hlgB, hlgC, lukF-PVL and splA was slightly reduced in the absence of agr (fig. 3). Furthermore, to confirm that the noted changes were specific to saeR/S, we also assessed gene expression during neutrophil interaction using a USA300ΔsaeR/S strain complemented with a plasmid containing saeR/S (online suppl. fig. 1). Complementation restored the wild-type gene expression levels, confirming that transcriptional changes in response to neutrophils are saeR/S-mediated (online suppl. fig. 1). These findings suggest that while the SaeR/S system is the major regulator of the relevant genes, Agr has moderate influence over saeR/S-regulated gene expression in the presence of human neutrophils.
saeR/S Targets Are Activated following Murine Skin Infection
Next, we expanded on the aforementioned findings by investigating expression of saeR/S-regulated genes in vivo. Since saeR/S and agr are essential to successful initiation of S. aureus skin infection [3, 19, 20], we hypothesized that the skin environment will activate saeR/S targets. To this end, we isolated S. aureus RNA from skin abscesses of mice 8 h following infection with USA300, USA300ΔsaeR/S or USA300Δagr and measured gene expression using the QuantiGene 2.0 assay. To assess changes in saeR/S target expression elicited by the skin environment, we compared gene transcription in S. aureus strains from the abscess to transcript levels in corresponding strains at midexponential growth, representing the initial inoculum. With the exception of sbi, saeR/S targets hlgA, hlgB, hlgC and lukA, previously shown to be activated by mouse neutrophils (fig. 2a, b), were also highly expressed in USA300 and USA300Δagr but abolished in the USA300ΔsaeR/S strain (fig. 4a; online suppl. table 3). On the other hand, only the skin environment triggered robust transcription (fold change in parentheses) of hla (6.4), splA (7.65) and ssl7 (3.95) in USA300 (fig. 4b; online suppl. table 3) when compared to mouse neutrophil-induced expression of hla (1.52), splA (2.43) and ssl7 (1.16) (fig. 2a, b; online suppl. table 2), further emphasizing that the specificity of the transcriptional response in S. aureus is dependent on the external environment. Our results also confirm previous studies that showed the saeR/S mutant to have increased levels of mecA expression, which are likely due to an indirect effect since mecA does not have the SaeR binding site [3].
Fig. 4.
The skin environment promotes saeR/S target transcription. QuantiGene 2.0 assays were used to assess transcript levels of the genes shown in skin abscesses 8 h after subcutaneous inoculation with USA300, USA300ΔsaeR/S or USA300Δagr.a Genes previously shown to be activated by other stimuli (fig. 1, 2). b Additional targets activated by the skin environment. Transcript levels were normalized to gyrB and calibrated to those from corresponding S. aureus strains at midexponential growth phase. Data are reported as the mean fold change of 2 separate experiments with 2 mice per group (values shown in online suppl. table 3).
In order to determine the influence of saeR/S and agr on expression of the relevant targets, we compared transcript levels of the wild type to those in the mutant strains 8 h after skin infection. Genes that were downregulated in USA300ΔsaeR/S during neutrophil interaction (fig. 3) were also substantially repressed in USA300ΔsaeR/S in the skin, and lukF-PVL and splA were downregulated in USA300Δagr (fig. 5). Surprisingly, hlgA, hlgB, hlgC and ssl7, which were repressed in the presence of neutrophils in USA300Δagr (fig. 3), were upregulated during skin infection (fig. 5). While higher transcript levels of the virulence factors in USA300Δagr were unexpected, they correlate with increased expression of saeR/S in the USA300Δagr strain during skin infection (fig. 5). Additionally, increased transcription of agrA in the saeR/S mutant during skin infection implies that saeR/S and agr might compensate for each other's absence to promote transcription of essential virulence factors (fig. 5).
Discussion
The ability of S. aureus to evade neutrophils as well as establish invasive and skin infections has been linked to the two-component system SaeR/S that regulates expression of virulence factors and immunomodulatory proteins [3, 4, 5, 7, 20]. In this investigation, we studied physiological triggers of SaeR/S and how host stimuli affect saeR/S-regulated gene expression. We found that a robust transcriptional response of genes with the SaeR binding site in USA300 takes place as early as 5 min after human neutrophil interaction. The immediate activation of the specific saeR/S targets hlgA, hlgB, hlgC, lukA and sbi in response to human PMNs suggests that these effectors might be required for initiation of infection in humans. Our findings correspond with previous reports that showed hlgABC, lukAB/lukGH and sbi promote neutrophil cell death and/or S. aureus survival during neutrophil phagocytosis [21, 22, 23, 24]. Lack of pronounced transcription of hla during early neutrophil interaction also coincides with studies that demonstrated Hla does not promote cell death of human neutrophils; however, this toxin plays an important role in targeting lymphocyte populations [25]. Given that the SaeR/S system promotes PMN damage and is required for S. aureus survival following phagocytosis [4], our findings imply that saeR/S activation triggers a PMN-specific transcriptional response facilitating upregulation of virulence factors essential to evade neutrophil killing. This idea is supported by the observation that not all genes containing the SaeR binding site were activated in response to neutrophils even though their transcription is dependent on the SaeR/S system [3, 7]. While the quorum system Agr had influence over several saeR/S targets, its impact on expression of the virulence factors during early interaction with the neutrophils was not as robust. This finding was expected as transcription of agr is not stimulated by the neutrophil environment at the multiplicities of infection tested [[11]; this study] and is not essential to USA300 survival during PMN interaction [10]. Collectively, these findings highlight the specificity of the saeR/S-mediated transcriptional response to neutrophils.
Another goal of our study was to define expression of saeR/S targets during the initial interactions with neutrophil-derived components. We found that human α-defensin elicited transcriptional changes of saeR/S targets 10 and 30 min after treatment, in accordance with previous reports [8, 16, 17]. While we cannot rule out that α-defensin-mediated cell membrane disruption promotes activation of SaeS, especially since antimicrobial peptides elicit cell damage through various mechanisms [26], LL-37 did not activate saeR/S at sublethal nor at lethal concentrations. This further emphasizes that SaeR/S activation is dependent on particular stimuli as opposed to any type of microbial damage.
α-Defensin, contained within primary azurophilic granules of the human PMN, has been suggested to be the molecular trigger of SaeR/S [8, 16, 17]; however, the expression profile elicited by α-defensin was relatively different in magnitude from that of the human neutrophil. This observation prompted us to determine whether neutrophils had to contain α-defensin to promote saeR/S target activation by exposing S. aureus to mouse neutrophils. We chose mouse PMNs because they are similar in function to the human neutrophil but do not contain α-defensin [18]. Surprisingly, mouse PMNs upregulated transcription of saeR/S targets even more robustly than human neutrophils. Based on these observations, we propose that while α-defensin is an essential activator of the SaeR/S system, there are other neutrophil-derived components and processes such as phagocytosis, reactive oxygen species production and/or degranulation that might promote transcription of saeR/S targets.
Next, we wanted to define transcript changes of saeR/S targets in vivo. As expected, the skin environment promoted a robust transcription of saeR/S targets with γ-hemolysins and leukotoxin AB/GH transcript levels being substantially high 8 h after inoculation. Interestingly, lukAB and hlgABC have been shown to be dispensable for abscess formation during skin infection in rabbits and mice, respectively [27, 28]. This could be due to the redundancy of various toxins produced by S. aureus that might compensate for the lack of HlgABC or LukAB/GH. It remains to be determined whether these toxins play a role in modulating immune responses that could be important for the progression of infection.
The specificity of the saeR/S-mediated response during skin infection was further highlighted by increased expression of α-hemolysin transcript, which is essential to dermonecrosis formation [19, 27]. Given that skin abscesses are composed of various cell types, including lymphocytes that are susceptible to Hla, our results suggest there are additional skin-derived triggers of the SaeR/S system. To begin to address this, we measured mRNA levels of saeR/S targets following exposure to skin-derived peptides β-defensin and dermicidin; however, neither of the stimuli promoted SaeR/S activation (data not shown). While the infiltrated neutrophils at the site of infection and the long exposure to the skin environment likely contribute to the observed expression profile, future investigations are needed to identify triggers of the SaeR/S system in the skin.
To confirm the impact of SaeR/S on gene expression in vivo, we examined saeR/S target expression in USA300ΔsaeR/S and USA300Δagr. As in the case of human neutrophils, expression of the majority of the genes with the SaeR binding site was dependent on the SaeR/S system during skin infection. Although deletion of agr results in reduced pathogenesis of S. aureus in the skin [20, 29], the transcript levels of several virulence factors, including α-hemolysin, which contributes to dermonecrosis, were either equal or substantially higher in the agr mutant when compared to the wild-type strain. This discrepancy can be explained by the role of Agr in posttranscriptional regulation of toxins such as α-hemolysin which translation is dependent on agr[30]. Furthermore, genes that were upregulated in the agr mutant in the skin were downregulated in the same mutant during neutrophil interaction. The biphasic regulation could be due to a variety of factors including the difference in external stimuli, bacteria growth phase and/or timing (10 min with neutrophils vs. 8 h in the skin). These findings further underscore that regulation of transcription mediated by two-component systems is complex and highly specific to the environment sensed by S. aureus.
The finding that only select saeR/S-regulated genes, as opposed to all targets with the SaeR recognition sequence, were transcriptionally upregulated in response to saeR/S-activating stimuli was unexpected. We suggest that differential activation of saeR/S targets is dependent on specific stimuli, most likely in concert with other regulatory systems, RNA decay mechanisms [31] and/or transcription factors such as negative regulator of toxins, which has been shown to repress saeR/S-regulated genes [32]. In addition, the temporal expression of targets such as lukA and sbi, which declined over the 30-min exposure to human neutrophils, as opposed to hlgABC mRNA levels that increased over time, suggests that the transcription kinetics of saeR/S targets is important and should be investigated in respect to S. aureus pathogenesis. Finally, previous studies proposed that the availability of phosphorylated SaeR (SaeR-P) and arrangement of the SaeR binding sites influence the differential regulation of the saeR/S targets [33, 34]. While some genes might require low levels of SaeR-P to be transcriptionally active, others will be activated in the presence of high SaeR-P concentrations [33]. Since transcript levels of saeR/S do not correlate with the robustness of the downstream gene expression [[33]; this study], it is likely that SaeR-P levels might dictate the magnitude of the gene transcription downstream of the SaeR/S system. Thus, future studies are needed to determine the effect of external stimuli on SaeR-P levels along with the influence of differences within the SaeR binding site sequence on differential transcription of saeR/S targets.
To our knowledge, this is the first study to identify genes that are regulated by saeR/S and agr under physiologically relevant conditions and determine the effects of host-derived stimuli including skin on saeR/S target gene expression. Our findings suggest that the SaeR/S system is a major regulator of virulence factors in the initial stages of S. aureus interaction with the innate immune system, while the Agr system has a partial influence on saeR/S target expression. Given that SaeR/S is required for successful establishment of skin and invasive S. aureus infections, subsequent studies are necessary to determine which of the saeR/S targets are responsible for saeR/S-mediated pathogenesis. Moreover, with the recent findings that the WalKR system potentially influences SaeR/S activity [35], future analyses will include the contribution of additional regulatory networks to gene expression of saeR/S targets.
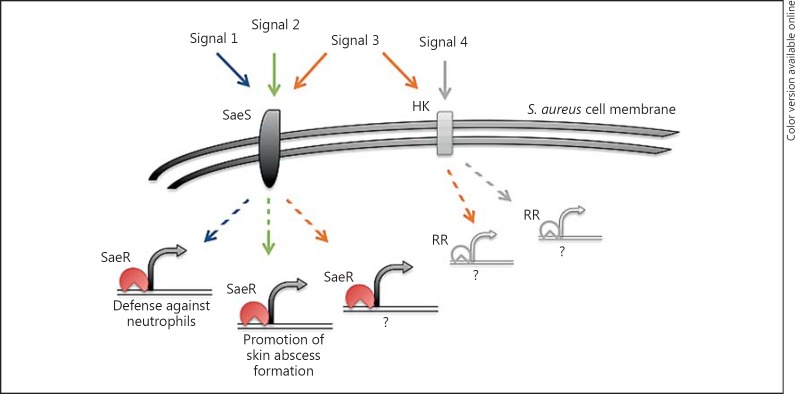
Collectively, our findings provide insight into gene regulation in S. aureus and highlight the importance of examining downstream events of gene-regulatory systems upon their stimulation. To that end, we hypothesize that activation of SaeR targets depends on the stimulus and that only specific subsets of genes respond on a transcriptional level, depending on the environment (fig. 6). This viable strategy allows the pathogen to tailor its response by expending energy only on factors necessary to survive the pending host challenge.
Fig. 6.
A model representing the dependence of saeR/S-mediated transcription on host-derived signals. Depending on the source of SaeR/S system activation, only specific subsets of genes with the SaeR binding site are activated during early interactions with host components and subcutaneous skin infection. In the presence of neutrophils and α-defensin (signal 1), activation of the SaeR/S system leads to promotion of gene transcription necessary to evade neutrophils. When stimulated by the skin environment (signal 2), specific saeR/S targets, including genes that promote skin abscess formation, are transcriptionally activated and vary in type and robustness of response from those stimulated by signal 1. Given that mouse neutrophils promote SaeR/S activation despite lacking α-defensin, there are additional host-derived components (signal 3) that can activate the SaeR/S system and potentially stimulate other gene-regulatory networks promoting pathogen survival. Additionally, activation of the SaeR/S system is highly specific to the external environment since not all host-derived factors (signal 4) promote transcription of SaeR/S targets. HK = Histidine kinase; RR = response regulator.
Disclosure Statement
The authors declare that there are no competing financial interests.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgments
We would like to thank Dr. Michael Muchow for the technical help with QuantiGene 2.0 Affymetrix assays and Caralyn Flack (University of Iowa) for critical review of the manuscript.
This work was supported by National Institute of Health grants (NIH-RR020185, NIH-R01 award A1090046-01, NIH-R21 award R21 A1088041-01) and a Molecular Biosciences Fellowship (P20RR16455-07 to O.W.Z. and R.L.W.) as well as the Montana State University Agriculture Experiment Station and an equipment grant from the Murdoch Charitable Trust. Additional support was provided by the New York University School of Medicine Development Funds (V.J.T.), an American Heart Association Scientist Development Grant (09SDG2060036 to V.J.T.) and National Institute of Health grants (R56-AI091856-01A1 to V.J.T. and NIH-AI083211 to A.R.H.).
References
- 1.Fowler VG, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, et al. Staphylococcus aureus endocarditis: a consequence of medical progress. JAMA. 2005;293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 2.McCaig LF, McDonald LC, Mandal S, Jernigan DB. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg Infect Dis. 2006;12:1715–1723. doi: 10.3201/eid1211.060190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis. 2010;201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis. 2009;199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins RL, Pallister KB, Voyich JM. The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during Staphylococcus aureus infection. PLoS One. 2011;6:e19939. doi: 10.1371/journal.pone.0019939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montgomery CP, Daum RS. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: a role for panton-valentine leukocidin? Infect Immun. 2009;77:2159–2167. doi: 10.1128/IAI.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192:2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong D-W, Cho H, Jones MB, Shatzkes K, Sun F, Ji Q, et al. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol Microbiol. 2012;86:331–348. doi: 10.1111/j.1365-2958.2012.08198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick RP. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 10.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Saïd-Salim B, Porcella SF, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 12.Siemsen DW, Schepetkin IA, Kirpotina LN, Lei B, Quinn MT. Neutrophil isolation from nonhuman species. Methods Mol Biol. 2007;412:21–34. doi: 10.1007/978-1-59745-467-4_3. [DOI] [PubMed] [Google Scholar]
- 13.Voyich JM, Sturdevant DE, DeLeo FR. Analysis of Staphylococcus aureus gene expression during PMN phagocytosis. Methods Mol Biol. 2008;431:109–122. doi: 10.1007/978-1-60327-032-8_9. [DOI] [PubMed] [Google Scholar]
- 14.Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 15.Affymetrix: QuantiGene 2.0 Assay User Manual https://www.panomics.com/downloads/UM13074_QG2Manual_RevB_080102.pdf (accessed January 20, 2013)
- 16.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol. 2008;190:3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palazzolo-Ballance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, Kreiswirth BN, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180:500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 18.Eisenhauer PB, Lehrer RI. Mouse neutrophils lack defensins. Infect Immun. 1992;60:3446–3447. doi: 10.1128/iai.60.8.3446-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy AD, Wardenburg JB, Gardner DJ, Long D, Whitney AR, Braughton KR, et al. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis. 2010;202:1050–1058. doi: 10.1086/656043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One. 2010;5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DuMont AL, Nygaard TK, Watkins RL, Smith A, Kozhaya L, Kreiswirth BN, et al. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol. 2010;79:814–825. doi: 10.1111/j.1365-2958.2010.07490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ventura CL, Malachowa N, Hammer CH, Nardone GA, Robinson MA, Kobayashi SD, et al. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One. 2010;5:e11634. doi: 10.1371/journal.pone.0011634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malachowa N, Whitney AR, Kobayashi SD, Sturdevant DE, Kennedy AD, Braughton KR, et al. Global changes in Staphylococcus aureus gene expression in human blood. PLoS One. 2011;6:e18617. doi: 10.1371/journal.pone.0018617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith EJ, Visai L, Kerrigan SW, Speziale P, Foster TJ. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun. 2011;79:3801–3809. doi: 10.1128/IAI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nygaard TK, Pallister KB, DuMont AL, DeWald M, Watkins RL, Pallister EQ, et al. Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 Infection. PLoS One. 2012;7:e36532. doi: 10.1371/journal.pone.0036532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Midorikawa K, Ouhara K, Komatsuzawa H, Kawai T, Yamada S, Fujiwara T, et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect Immun. 2003;71:3730–3739. doi: 10.1128/IAI.71.7.3730-3739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, et al. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis. 201(204):937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malachowa N, Kobayashi SD, Braughton KR, Whitney AR, Parnell MJ, Gardner DJ, et al. Staphylococcus aureus leukotoxin GH promotes inflammation. J Infect Dis. 2012;206:1185–1193. doi: 10.1093/infdis/jis495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright JS, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc Natl Acad Sci USA. 2005;102:1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marincola G, Schäfer T, Behler J, Bernhardt J, Ohlsen K, Goerke C, et al. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol. 2012;85:817–832. doi: 10.1111/j.1365-2958.2012.08144.x. [DOI] [PubMed] [Google Scholar]
- 32.Said-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, et al. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol. 2003;185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol. 2010;192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H, Jeong DW, Li C, Bae T. Organizational requirements of the SaeR binding sites for a functional P1 promoter of the sae operon in Staphylococcus aureus. J Bacteriol. 2012;194:2865–2876. doi: 10.1128/JB.06771-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaune A, Dubrac S, Blanchet C, Poupel O, Mader U, Hiron A, et al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun. 2012;80:3438–3453. doi: 10.1128/IAI.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data