ABSTRACT
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (CRISPR/Cas) system is an adaptive immune system present in many archaea and bacteria. CRISPR/Cas systems are incredibly diverse, and there is increasing evidence of CRISPR/Cas systems playing a role in cellular functions distinct from phage immunity. Previously, our laboratory reported one such alternate function in which the type 1-F CRISPR/Cas system of the opportunistic pathogen Pseudomonas aeruginosa strain UCBPP-PA14 (abbreviated as P. aeruginosa PA14) inhibits both biofilm formation and swarming motility when the bacterium is lysogenized by the bacteriophage DMS3. In this study, we demonstrated that the presence of just the DMS3 protospacer and the protospacer-adjacent motif (PAM) on the P. aeruginosa genome is necessary and sufficient for this CRISPR-dependent loss of these group behaviors, with no requirement of additional DMS3 sequences. We also demonstrated that the interaction of the CRISPR system with the DMS3 protospacer induces expression of SOS-regulated phage-related genes, including the well-characterized pyocin operon, through the activity of the nuclease Cas3 and subsequent RecA activation. Furthermore, our data suggest that expression of the phage-related genes results in bacterial cell death on a surface due to the inability of the CRISPR-engaged strain to downregulate phage-related gene expression, while these phage-related genes have minimal impact on growth and viability under planktonic conditions. Deletion of the phage-related genes restores biofilm formation and swarming motility while still maintaining a functional CRISPR/Cas system, demonstrating that the loss of these group behaviors is an indirect effect of CRISPR self-targeting.
IMPORTANCE
The various CRISPR/Cas systems found in both archaea and bacteria are incredibly diverse, and advances in understanding the complex mechanisms of these varied systems has not only increased our knowledge of host-virus interplay but has also led to a major advancement in genetic engineering. Recently, increasing evidence suggested that bacteria can co-opt the CRISPR system for functions besides adaptive immunity to phage infection. This study examined one such alternative function, and this report describes the mechanism of type 1-F CRISPR-dependent loss of the biofilm and swarming in the medically relevant opportunistic pathogen Pseudomonas aeruginosa. Since both biofilm formation and swarming motility are important in the virulence of P. aeruginosa, a full understanding of how the CRISPR system can regulate such group behaviors is fundamental to developing new therapeutics.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR-associated (Cas) proteins represent an elegant defense system against bacteriophage, plasmids, and transposable elements in which the incorporation of a small section of foreign DNA at the CRISPR locus in the bacterial or archaeal host genome provides adaptive immunity to the mobile genetic element (MGE) from which the DNA was acquired (1, 2). In general, the CRISPR system functions by transcribing the entire CRISPR locus into a long precursor RNA molecule which is further processed into multiple RNA molecules, each containing a single spacer flanked by partial repeats, known as mature CRISPR RNA (crRNA). This mature crRNA molecule then associates with Cas proteins to form a crRNA riboprotein complex that targets and inhibits the invading MGE, effectively protecting the host and preventing future infections from any MGE containing the protospacer (3–5).
CRISPR/Cas systems are widespread, present in over 84% and 47% of sequenced archaeal and bacterial genomes, respectively (6). Additionally, CRISPR/Cas systems are incredibly diverse and currently grouped into three main types: the distantly related type I and type III systems found in a wide range of bacteria and archaea and the phylogenetically distinct type II systems found solely in bacteria (7). These three types of systems are further divided into numerous subtypes that differ in many aspects, including differing cas genes, modes of action, and CRISPR repeat sequences. With the level of diversity found across these CRISPR systems, it is no surprise that there is increasing evidence of CRISPR involvement in other cellular processes besides adaptive immunity, including regulation of bacterial virulence in Campylobacter jejuni (8), group behavior in Myxococcus xanthus (9), and increased pathogenicity and antibiotic resistance in Francisella novicida (10, 11).
Previous work in our laboratory characterized an additional alternative function of the CRISPR system in which the type 1-F CRISPR system of the opportunistic pathogen Pseudomonas aeruginosa UCBPP-PA14 (abbreviated as P. aeruginosa PA14) inhibits biofilm formation and swarming motility, two group behaviors important in virulence, when lysogenized by the bacteriophage DMS3 (12, 13). This CRISPR-dependent loss of biofilm formation and swarming motility was dependent on the interaction of a specific spacer, CRISPR2 spacer 1 (CR2_sp1), with a specific protospacer located on the DMS3 lysogen genome that partially matches CR2_sp1. In this report, we describe the mechanism of this CRISPR-dependent loss of biofilm formation and swarming. We demonstrate that the interaction of CR2_sp1 with the partial matching protospacer present on the bacterial genome activates RecA through Cas3 nuclease activity. Furthermore, we show that RecA activation leads to the induction of SOS-regulated, phage-related genes that induce cell death in surface-attached cells and yet has minimal impact on planktonic cells. These findings suggest yet another mechanism by which a CRISPR/Cas system can have unexpected impacts on bacterial biology that are independent of immunity to the MGE.
RESULTS
Point mutations reveal distinct sequence requirements of the DMS3-42 protospacer for CRISPR-induced loss of biofilm formation and swarming.
Previously, we characterized the cas genes responsible for the loss of swarming and biofilm formation observed in the P. aeruginosa PA14 laboratory strain lysogenized with bacteriophage DMS3 and determined CRISPR2 spacer 1 (CR2_sp1) and the protospacer present within DMS3 gene 42 (DMS3-42) to be the required spacer and target, respectively (13). Additionally, we previously characterized several nucleotides within the spacer region required for this interaction (13). Here, we sought to further characterize this interaction between the CRISPR system and lysogenic phage DMS3 by determining the exact nucleotides required for CR2_sp1- and DMS3-42-mediated loss of biofilm formation. In addition to the 5 mismatches naturally present between CR2_sp1 and the DMS3-42 protospacer, an additional mismatch was generated for each predicted interaction site through a point mutation on the protospacer, including the protospacer-adjacent motif (PAM; see Fig. S1 in the supplemental material).
Of the 29 generated point mutations, a total of 25 resulted in complete restoration of biofilm formation and one mutation resulted in partial restoration, indicative of the nucleotides required for a productive interaction between CR2_sp1 and the DMS3-42 protospacer. A total of 3 mutations had no effect on biofilm formation, indicative of a dispensable interaction (see Fig. S1 in the supplemental material). Together, these findings defined the nucleotide sequence required for CRISPR system engagement and provided the control strains used for the studies described below.
The presence of the DMS3-42 protospacer and PAM is necessary and sufficient for CRISPR-induced loss of biofilm formation and swarming.
One explanation for the swarming and loss of the biofilm observed when strain P. aeruginosa PA14 is lysogenized by phage DMS3 is that the partial match between DMS3-42 and CR2_sp1 results in aberrant expression of DMS3 genes downstream of DMS3-42. We hypothesized that expression of these genes may result in a general stress response, leading to the observed loss of biofilm formation and swarming motility. To test this idea, we generated a deletion of the DMS3 lysogen starting 5 bp downstream from the +1 position of the DMS3-42 protospacer and ending at the end of the DMS3 genome, removing all potentially misregulated phage genes downstream of DMS3-42; this deletion had no significant effect on biofilm loss (see Fig. S2 in the supplemental material). These data suggested that genes downstream of the site of the DMS3-42 protospacer were not responsible for the observed loss of biofilm formation and swarming.
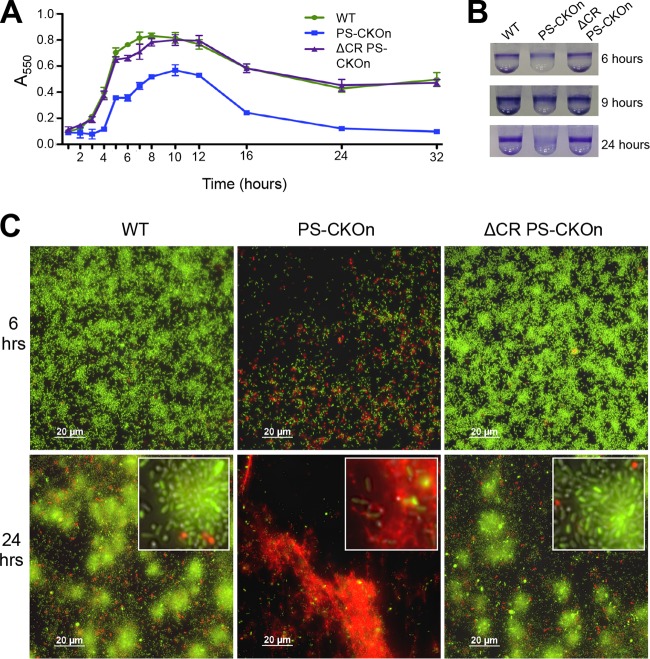
An alternative hypothesis is that the CR2_sp1 interaction with the protospacer in DMS3-42 resulted in changes to the cell that were independent of the presence of phage DMS3 and required only the presence of the protospacer and the protospacer-adjacent motif (PAM). A strong prediction of such a hypothesis is that introducing the protospacer plus PAM alone onto the chromosome of P. aeruginosa PA14 should be sufficient to recapitulate the phenotypes observed by phage DMS3 lysogeny. To test the hypothesis that the sequence consisting of the DMS3-42 protospacer plus the PAM sequence is necessary and sufficient to block biofilm formation and inhibit swarming motility, we inserted the 32-nucleotide (nt) DMS3 protospacer plus 5 bp upstream and downstream (which includes the PAM) into the P. aeruginosa att::Tn7 site (see Fig. S3 in the supplemental material), a commonly used neutral site for genomic insertions (14), in both a wild-type (WT) background and a CRISPR-deficient (ΔCR) background. Note that, even though the P. aeruginosa att::Tn7 site was used as an insertion site, the construct was inserted by using allelic exchange and not by using the Tn7 system, so no constitutively expressed drug marker or Tn7 element was inserted with the protospacer. The presence of just this 42-bp insertion (designated the PS-CKOn, for “protospacer chromosomal knock-on”) inhibited biofilm formation in a CRISPR-dependent manner since the PS-CKOn insertion in the ΔCR background had no effect, perfectly phenocopying a DMS3 lysogen and thus indicating no requirement of bacteriophage genes to confer these phenotypes (Fig. 1A). Scrambling the spacer sequence and inserting this scrambled sequence at the att site resulted in no impact on biofilm formation compared to the wild-type strain (Fig. 1A; scrambled CKOn).
FIG 1 .
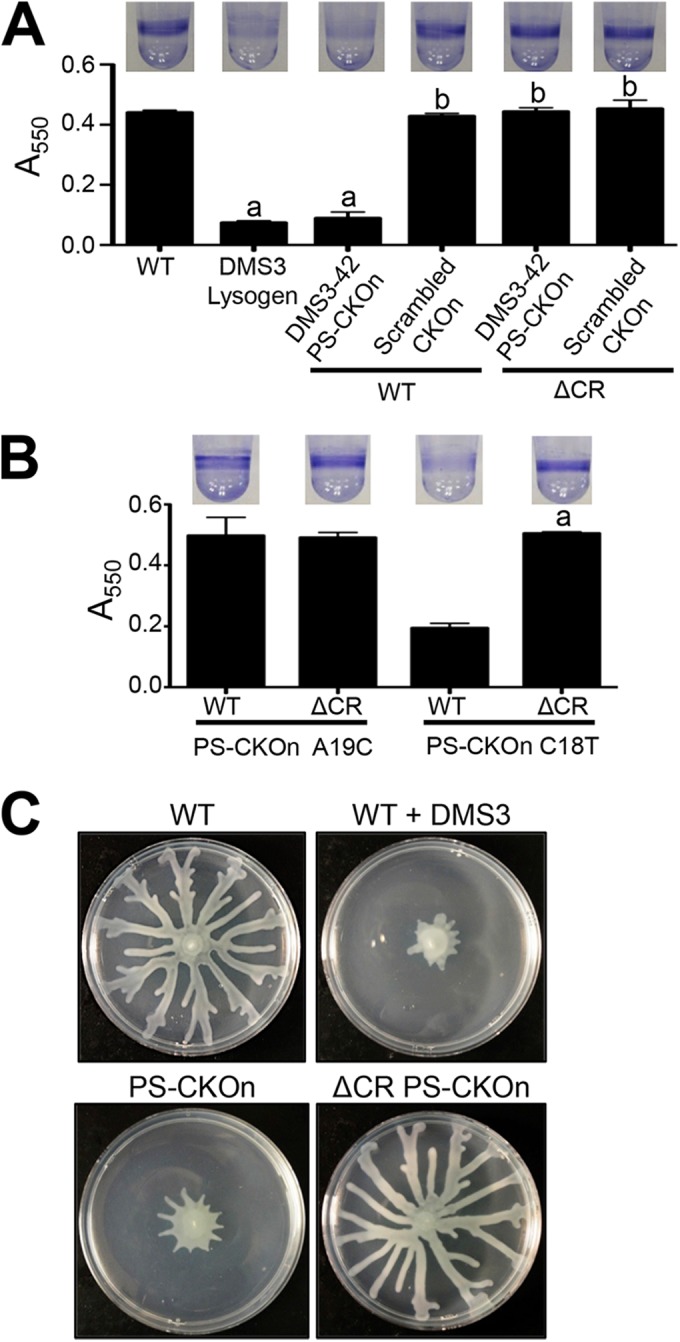
The DMS3-42 protospacer is necessary and sufficient for CRISPR-induced loss of biofilm formation and swarming. (A and B) Biofilm formation assays were performed on the indicated strains as described in Materials and Methods, with CKOn representing the chromosomal knock-on of the DMS3-42 protospacer (PS-CKOn) or the scrambled sequence of the DMS3-42 protospacer (Scrambled CKOn) or the DMS3-42 protospacer with a point mutant (PS-CKOn A19C and PS-CKOn C18T). Error bars represent standard deviations of the results of at least three biological replicates. “a” indicates a result significantly different from the WT result at a P value of <0.05; “b” indicates a result significantly different from the DMS3 lysogen result at a P value of <0.05. (C) The indicated strains were assayed for swarming motility as described in Materials and Methods.
Additionally, two point mutations were introduced into the PS-CKOn construct at nucleotides that were shown to be required (A19C) or dispensable (C18T) for loss of biofilm formation; these mutant PS-CKOn constructs were inserted into both a WT and a CRISPR-deficient (ΔCR) background. Similarly to the point mutations engineered into the bacteriophage, as illustrated in Fig. S1 in the supplemental material, A19C (equivalent to A245C) restored biofilm formation whereas C18T (equivalent to C246T) had no effect (Fig. 1B). In addition to the loss of biofilm formation, the 42-nt PS-CKOn insertion inhibited swarming motility in a CRISPR-dependent manner to a degree indistinguishable from that seen with lysogeny by phage DMS3 (Fig. 1C). These data show that mutations in the full-length phage and the protospacer sequence confer the same phenotypes.
Interestingly, the presence of the DMS3-42 protospacer and PAM on a stably maintained plasmid had no effect on biofilm formation or swarming (data not shown), indicating that this 42-nt sequence needed to be inserted onto the chromosome to confer the observed biofilm and swarming phenotypes. Taken together, these data indicate that the presence of the 32-nt DMS3-42 protospacer plus the 5 flanking nucleotides on each side (including the PAM) on the P. aeruginosa chromosome is necessary and sufficient for CRISPR-dependent loss of biofilm formation and swarming, with no requirement of any additional bacteriophage-derived sequences.
The CR2_sp1-guided CRISPR interaction with PS-CKOn reduces biofilm formation in P. aeruginosa by triggering cell death upon surface attachment.
After establishing that P. aeruginosa harboring a DMS3 lysogen, or the 42-nt PS-CKOn construct, did not form a biofilm at 24 h, we wanted to further characterize this phenotype by assaying biofilm formation at earlier time points. Biofilm formation was measured for wild-type P. aeruginosa PA14, the PS-CKOn strain, and the CRISPR-deficient strain carrying the PS-CKOn insertion (ΔCR PS-CKOn) every hour for the first 8 h and then at 10, 12, 16, 24, and 32 h postinoculation. As illustrated in Fig. 2A and B, the PS-CKOn strain attached and formed a biofilm with a lag greater than that observed for the WT, but once biofilm formation was detected, the PS-CKOn strain formed biofilms with kinetics similar to that of wild-type P. aeruginosa PA14 over the first few hours; however, significantly lower levels of biofilm formation were observed for the PS-CKOn strain from 3 h onward (P < 0.01 for all time points >3 h). Furthermore, by 24 and 32 h, the PS-CKOn strain showed significantly reduced levels of biofilm formation which were close to background levels (i.e., biofilm levels below the limit of detection), but for the parental P. aeruginosa PA14 strain, a robust biofilm was still detected (Fig. 2A and B). Notably, this phenotype is CRISPR dependent as the ΔCR PS-CKOn strain was indistinguishable from the wild type at all time points assessed.
FIG 2 .
The CR2_sp1-guided CRISPR interaction with PS-CKOn induces cell death on a surface and a reduction in biofilm formation. (A) Biofilm formation was assayed and quantified at the indicated time points. Error bars represent standard deviations. (B) Representative crystal violet (CV) staining images of biofilm assays from the three time points indicated. (C) The indicated strains were grown on a glass coverslip under biofilm-inducing conditions, stained using a BacLight cell viability kit, and visualized at the air-liquid interface (ALI) at 6 or 24 h. Green cells are considered viable and red cells nonviable. The diffuse red staining is likely to represent extracellular DNA. The entire field of view is displayed, with white-outlined insets showing a magnified section of the image.
Inspection of the earliest time points (Fig. 2A, 0 to 5 h) revealed that the PS-CKOn strain showed a lag in biofilm formation but that, once initiated, the kinetics of biofilm formation was similar to the WT kinetics during this early time period, indicating that the biofilm defect observed at 24 h was not due solely to a defect in the initial attachment of the bacteria. Furthermore, the kinetics of biofilm formation observed at 5 h slowed for the PS-CKOn strain and never attained the full extent of biofilm formation observed for the WT, with the PS-CKOn strain actually showing a reduction in biofilm formation beginning at ~10 h. These data suggested that the PS-CKOn strain could initiate attachment but that the cells in the biofilm might have been losing viability over time. To test this idea, the wild-type P. aeruginosa PA14, PS-CKOn, and ΔCR PS-CKOn strains were grown using the air-liquid-interface (ALI) assay previously described by our group (15). Briefly, cells were grown in a 12-well dish, a glass coverslip was submerged in each well, and the coverslip was removed at the indicated time point, washed, and stained using a Molecular Probes LIVE/DEAD BacLight kit to assay cell viability of the biofilm population at the air-liquid interface of the coverslip. In this assay, cells with intact membranes (indicating viability) stain green, while cells with compromised membranes, and thus considered nonviable, stain red with propidium iodide (PI).
At 6 h, the biofilm formed by the strain carrying the PS-CKOn insertion was visibly less dense than the biofilm formed by the wild-type parent strain or the biofilm formed by the ΔCR PS-CKOn strain (Fig. 2C). Furthermore, the biofilm formed by the strain carrying the PS-CKOn construct demonstrated much-more-observable PI-staining cells compared to the wild-type strain or the CRISPR-deficient strain carrying the protospacer insertion (ΔCR PS-CKOn; Fig. 2C). At 24 h, few viable cells were visible among the biofilm cells formed by the strain carrying the PS-CKOn construct and a strong, diffuse PI staining pattern was observed, presumably representing binding of the DNA from lysed cells, indicative of widespread cell death. This biofilm reduction through a cell death phenotype was specific to the stain carrying the PS-CKOn and was not common in other biofilm-defective mutants, as the strain carrying a mutation in the gene coding for the SadC di-guanylate cyclase, which has a biofilm defect similar to the PS-CKOn strain at 6 h (16, 17), showed no difference from the WT in viability staining (see Fig. S4A to C in the supplemental material). Together, these data indicate that strains carrying the protospacer and a functional CRISPR system showed increased cell death leading to a reduction in the biofilm biomass, a finding in strong contrast to the results seen with the wild-type strain or the ΔCR PS-CKOn strain, both of which formed a robust, viable biofilm at 24 h (Fig. 2).
Note that the cell death observed for the strain carrying the PS-CKOn insertion was specific to the biofilm population, as no difference in viability staining was observed for these strains in the planktonic population (see Fig. S4D and E in the supplemental material) and a minimal difference in planktonic growth was observed for the planktonic cells from these strains during the 24-h biofilm assay (see Fig. S4F). Together, these data support a model in which biofilm formation is inhibited by CRISPR-mediated, biofilm-dependent cell death and not by attachment defects or early dispersal events.
The interaction of the CRISPR system with the DMS3 protospacer results in the overexpression of SOS-induced genes, including pyocins and the predicted phage-related PA14_52530 operon.
To explore the mechanism by which the strain carrying the DMS3-42 spacer and PAM showed enhanced cell death when growing as a biofilm, we explored differences in transcription between the wild-type and PS-CKOn-carrying strains. Whole-cell RNA was harvested from either wild-type or PS-CKOn strains grown on swarm agar for 16 h and submitted for high-throughput RNA sequencing (RNA-Seq). We used RNA isolated from cells grown on swarm agar because this was a growth condition under which we observed a clear phenotypic difference between these strains (Fig. 1C) and that also allowed us to collect sufficient cellular material to isolate the RNA required for the transcriptional analysis. CLC Genomics Workbench and edgeR were used to assemble and analyze the data obtained from the transcriptional profiling, using a cutoff of a change of 2-fold or greater and a P value of less than 0.05. As displayed in Table S1 in the supplemental material, a total of 65 genes were found to be significantly and differentially regulated between these strains using these criteria, with 60 genes upregulated and 5 downregulated in the PS-CKOn strain compared to the wild type.
Thirty-five of the upregulated genes identified by RNA-Seq in the PS-CKOn strain, which include prtN (the gene coding for the transcriptional activator of pyocin production) and all 34 predicted genes of the R pyocin, the F pyocin, and the associated lambda-like lysis cassette, map to the R2 and F2 pyocins of P. aeruginosa. The R and F pyocins, which resemble bacteriophage tails, are normally produced in P. aeruginosa under the control of the SOS response (18), which is induced by stressors such as mitomycin C (19), hydrogen peroxide (20), or nitric oxide (21). The R and F pyocins show bactericidal activity against closely related P. aeruginosa strains by adsorption via lipopolysaccharide (LPS), creating a pore and permeabilizing the inner membrane and thus effectively depolarizing the cell (22). Relevant to our observation that the strain carrying the PS-CKOn insertion showed increased cell death, the pyocins are released via autolysis induced by the phage-like lysis cassette encoded by this genetic locus (22, 23); thus, we hypothesized that overexpression of pyocin genes could account for the observed cell death.
Another five of the upregulated genes belong to the PA14_52530-repressed operon (see Table S1 in the supplemental material). This operon has been characterized in P. aeruginosa PAO1 and includes five genes that are normally repressed by PA14_52530, a LexA-like repressor, but are highly expressed during the SOS response due to the induced proteolysis of the PA14_52530 transcriptional regulator by activated RecA (23, 24). Four of these genes (PA14_52480 to PA14_52510) encode predicted phage-related proteins, with two (PA14_52510 and PA14_52500) showing sequence similarity with phage holins, suggesting a role in cell lysis. The remaining differentially regulated genes are described in Text S1.
CRISPR targeting of the DMS3 protospacer leads to overexpression of R/F pyocin-encoding genes and the PA14_52530 operon, and this overexpression is enhanced in biofilm-grown bacteria.
To confirm that the R and F pyocins are overexpressed in the PS-CKOn strain growing on a surface, expression of PA14_07990 (the predicted R/F pyocin holin; Fig. 3A) and of PA14_08300 (a predicted F pyocin tail protein; see Fig. S5A in the supplemental material) was assayed in swarm-grown cells as representative pyocin-related genes using quantitative reverse transcriptase PCR (qRT-PCR). These two genes were chosen due to their respective locations near the beginning and the end of the 26.9-kb pyocin R/F-encoding operon. RNA was harvested from the wild type, the strain carrying the PS-CKOn insertion, and a CRISPR-deficient strain carrying this same protospacer/PAM insertion (ΔCR PS-CKOn) grown for 16 h either on M8 soft agar (swarm agar conditions) or in M8 broth (planktonic conditions) at 37°C and subsequently reverse transcribed to cDNA for qRT-PCR analysis.
FIG 3 .
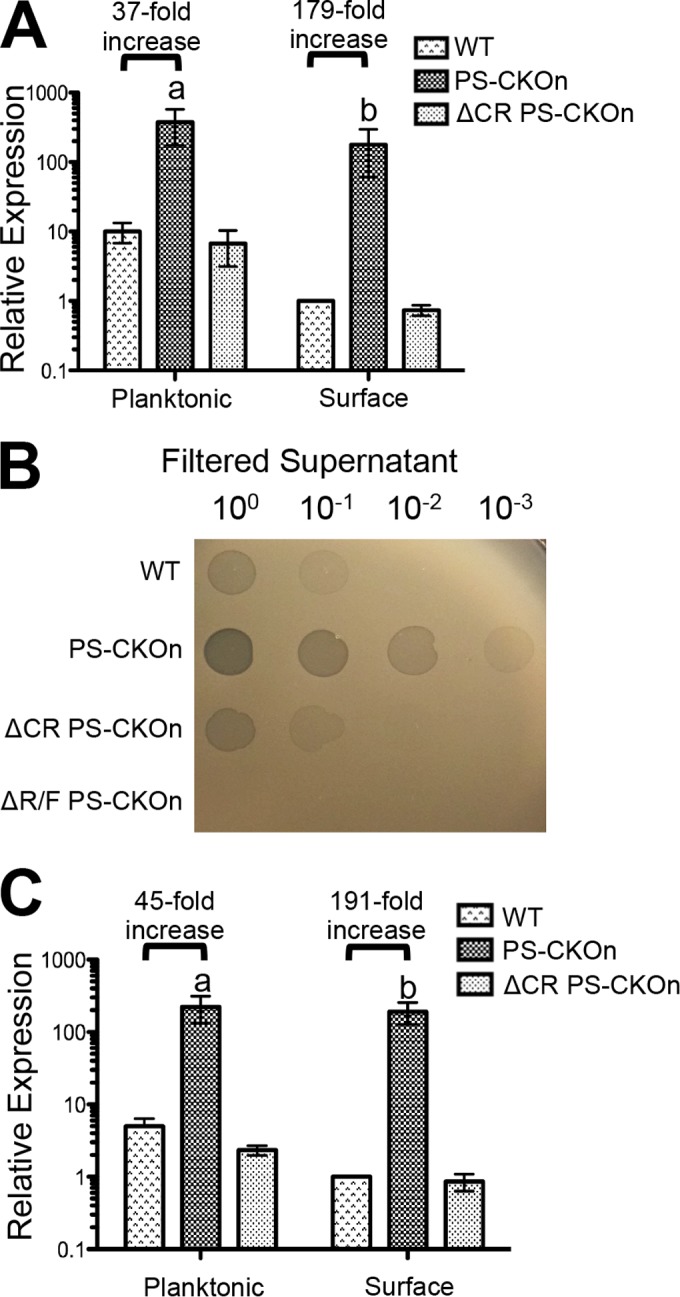
The interaction of the CRISPR system with the DMS3 protospacer induces the expression of SOS-regulated phage-related genes normally repressed on a surface. (A) qRT-PCR assays were performed to measure the transcript of PA14_07990 for planktonic and surface populations for the indicated strains normalized to rplU and displayed relative to the WT surface-grown population. (B) Ten-fold serial dilutions of filtered supernatant from the indicated strains grown overnight in LB at 37°C were spotted onto a lawn of pyocin-sensitive P. aeruginosa strain PAK. Plaques indicating lysis of the bacterial cells are shown. (C) qRT-PCR assays were performed to measure the transcript of PA14_52480 for planktonic and surface populations for the indicated strains normalized to rplU and are displayed relative to the WT surface-grown population. For panels A and C, “a” indicates a P value of <0.05 compared to the WT grown planktonically and “b” indicates a P value of <0.05 compared to the WT grown on a surface. Black horizontal bars display the fold increase in relative expression of the indicated transcript from the wild type compared to the results from the PS-CKOn strains under both planktonic and surface conditions. Error bars represent standard deviations of the results from at least three biological replicates.
Strikingly, for the cells grown in M8 broth (e.g., planktonic conditions), the pyocin transcripts were ~40-fold more abundant in the strain carrying the PS-CKOn construct than in the wild type. Furthermore, this expression phenotype was CRISPR dependent, as the upregulation of these genes was lost in the CRISPR-deficient background (ΔCR PS-CKOn) (Fig. 3A; see also S5A in the supplemental material). Because the R/F pyocin operon includes a lysis cassette and we observed enhanced killing of biofilm-grown cells but not planktonic cells, we hypothesized that the CRISPR-dependent overexpression of the R/F pyocin operon would be more pronounced in surface-grown cells than in planktonic cells. As predicted, for cells grown on a surface, the pyocin transcripts were >150-fold more abundant in the strain carrying the PS-CKOn insertion than in the wild type (Fig. 3A; see also Fig. S5A in the supplemental material). Interestingly, this difference in expression was due to an ~10-fold decrease in pyocin expression by wild-type, surface-grown cells compared to wild-type planktonic cells (Fig. 3A; see also Fig. S5A). These data suggest that P. aeruginosa normally downregulates pyocin expression on a surface, and the inability of the PS-CKOn strain to do so could account for the surface-specific cell death phenotype that we observed. This downregulation of pyocin expression by the wild-type cells growing on the surface also suggested the possibility that P. aeruginosa PA14 may be more susceptible to pyocin-mediated killing when growing as a biofilm, a point we address further in the Discussion. It should be noted that if the expression of our qRT-PCR control gene, rplU, were increased in surface-grown cells, the levels of pyocin transcript in these surface-grown cells would be underrepresented. To control for this possibility, a second and functionally distinct control gene, proC, was selected as a normalization control, and a similar trend was observed (see Fig. S5B). To our knowledge, this is the first study to have assayed pyocin production in surface-grown cells versus planktonically-grown cells.
To investigate whether these overexpressed pyocins are produced and released and are thus functional, a pyocin killing assay was performed. Briefly, the wild-type, PS-CKOn, and ΔCR PS-CKOn strains, as well as the PS-CKOn strain in which the 26.7-kb·R/F operon was deleted (ΔR/F PS-CKOn), were grown in LB at 37°C for 16 h. The supernatants were then centrifuged to remove the bacterial cells, filter sterilized through a 0.22 µm-pore-size filter, and serially diluted (1:10), and 5-µl volumes of the dilutions were spotted onto a lawn of P. aeruginosa strain PAK, which is a strain sensitive to P. aeruginosa PA14-produced pyocins (25). In analogy to the qRT-PCR data, the PS-CKOn strain produced ~100-fold more pyocin particles than the wild-type strain and did so in a CRISPR-dependent manner. As a control, no zone of clearing was present with the supernatant prepared from the ΔR/F PS-CKOn strain, indicating that the zones of clearing were R and F pyocin specific (Fig. 3B).
To also confirm the RNA-Seq data showing upregulation of the phage-related PA14_52530 operon in the PS-CKOn background, qRT-PCR was used to assay expression of two genes in the PA14_52530 operon, PA14_52480 (Fig. 3C) and PA14_52490 (see Fig. S5C in the supplemental material). The growth conditions discussed above were used for these experiments. As seen with the pyocin genes, the transcripts of both PA14_52480 and PA14_52490 were found to be >140-fold higher in the strain containing the PS-CKOn insertion than in the wild-type strain when grown on a surface but were only ~40-fold higher in the PS-CKOn strain than in wild-type strain when grown planktonically (Fig. 3C; see also Fig. S5C). Similar data were also observed using proC as a normalization control (see Fig. S5D). Similarly to the pyocin transcripts, these data suggest that the predicted phage-related PA14_52530 operon genes are normally repressed on a surface and that the inability of the PS-CKOn strain to downregulate their expression could contribute to the surface-specific death observed. To ensure that the surface-dependent repression of the pyocins and PA14_52530-repressed genes in the WT background was not an artifact of the rplU and proC normalization control genes being differentially regulated under surface versus planktonic conditions, a third distinct normalization control gene, fabD, was used to measure the expression of both rplU and proC in WT cells grown planktonically or on a surface. No difference in expression of either gene was observed (see Fig. S5E and F). Taken together, these data suggest that the CRISPR system targeting the DMS3 protospacer was inducing the SOS response, leading to >100-fold upregulation of the R/F pyocin and PA14_52530 operon genes when cells were grown on a surface, as well as increasing the functional production of pyocins to a similar degree.
The CRISPR-dependent overexpression of pyocins and PA14_52530 genes requires RecA and Cas3 nicking activity.
It has been reported that the R/F pyocin genes and the PA14_52530 operon are derepressed during the SOS response through RecA-mediated proteolysis of LexA-like repressors PrtR and PA14_52530, respectively (22, 23). To test if the overexpression of the R/F pyocins and the PA14_52530 operon in the PS-CKOn background was due to RecA activity, the recA gene was deleted in both the wild-type and PS-CKOn genetic backgrounds. The wild-type strain, the ΔrecA mutant, the strain carrying the PS-CKOn construct, and the ΔrecA PS-CKOn strain were grown for 16 h on M8 soft agar at 37°C (swarming conditions) before RNA was harvested for qRT-PCR studies, as described above. The CRISPR-induced pyocin overexpression was completely abolished in the ΔrecA mutant background (Fig. 4A), confirming that the pyocin overexpression was indeed likely the result of the SOS response. Additionally, deleting recA in the PS-CKOn background resulted in a severe growth defect compared to cells in which recA was deleted in the wild-type background (see Fig. S6 in the supplemental material), indicating that RecA is required for normal planktonic growth in the PS-CKOn background.
FIG 4 .
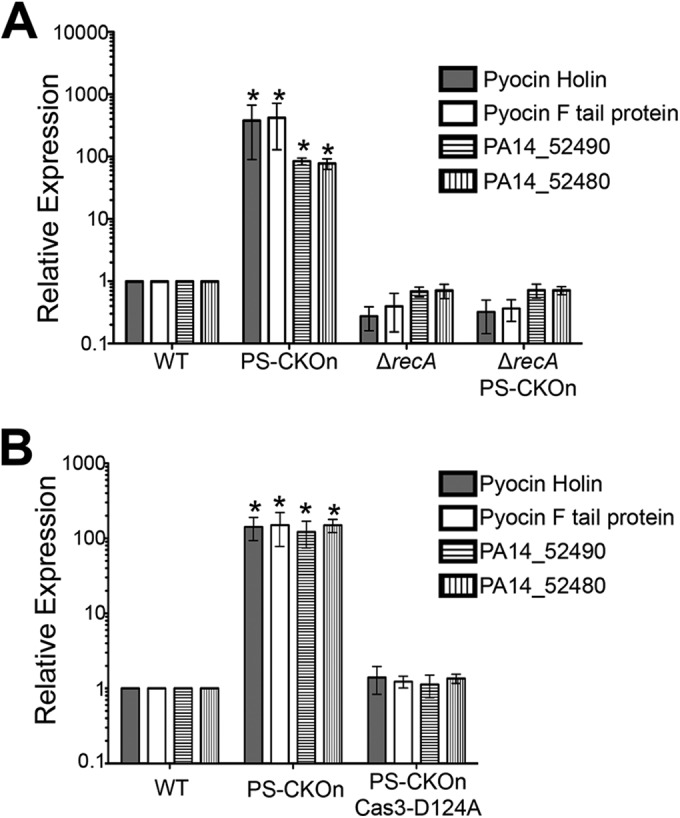
The CRISPR-dependent overexpression of phage-related genes requires RecA and the nicking activity of CRISPR-associated nuclease Cas3. (A and B) qRT-PCR assays were performed to measure the transcript of the indicated strain for swarm-grown cells normalized to rplU, and the results are displayed relative to the expression of the WT strain. Error bars represent standard deviations of the results from at least three biological replicates. *, P < 0.05 (compared to the WT).
Given that a functional CRISPR system is also required for the pyocin overexpression (Fig. 3A and B), and because RecA is activated by DNA damage (24, 26), we hypothesized that CRISPR-associated protein Cas3, a well-characterized nickase conserved in type 1 systems (4), might bind at the partially matching DMS3 protospacer (see Fig. S1 in the supplemental material) and activate RecA through the formation of single-strand nicking. To test this idea, we introduced the DMS3 protospacer and PAM into a P. aeruginosa strain carrying a nickase-defective allele of Cas3 (D124A). For this mutant, the aspartate at the HD-nuclease domain has been substituted with an alanine—the HD-nuclease domain is required for nuclease activity (27, 28). We previously demonstrated that the Cas3(D124A) mutant restores biofilm formation in a DMS3 lysogen (indicating a loss of CRISPR/Cas activity), is expressed as a stable protein, and complements mature crRNA generation lost in a Δcas3 mutant background (13). Thus, the Cas3(D124A) mutant appears to have a defect specific to the loss of its nicking activity. As expected, in the Cas3(D124A) mutant background, the high-level pyocin overexpression observed in the strain carrying the 42-nucleotide protospacer/PAM insertion, measured with qRT-PCR, returned to the wild-type level (Fig. 4B).
Together, these data suggest that the partial binding of the CRISPR system to the DMS3 protospacer recruits Cas3, which apparently nicks at a level low enough to not cause any noticeable growth defects in planktonic cells but generates sufficient DNA damage to activate RecA, which, in turn, based on published reports (22, 23), induces proteolysis of PrtR and PA14_52530, resulting in high-level induction of the genes encoding the pyocins and the PA14_52530 operon. Supporting this model is the observation that the strain carrying the ΔrecA mutation and the PS-CKOn insertion has a severe growth defect compared to the wild type or the ΔrecA mutant (see Fig. S6 in the supplemental material), indicating that the Cas3 nicking of the DMS3 protospacer is detrimental to the cell in the absence of a fully functioning SOS response system.
Deletion of the R/F pyocins and the PA14_52530 operon prevents the CRISPR-DMS3 protospacer interaction from inducing cell death for surface-grown bacteria.
We hypothesized that CRISPR-dependent overexpression of pyocins and the PA14_52530 operon, particularly, the inability to suppress the expression of these functions in biofilm-grown cells, is responsible for the surface-dependent death. To test this hypothesis, the entire R/F pyocin operon (PA14_07970 to PA14_08330, not including the prtR and prtN regulatory genes) and the PA14_52530 operon (PA14_52480 to PA14_52520, not including the PA14_52530 regulatory gene) were deleted in the wild-type and PS-CKOn backgrounds. Using the air-liquid-interface cell-viability assay, we examined CRISPR-induced, surface-dependent death after 6 h of biofilm growth. Additionally, we quantified the level of cell death by measuring the intensity of live-cell fluorescence (Syto-9) and dividing by the fluorescence of the membrane-compromised population (propidium iodide).
With the pyocins and PA14_52530 operon intact, PS-CKOn cells were significantly less viable (P < 0.001), with a decrease in viability staining of over 500-fold compared to the WT cells (Fig. 5). Deleting the R/F pyocin operon in the PS-CKOn background significantly reduced cell death by over 40-fold (P < 0.01), and deleting the PA14_52530 operon also significantly reduced cell death by over 20-fold (P < 0.01), indicating that both operons contribute to CRISPR-mediated cell death (Fig. 5). Furthermore, in a PS-CKOn background where both the pyocins and PA14_52530 operon were deleted, cell viability was restored to nearly WT levels, with a reduction in cell killing of over 200-fold, and the double mutant was significantly more viable than the mutants with a single R/F deletion (P < 0.01) or the PA14_52530 operon deletion (P < 0.01).
FIG 5 .
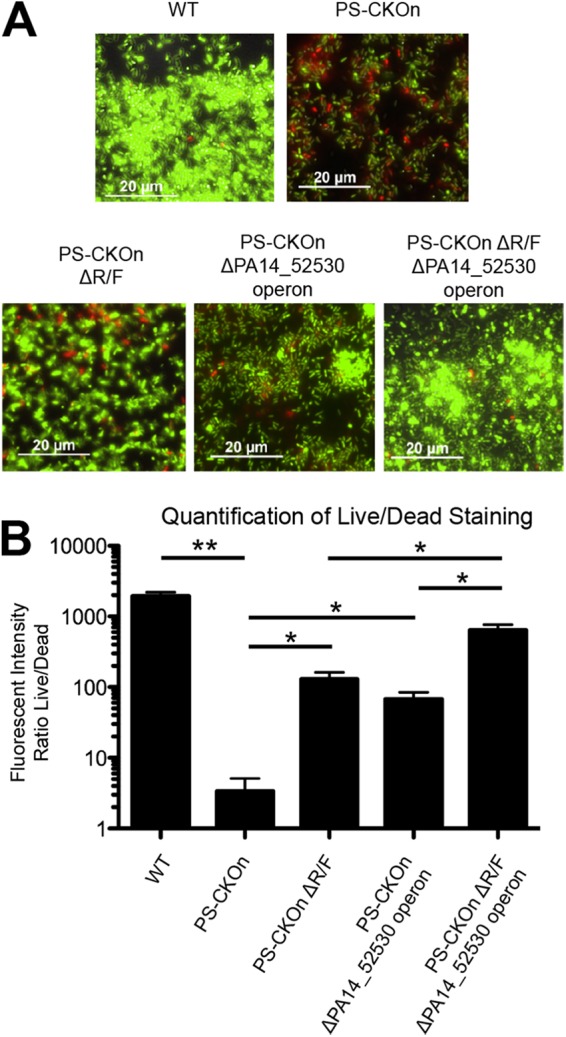
Deletion of the CRISPR-induced, phage-related genes restores viability in the biofilm population. (A) The indicated strains were grown on a glass coverslip under biofilm-inducing conditions, stained with a BacLight cell viability kit, and visualized at the air-liquid interface (ALI) at 6 h. (B) Quantification of the ALI images displayed as the mean intensity of viable cells (Syto-9; green cells in panel A) over the mean intensity of the membrane-compromised cells (propidium iodide; red cells in panel A) for the indicated strains. Error bars represent standard deviations of the results from three biological replicates. Horizontal black bars indicate a P value of either <0.01 (*) or <0.001 (**) as determined with a Student’s t test.
To confirm that the biofilm restoration in these mutants is not related to a loss in CRISPR functionality, all three mutants represented in Fig. 5B, along with the WT, PS-CKOn, and ΔCR PS-CKOn strains, were challenged with phage DMS3 or DMS3-T255C. We previously showed that DMS3-T255C is targeted by the P. aeruginosa CRISPR system (29), so we hypothesized that if the pyocin and PA14_52530 repressed-operon deletion mutants were CRISPR defective, these strains would support plaque formation by DMS3-T255C. As illustrated in Fig. S7A in the supplemental material, all three of the mutants assayed as described for Fig. 5 have a CRISPR system with functionality similar to that of the P. aeruginosa WT and PS-CKOn strains. Together, our data indicate that the observed cell death in the PS-CKOn background is not a direct result of CRISPR-protospacer interaction but, rather, is an indirect effect caused by the inability to suppress RecA-dependent overexpression of the R/F pyocins and the PA14_52530 operon on a surface.
Deletion of the R/F pyocins and the PA14_52530 operon restores swarming and biofilm formation in the PS-CKOn strain background.
Given that deleting the R/F pyocins and PA14_52530 operon in the PS-CKOn background prevented CRISPR-mediated killing on a surface, and because we hypothesized that this surface-specific killing was reducing biofilm formation and swarming motility, we predicted that, without the R/F pyocins and PA14_52530 operon, the CRISPR-DMS3 protospacer interaction would no longer impact biofilm formation and swarming motility.
We assayed biofilm formation and swarming motility at 24 and 16 h, respectively, since these group behaviors are completely inhibited at those time points in the PS-CKOn background (Fig. 1A and C). Interestingly, while deleting either the R/F pyocins or the PA14_52530 operon in the PS-CKOn background inhibited cell death only partially (Fig. 5), deleting the R/F pyocins restored biofilm formation to approximately wild-type levels and deleting the PA14_52530 operon restored biofilm levels such that they approached but were still significantly less (P < 0.05) than wild-type levels at 24 h (Fig. 6A). It is worth noting that P. aeruginosa PA14 has three additional S pyocins (S2, S3, and S5) that are also induced during the SOS response but that deleting these pyocins in addition to the R/F pyocins had no detectable effect on biofilm formation in the PS-CKOn background (see Fig. S7B in the supplemental material).
FIG 6 .
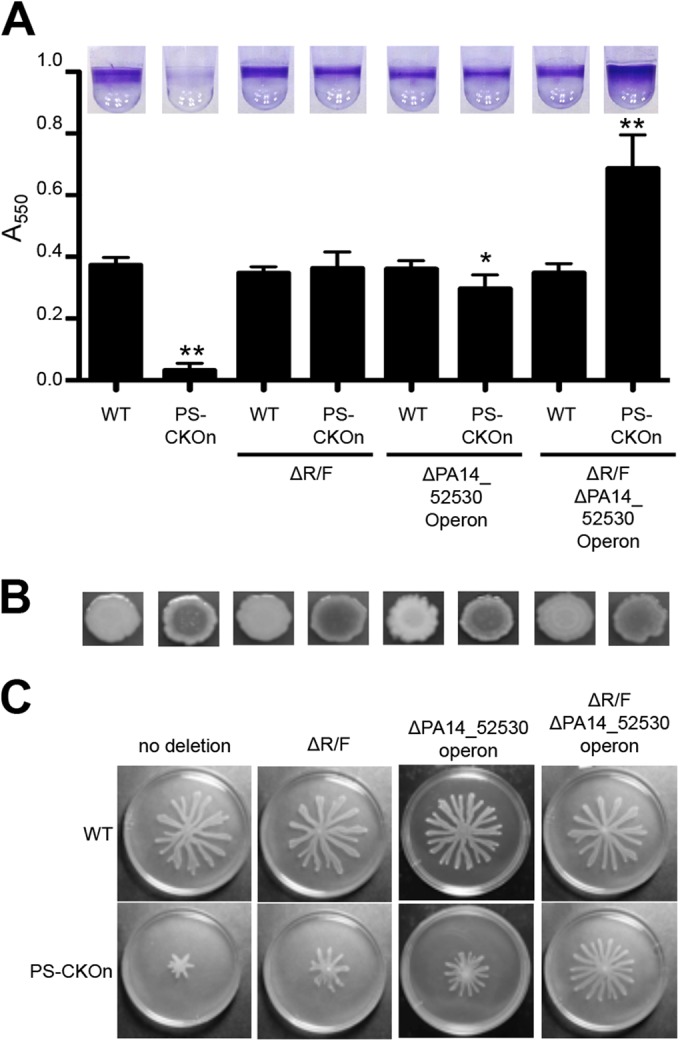
Deletion of the CRISPR-induced, phage-related genes restores biofilm formation and swarming motility in the PS-CKOn background. (A) Biofilm assays for the indicated strains are shown. Error bars represent standard deviations of the results of at least three biological replicates, with representative wells shown at the top of the panel and with a single asterisk (*) and two asterisks (**) representing P values of <0.05 and <0.01, respectively, compared to the WT as determined with a Student’s t test. (B) The strains indicated in panel A were inoculated on Congo red plates, grown for 16 h at 37°C followed by incubation at room temperature for 48 h, and imaged. A darker center is indicative of increased polysaccharide production. (C) The indicated strains were assessed for swarming as described in the text.
Additionally, the double deletion in which both the R/F pyocins and the PA14_52530 operon were deleted in the PS-CKOn background resulted in hyperbiofilm formation compared to the wild-type strain. Importantly, this hyperbiofilm phenotype was specific to the CRISPR-DMS3 protospacer interaction since deleting the R/F pyocin operon or the PA14_52530 operon, or both, in a wild-type background (in which the DMS3 protospacer was absent) had no effect on biofilm formation (Fig. 6A).
We had observed that P. aeruginosa DMS3 lysogens show a CRISPR-dependent increase in the Congo red binding phenotype, suggesting increased polysaccharide production, so we assayed the Congo red binding of the strains used in the biofilm assay represented in Fig. 6A. The PS-CKOn strains all had increased Congo red binding compared to the WT strain (Fig. 6B). Since increased polysaccharide production has previously been shown to correlate with hyperbiofilm formation in P. aeruginosa (30), we hypothesized that the hyperbiofilm phenotype observed with the PS-CKOn ΔR/F ΔPA14_52530 operon strain was likely due to increased expression of polysaccharide in the CRISPR-DMS3 protospacer-interacting backgrounds. This hyperbiofilm phenotype would be revealed in the PS-CKOn strain only when the pyocin and PA14_52530 operons were deleted, eliminating surface-associated killing of the cells. The basis of the CRISPR/Cas-induced increase in Congo red binding is not understood.
The same strains used in the biofilm assay were inoculated on swarm agar and incubated at 37°C for 16 h. With the pyocins and PA14_52530 operon intact, the presence of the DMS3 protospacer on the chromosome in the PS-CKOn strain inhibited swarming motility compared to the WT strain (Fig. 6C). However, deleting either the R/F pyocins or the PA14_52530 operon in the PS-CKOn strain resulted in a partial increase in swarming motility, while deleting both the R/F pyocins and the PA14_52530 operon restored swarming to near-WT levels (Fig. 6C). Additionally, these deletions had no effect on swarming without the DMS3 protospacer on the chromosome. Together, these data suggest that, similarly to the surface-associated cell death phenotype, the observed loss of swarming and biofilm formation in the PS-CKOn background is not a direct result of the CRISPR-DMS3 protospacer interaction but is likely rather an indirect effect caused by the inability to suppress overexpression of the R/F pyocins and the PA14_52530 operon on a surface.
DISCUSSION
The CRISPR/Cas system has been well characterized as an adaptable microbial immune system against phage and other mobile genetic elements (31), including in P. aeruginosa (29), but it has become evident that bacteria, archaea, and phage can co-opt the CRISPR/Cas system for alternative functions, including regulation of group behavior (9), gene regulation (32), or avoiding phage defense systems (33). Our first observations of CRISPR function in P. aeruginosa suggested an alternative function for this system, since CRISPR targeting of the phage DMS3 protospacer inhibited biofilm formation and swarming motility while imposing no obvious planktonic growth defect on the bacteria and no decrease in the efficiency of plaquing of phage DMS3 (12, 29). Further investigation determined that these biofilm- and swarm-inhibited phenotypes were dependent on the two mismatches between CR2_sp1 and DMS3 at positions +9 and +11 from the 5′ end of the spacer. If the mismatch at either position were corrected, the CRISPR system would switch to the more typical defense system, and the DMS3 bacteriophage would not be able to infect P. aeruginosa (29).
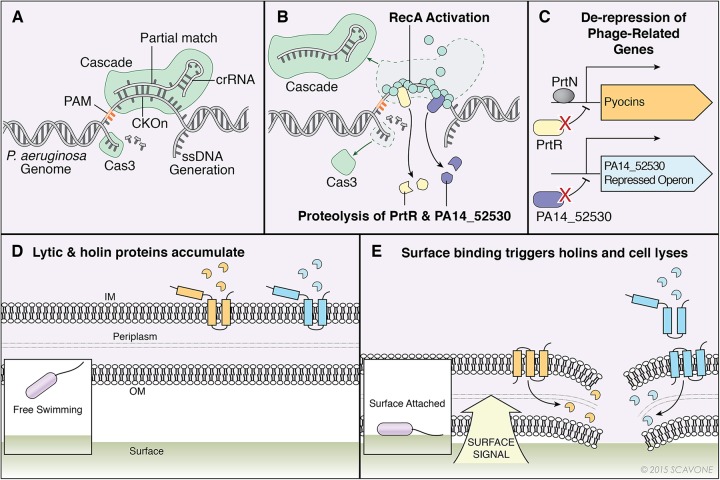
How is this CRISPR-protospacer interaction leading to loss of biofilm formation and swarming? Our work supports a model, illustrated in Fig. 7, which requires the P. aeruginosa CRISPR system to self-target the P. aeruginosa chromosome though CRISPR2 spacer 1-guided Cascade (CRISPR-associated complex for antiviral defense) binding of the DMS3-42 protospacer and PAM present on the chromosome either in the DMS3 lysogen or introduced directly onto the chromosome (PS-CKOn). This interaction is not lethal, due to the mismatches in the crRNA present at the +9 and +11 positions, but features enough complementarity to recruit the single-stranded nuclease Cas3 and induce DNA damage at levels high enough to activate RecA and the subsequent SOS response (Fig. 7A and B). A consequence of activated RecA is induced proteolysis of the PrtR pyocin repressor and the PA14_52530 transcriptional repressor, which in turn allows expression of the pyocins and PA14_52530-repressed genes, respectively, including predicted holin-like and endolysin-like genes (Fig. 7C). In WT P. aeruginosa, these lysis genes are normally repressed when the bacterial cell is growing on a surface compared to a planktonic population, but due to the RecA activation in the PS-CKOn strain described above, the PS-CKOn strain is unable to repress these lysis genes when growing on a surface, resulting in cell death, presumably through activation of the lysis proteins (Fig. 7D and E), which in turn reduces both biofilm formation and swarming motility but maintains a viable planktonic population.
FIG 7 .
Model of CRISPR-dependent, biofilm-specific death of Pseudomonas aeruginosa. (A) Cascade (CRISPR-associated complex for antiviral defense) was targeted to the DMS3-42 PS-CKOn using CRISPR2 spacer 1 crRNA. The partial match between CRISPR2 spacer 1 and the PS-CKOn recruits nuclease Cas3, which cleaves the single strand of DNA displaced in the R-loop, but the mismatches at positions +9 and +11 in the crRNA prevent lethal nuclease activity. ssDNA, single-stranded DNA. (B) The single-stranded DNA generated from Cas3 nuclease activity recruits RecA, which polymerizes, forms a helical filament, and is activated. The activated RecA induces proteolysis of pyocin repressor PrtR and transcriptional repressor PA14_52530. (C) The proteolysis of PrtR allows binding of transcriptional activator PrtN, which in turn induces pyocin gene expression, including that of the associated lambda-like lysis cassette. The proteolysis of PA14_52530 allows expression of the PA14_52530-repressed operon, including putative phage-related lysis genes. (D) The highly expressed lambda-like lysis cassette encoded in the pyocin operon in addition to the highly expressed putative phage-related lysis genes in the PA14_52520-repressed operon leads to accumulation lysis proteins in planktonic cells. IM, inner membrane; OM, outer membrane. (E) Bacteria interacting with a surface trigger the activation of lysis proteins, leading to cell lysis and death.
Previously, we had speculated that the loss of the biofilm and swarming in the DMS3 lysogens was the result of CRISPR-dependent expression of genes expressed by phage DMS3 as a consequence of the interaction between the CRISPR system and the protospacer. This hypothesis was proposed, in part, due to the observation that a P. aeruginosa PA14 bacteriophage MP29 lysogen did not inhibit biofilm or swarming, despite MP29 containing the exact same protospacer as DMS3. We now know that this lack of impact on biofilm formation and swarming is likely due to MP29 containing a recently described anti-CRISPR system (34).
We show that the presence of the DMS3 protospacer plus the PAM sequence on the chromosome (but not on an extrachromosomal element) is both necessary and sufficient for loss of the biofilm and swarming. The PS-CKOn construct, which carries the 32-nt target of the CRISPR spacer plus 5 flanking nucleotides, including the PAM sequence, in an otherwise wild-type genetic context, served as an excellent tool in our studies, as the insertion of this sequence into the genome of P. aeruginosa phenocopied the effects of the DMS3 lysogen without potential confounding factors caused by the presence of bacteriophage genes. Thus, our data support the conclusion that it is engagement of the CRISPR system with its target, and not changing expression of phage genes secondary to such engagement, that results in the alteration of P. aeruginosa group behaviors. We confirmed this conclusion by deleting all the genes downstream of the DMS3-42 protospacer, which showed no impact on the biofilm or swarming phenotypes of this lysogen.
We found every 6th nucleotide of the spacer starting from the 5′ end was dispensable for the CRISPR-spacer interaction, which agrees with evidence in the type 1-E CRISPR/Cas system, in which these particular nucleotides are hidden due to the crRNA underwound ribbon structure (5, 35, 36). Interestingly, we found that generating a mismatch at position +32 restored biofilm formation only partially. Based on the 5′ seed-sequence targeting model in which complementarity at the 5′ end of the spacer is the most important feature for interaction and CRISPR interference (37), we propose that complementarity at position +32 is partially dispensable for interaction due to its location at the 3′-terminal site of the spacer, away from the seed sequence. The +27 position is not predicted to be hidden and yet is dispensable for loss of the biofilm as well as interference (29), suggesting a potential difference between the type 1-E and type 1-F systems. Such a difference is not surprising, as there is already a distinct difference between type 1-E and type 1-F systems regarding the stringency of PAM sequences and CRISPR priming (38) as well as the structure of the targeting crRNA-riboprotein complex (39). Overall, our data help define the nucleotide requirements of these two closely related CRISPR systems and suggest that the partial binding generated through mismatches at sites +9 and +11 of the spacer in the type 1-F system could result in additional alternative CRISPR functions. Finally, we utilized selected mutations from this analysis in subsequent experiments presented here.
Using the strain carrying the PS-CKOn insertion and the information gained from our mutagenesis studies, we were able to determine that the biofilm reduction phenotype did not appear to be the result of an initial attachment defect or early dispersal event but was, rather, a surface-dependent killing phenotype. Once initiated, the kinetics of biofilm formation of the wild-type strain and the strain carrying the PS-CKOn insertion were similar over the first several hours, but the strain carrying the protospacer and PAM (i) never attained the same maximal level of biofilm formation as the wild-type strain, (ii) showed declining biofilm biomass between 10 and 24 h judged by the crystal violet assay, and (iii) exhibited reduced viability on the basis of propidium iodide staining. These observations were particularly interesting given that the viability of the PS-CKOn-carrying strain growing planktonically in the biofilm assay plates was identical to that of the wild-type strain grown planktonically, indicating that the cells needed to be on a surface to experience CRISPR-mediated killing.
To probe any transcriptional differences between the wild type and the strain carrying the PS-CKOn insertion grown on a surface, and to gain insight into the CRISPR-specific killing mechanism, RNA was harvested from swarm-grown cells and subjected to RNA-Seq analysis. While there were no global transcriptional differences between the two samples, 40 of the 65 identified differentially upregulated genes belonged to the R/F pyocin operon and the PA14_52530 operon—both sets of genes are known to be SOS induced (18, 24). Interestingly, using qRT-PCR to confirm these results indicated that the aforementioned genes were induced over 100-fold in the PS-CKOn strain compared to the wild type, suggesting that our RNA-Seq studies might underestimate the magnitude of the transcriptional data and potentially explaining why we did not find other, more moderately expressed SOS-regulated genes in our RNA-Seq data set. Note that the large transcriptional upregulation of the pyocin genes was accompanied by a comparable increase in pyocin activity.
We investigated whether the SOS response was indeed needed for the overexpression of these genes and whether this overexpression was dependent on the presence of a functional CRISPR system. Deleting the recA gene or introducing a nicking-deficient Cas3 allele into the PS-CKOn strain completely abolished high-level pyocin expression. Thus, our data indicate that induction of the SOS response occurs upon self-targeting and engaging the CRISPR system and is likely the consequence of Cas3-mediated nicking, as illustrated in Fig. 7A and B. We were surprised by the lack of growth defect observed due to this self-targeting event in the planktonic population of bacteria. This observation indicates that the partial match of CR2_sp1 to the DMS3 protospacer, while sufficient to elicit a SOS response via the Cas3 nuclease activity, does not have an overtly negative impact on the cell, likely due to the action of the SOS system in repairing the nicked DNA. It was only when the recA gene was removed, and the SOS-mediated repair system compromised, that this self-targeting event became detrimental, as indicated by a marked decrease in even the planktonic growth of strains carrying the PS-CKOn in a recA mutant background. Thus, the biofilm and swarming phenotypes appear to be an indirect effect of the CRISPR self-targeting since the PS-CKOn strain carrying the ΔR/F and ΔPA14_52530 operon mutants regained wild-type phenotypes despite the presence of the protospacer and a functioning CRISPR system.
Why do we observe preferential killing of surface-associated bacteria when the CRISPR system is engaged? The results of our studies indicate that the wild-type strain significantly downregulates expression of the genes encoding the pyocins and the PA14_52530 operon when growing on a surface; such downregulation may be important in the context of the close-packed, high-cell-density environment of biofilm- or swarm-grown microbes. In Fig. 7D and E, the lysis genes are illustrated as lambda holins, antiholins, and endolycins due to the genetic and functional similarity between the pyocin lysis cassette and the lambda lysis cassette (40), the genetic similarity between PA14_52500 and PA14_52510 with putative phage holins, and the requirement of these genes for the cell lysis phenotype in the PS-CKOn-containing strain. Based on this model, expression of these phage-related lysis genes would be harmless until triggering of the holins occurred, which might occur when cells are switching from planktonic to surface growth. Therefore, adaptations that occur during surface growth may sensitize the cell to the accumulation of the pyocin- and PA14_52530 operon lysis-related proteins normally tolerated in planktonically growing cells. However, when the CRISPR system is engaged, the typical downregulation of the genes encoding the R/F pyocins and of the PA14_52530 genes no longer occurs, resulting in high-level expression of both sets of these genes in the surface populations and thus in the enhanced killing of the bacterial population. The mechanism by which the wild-type strain typically downregulates the expression of the pyocin genes and PA14_52530 operon is not currently understood, but we have observed (~6-fold) reduced recA gene expression in surface-grown versus plankton-grown bacteria, suggesting that the SOS response is mitigated in these surface-grown bacteria (data not shown).
We note that CRISPR-related self-targeting resulting in genome rearrangement has previously been observed in a type 1-F system (41). Our data suggest a self-targeting event that modulates group behaviors of P. aeruginosa, without being toxic with respect to the growth of the planktonic population. Given the number and diversity of spacers identified to date, and the observation that an exact match between the spacer and target was not required to engage the CRISPR system in our system, it is likely that such self-targeting events are not uncommon. An open issue remains, however, regarding whether such self-targeting is an occasional accident or whether bacteria have harnessed CRISPR self-targeting as yet one more means by which they can regulate environmental responsiveness. For example, one could speculate that self-targeting in the context of lysogenized P. aeruginosa serves to preferentially eliminate phage-infected cells from a biofilm, an environment in which an infectious agent could rapidly spread through the population. In addition, it has been demonstrated that dispersal from a biofilm in the presence of bacteriophages is advantageous in P. aeruginosa (42), so it is possible that CRISPR self-targeting could be selected in this context. Alternatively, the self-targeting may be advantageous for the bacteriophage. For example, identical DMS3-42 protospacers are found on several Pseudomonas phages, including a P. aeruginosa strain LESB-58 prophage, MP29, and D3112. Furthermore, pyocins are widespread, being found in ~90% of P. aeruginosa strains (22), raising the possibility that self-targeting through the CRISPR system in a manner similar to the C2_sp1–DMS3-42 interaction could occur in other P. aeruginosa strains and benefit phages via their enhanced release by pyocin-mediated cell lysis. Such assertions are compelling but will likely be difficult to support until additional examples of such CRISPR-mediated control of bacterial biology are uncovered.
MATERIALS AND METHODS
Strains and media.
Strains, plasmids, and primers used in this study are listed in Table S2 in the supplemental material. P. aeruginosa strain UCBPP-PA14 (abbreviated as P. aeruginosa PA14) was used in this study. P. aeruginosa and Escherichia coli strains were routinely cultured in lysogeny broth (LB) at 37°C. Growth media were supplemented with antibiotics at the following concentrations: for gentamicin (Gm), 10 µg·ml−1 (E. coli) and 50 µg·ml−1 (P. aeruginosa). The construction of strains, as well as the swarm assay, growth curve conditions, the Congo red assay, and the pyocin killing assay, are described in detail in Text S1 in the supplemental material.
Static biofilm assay and quantification.
Biofilm formation at the indicated time points was assayed on polyvinyl chloride (PVC) as previously described (13, 43) using M63 supplemented with 0.4% arginine and 1 mM MgSO4. Quantification of biofilm formation was performed as previously described (13). To measure planktonic growth during the biofilm assay, a 50-µl volume of cells was directly removed from a single well at indicated time periods, serially diluted, and plated on LB agar to determine CFU.
Fluorescence microscopy viability assay.
Cells were grown in M63 medium supplemented with 0.4% arginine and 1 mM MgSO4 at 37°C for the indicated time periods in a 12-well dish with a glass coverslip partially submerged in each well. The coverslip was then subjected to a Molecular Probes LIVE/DEAD BacLight viability assay and visualized at the air-liquid interface with a Nikon eclipse Ti inverted microscope. Details of this assay are described in Text S1 in the supplemental material.
RNA isolation and quantitative reverse transcriptase PCR.
Strains were grown with M8 media supplemented with MgSO4 (1 mM), glucose (0.2%), and Casamino Acids (CAA; 0.5%) either in broth with shaking (planktonic conditions) or on 0.5% soft agar (swarm conditions) at 37°C for 16 h. For broth-grown cells, RNA was harvested from 1 ml of culture grown to an optical density at 600 nm (OD600) of ~1.0. For soft-agar-grown samples, cells were scraped from 4 total plates after 16 h of incubation and resuspended in 1 ml phosphate-buffered saline. Samples were pelleted and resuspended in 100 µl of 2 mg of lysozyme/ml–TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), followed by incubation at room temperature for 3 min to lyse the cells. The RNA was extracted using an RNeasy kit (Qiagen) according to the manufacturer’s instructions and tested for DNA contamination by PCR.
DNA was synthesized from 1 µg of total RNA using a qScript reverse transcription kit (Quanta Biosciences) according to the supplied protocols. Quantitative reverse transcriptase PCR (qRT-PCR) was performed using cDNA, primers designed for the indicated gene, and PerfeCTa SYBR green FastMix quantitative PCR (qPCR) master mix (Quanta Biosciences). A Bio-Rad iCycler was used to perform the reactions, and data analysis was performed using CFX manager software (Bio-Rad). The data for each gene of interest were normalized to the rplU or proC gene transcript control as previously described (44). The data are displayed as arbitrary expression units relative to an indicated wild-type condition.
RNA sequencing and identification of differentially expressed genes.
Total RNA purified as described above was sent to Helmholtz Centre for Infection Research (HZI) in Braunschweig, Germany, for mRNA isolation, cDNA preparation, and RNA sequencing using Illumina HiSeq SR50. Details of RNA-Seq data analysis are included in Text S1 in the supplemental material.
Plaque assays.
Plaque assays were completed as described by Budzik et al. (45). Briefly, 200 µl of the indicated bacterial strain was added to 4 ml molten top agar (0.5%) and poured over a prewarmed LB agar plate. After solidification of the top agar lawns, 5-µl volumes of 10-fold serially diluted DMS3 and DMS3-T255C were spotted onto the top agar lawn and incubated at 37°C overnight.
Statistical analysis.
All data were analyzed using Graph Pad Prism 6. The data represent the means and standard deviations of the results of at least three independent experiments with multiple replicates unless stated otherwise. All data were treated as normally distributed, and comparisons were tested with Student’s t test.
SUPPLEMENTAL MATERIAL
Supporting information. Download
Nucleotides required for CRISPR/DMS3-mediated loss of biofilm formation. (A) Red nucleotides inside the horizontal black bar represent the DMS3-42 protospacer, yellow nucleotides represent the PAM, and the gray nucleotides represent the nucleotides found in an adjacent position on the DMS3 genome. The nucleotides below the horizontal black bar represent partial matching CRISPR 2 spacer 1, with thin vertical black bars indicating complementarity. The nucleotides above the arrows represent the generated point mutants resulting in restoration of biofilm formation (black arrows) or a partial restoration of biofilm formation (striped arrow) or no restoration of biofilm formation (white arrows). The graph below the nucleotides represents the quantified biofilm formation at 24 h as measured by absorbance of the crystal violet stain, with a representative image of the stained well displayed above. The boxed designations represent control strains. Black bars indicate wild-type levels of biofilm formation, while white bars indicate CRISPR-mediated loss of the biofilm. Interestingly, one point mutation (A232T) at the +32 position on the protospacer resulted in partial restoration of biofilm formation, although the reason for this partial phenotype is not clear. Error bars represent standard deviations of the results of at least three replicates. (B) An illustration of the underwound ribbon structure, with squares representing nucleotides and vertical bars indicating binding between the crRNA and complementary protospacer. Overall, these data are consistent with cryo-electron microscopy (cryo-EM) and crystallography data from the E. coli type 1E CRISPR/Cas system in which every 6th nucleotide is rotated out of the crRNA-target RNA-DNA hybrid and is thus dispensable for interference. However, the 6th-nucleotide rotation model does not explain the mismatch at the +27 position, which we previously reported as dispensable for bacteriophage interference (K. C. Cady et al., J Bacteriol 193:3433–3445, 2011, http://dx.doi.org/10.1128/JB.01411-10). Download
Deletion of the DMS3 genes downstream of the DMS3-42 protospacer does not restore biofilm formation in a DMS3 lysogen. The indicated strains were grown in M63 supplemented with 0.4% arginine and 1 mM MgSO4 and stained with crystal violet (CV). The quantification of the CV stain as measured by absorbance at 550 nm is shown with a representative image displayed above. Error bars represent standard deviations of the results of at least three biological replicates. The horizontal black bar indicates no statistical significance as assessed with a Student’s t test. Download
The location of PS-CKOn. The DMS3-42 protospacer and the 5 upstream and downstream nucleotides are displayed inside the horizontal black bar. The nucleotides directly above the black bar represent partial matching CRISPR 2 spacer 1 (C2S1 crRNA), with thin vertical bars indicating complementarity. The PS-CKOn construct contains the DMS3-42 protospacer along with the 5 upstream and downstream nucleotides inserted on the P. aeruginosa chromosome between PA14_73160 and glmS as indicated at the bottom of the panel. Download
The cell death phenotype is specific to the strain carrying the PS−CKON with minimal viability loss observed in the PS-CKOn planktonic population. (A) Biofilm formation assays were performed on the indicated strains with an incubation time of 6 h as described in Materials and Methods. Error bars represent standard deviations of the results of three biological replicates, with an asterisk (*) representing a P value of <0.05 compared to the WT as determined with a Student’s t test. (B) The indicated strains were grown on a glass coverslip under biofilm-inducing conditions, stained with the BacLight cell viability kit, and visualized at the air-liquid interface (ALI) at 6 h. (C) Quantification of the ALI images displayed as the mean intensity of viable cells (Syto-9; green cells in panel B) over the mean intensity of the membrane-compromised cells (propidium iodide; red cells in panel B) for the indicated strains. Error bars represent standard deviations of the results from three biological replicates. (D) The indicated strains were grown in 2 ml M63 supplemented with 0.4% arginine and 1 mM MgSO4 (biofilm-inducing media) in a 12-well plate, and the planktonic cells were removed directly from the well at 6 h, stained using a BacLight cell viability kit, washed, and transferred to a glass coverslip for visualization. (E) Quantification of the images displayed as the direct counts of viable cells (Syto-9 stained) over direct counts of the membrane-compromised cells (propidium iodide stained) of the indicated strains. Error bars represent standard deviations of the results from three biological replicates with 10 randomly acquired images each. There is no significant difference between the strains as concluded from a Student’s t test. (F) The indicated strains were grown in a 96-well plate in 100 µl of biofilm-inducing media with 50 µl of the planktonic cells removed directly from each well and plated for determinations of levels of colony-forming units at the indicated time points. Error bars represent standard deviations of the results from three biological replicates, with an asterisk (*) indicating a P value of <0.05 compared to the WT as determined with a Student’s t test. Download
Quantitative RT-PCR of pyocin and PA14_52530-repressed genes using alternative normalization controls. RNA was harvested from both planktonic and surface (swarm) populations of the indicated strains and used for quantitative reverse transcriptase PCR to measure the transcript of PA14_08300 (a predicted F pyocin tail protein) normalized to rplU (A), PA14_07990 (a predicted pyocin-encoded holin) normalized to proC (B), PA14_52490 normalized to rplU (C), PA14_52480 normalized to proC (D), rplU normalized to fabD (E), and proC normalized to fabD (F). All values are displayed relative to the surface population wild-type expression. Black horizontal bars display the fold increase in relative expression of the indicated transcript from the wild type compared to the PS-CKOn strains under both planktonic and surface conditions. Error bars represent standard deviations of the results from at least three biological replicates. For all panels, “a” indicates a P value of <0.05 compared to the WT grown planktonically and “b” indicates a P value of <0.05 compared to the WT grown on a surface. Download
RecA is required for normal growth in the PS-CKOn construct. The indicated strains were grown in LB with shaking at 37°C for 8 h, with OD600 readings measured every hour. Error bars represent standard deviations of the results from three biological replicates. Download
The CRISPR system is functional in the PS-CKOn ΔR/F mutant, the PS-CKOn ΔPA14_52530 operon mutant, and the PS-CKOn ΔR/F ΔPA14_52530 operon double mutant. ΔPS-CKOn operon mutants and deletion of the S-pyocins do not increase biofilm formation in the PS-CKOn ΔR/F background. (A) Lawns of the indicated bacterial strains were prepared in 0.5% LB top agar as described in Materials and Methods. Either WT (CRISPR-insensitive) or T255C (CRISPR-sensitive) DMS3 was 10-fold serially diluted with 5 µl of each dilution prepared as plaques on the indicated bacterial lawn and incubated at 37°C for 16 h. (B) The indicated strains were grown in M63 supplemented with 0.4% arginine and 1 mM MgSO4 and stained with CV. The quantification of the CV stain as measured by absorbance at 550 nm is shown with a representative image displayed above. Error bars represent standard deviations of the results of three biological replicates. Download
Differentially expressed genes as assessed by RNA-Seq. The PAO1 ortholog is the homolog of the gene in P. aeruginosa PAO1. LogFC is the log2 fold change of the level of PS-CKOn compared to the WT level. The P value is the P value calculated with an exact test for the negative binomial distribution (see Materials and Methods). CPM WT and CPM PS-CKOn are the specific reads of the transcript (in counts per million) after edgeR normalization in the WT strain sequence and the PS-CKOn sequence, respectively. Functions and predicted functions were assigned based on information from http://www.pseudomonas.com
Strains, plasmids, and primers used in this study. All strains, plasmids, and primers used in this study are presented with name, genotype or relevant characteristic(s), and source.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 2 R37 AI83256-06 (G.A.O.), National Institutes of Health grant R01-HL074175 (B.A.S.), P20-RR018787/GM103413 (B.A.S.), the Cystic Fibrosis Foundation (STANTO14IO and STANTO11R0 to B.A.S.), and Dartmouth SYNERGY.
We thank Amanda Socha for her assistance in the construction of point mutations in the DMS3 protospacer. We also thank Michael Jarek for his help with RNA sequencing.
Footnotes
Citation Heussler GE, Cady KC, Koeppen K, Bhuju S, Stanton BA, O’Toole GA. 2015. Clustered regularly interspaced short palindromic repeat-dependent, biofilm-specific death of Pseudomonas aeruginosa mediated by increased expression of phage-related genes. mBio 6(3):e00129-15. doi:10.1128/mBio.00129-15.
REFERENCES
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJ, van der Oost J. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by cascade and Cas3. Mol Cell 46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, Heikema AP, Timms AR, Jacobs BC, Wagenaar JA, Endtz HP, van der Oost J, Wells JM, Nieuwenhuis EE, van Vliet AH, Willemsen PT, van Baarlen P, van Belkum A. 2013. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis 32:207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- 9.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol 189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin C-, Ratner HK, Llewellyn AC, Jones CL, Laroui H, Merlin D, Zhou P, Endtz HP, Weiss DS. 2014. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci U S A 111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cady KC, O’Toole GA. 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol 193:3433–3445. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambertsen L, Sternberg C, Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol 6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 15.Merritt JH, Kadouri DE, O’Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O’Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha DG, Richman ME, O’Toole GA. 2014. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl Environ Microbiol 80:3384–3393. doi: 10.1128/AEM.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsui H, Sano Y, Ishihara H, Shinomiya T. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175:1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farmer JJ III, Herman LG. 1969. Epidemiological fingerprinting of Pseudomonas aeruginosa by the production of and sensitivity of pyocin and bacteriophage. Appl Microbiol 18:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang W, Small DA, Toghrol F, Bentley WE. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyofuku M, Zhou S, Sawada I, Takaya N, Uchiyama H, Nomura N. 2014. Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ Microbiol 16:2927–2938. doi: 10.1111/1462-2920.12260. [DOI] [PubMed] [Google Scholar]
- 22.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 23.Penterman J, Singh PK, Walker GC. 2014. Biological cost of pyocin production during the SOS response in Pseudomonas aeruginosa. J Bacteriol 196:3351–3359. doi: 10.1128/JB.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Köhler T, Donner V, van Delden C. 2010. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192:1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little JW, Mount DW. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11–22. doi: 10.1016/0092-8674(82)90085-X. [DOI] [PubMed] [Google Scholar]
- 27.Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. 2011. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J 30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulepati S, Bailey S. 2011. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J Biol Chem 286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caiazza NC, Merritt JH, Brothers KM, O’Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet 46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 32.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulepati S, Héroux A, Bailey S. 2014. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fineran PC, Gerritzen MJ, Suárez-Diez M, Künne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ. 2014. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A 111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. 2014. Priming in the type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res 42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. 2014. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 41.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet 9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor TB, Buckling A. 2013. Bacterial motility confers fitness advantage in the presence of phages. J Evol Biol 26:2154–2160. doi: 10.1111/jeb.12214. [DOI] [PubMed] [Google Scholar]
- 43.O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuchma SL, Ballok AE, Merritt JH, Hammond JH, Lu W, Rabinowitz JD, O’Toole GA. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol 192:2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Budzik JM, Rosche WA, Rietsch A, O’Toole GA. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J Bacteriol 186:3270–3273. doi: 10.1128/JB.186.10.3270-3273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information. Download
Nucleotides required for CRISPR/DMS3-mediated loss of biofilm formation. (A) Red nucleotides inside the horizontal black bar represent the DMS3-42 protospacer, yellow nucleotides represent the PAM, and the gray nucleotides represent the nucleotides found in an adjacent position on the DMS3 genome. The nucleotides below the horizontal black bar represent partial matching CRISPR 2 spacer 1, with thin vertical black bars indicating complementarity. The nucleotides above the arrows represent the generated point mutants resulting in restoration of biofilm formation (black arrows) or a partial restoration of biofilm formation (striped arrow) or no restoration of biofilm formation (white arrows). The graph below the nucleotides represents the quantified biofilm formation at 24 h as measured by absorbance of the crystal violet stain, with a representative image of the stained well displayed above. The boxed designations represent control strains. Black bars indicate wild-type levels of biofilm formation, while white bars indicate CRISPR-mediated loss of the biofilm. Interestingly, one point mutation (A232T) at the +32 position on the protospacer resulted in partial restoration of biofilm formation, although the reason for this partial phenotype is not clear. Error bars represent standard deviations of the results of at least three replicates. (B) An illustration of the underwound ribbon structure, with squares representing nucleotides and vertical bars indicating binding between the crRNA and complementary protospacer. Overall, these data are consistent with cryo-electron microscopy (cryo-EM) and crystallography data from the E. coli type 1E CRISPR/Cas system in which every 6th nucleotide is rotated out of the crRNA-target RNA-DNA hybrid and is thus dispensable for interference. However, the 6th-nucleotide rotation model does not explain the mismatch at the +27 position, which we previously reported as dispensable for bacteriophage interference (K. C. Cady et al., J Bacteriol 193:3433–3445, 2011, http://dx.doi.org/10.1128/JB.01411-10). Download
Deletion of the DMS3 genes downstream of the DMS3-42 protospacer does not restore biofilm formation in a DMS3 lysogen. The indicated strains were grown in M63 supplemented with 0.4% arginine and 1 mM MgSO4 and stained with crystal violet (CV). The quantification of the CV stain as measured by absorbance at 550 nm is shown with a representative image displayed above. Error bars represent standard deviations of the results of at least three biological replicates. The horizontal black bar indicates no statistical significance as assessed with a Student’s t test. Download
The location of PS-CKOn. The DMS3-42 protospacer and the 5 upstream and downstream nucleotides are displayed inside the horizontal black bar. The nucleotides directly above the black bar represent partial matching CRISPR 2 spacer 1 (C2S1 crRNA), with thin vertical bars indicating complementarity. The PS-CKOn construct contains the DMS3-42 protospacer along with the 5 upstream and downstream nucleotides inserted on the P. aeruginosa chromosome between PA14_73160 and glmS as indicated at the bottom of the panel. Download
The cell death phenotype is specific to the strain carrying the PS−CKON with minimal viability loss observed in the PS-CKOn planktonic population. (A) Biofilm formation assays were performed on the indicated strains with an incubation time of 6 h as described in Materials and Methods. Error bars represent standard deviations of the results of three biological replicates, with an asterisk (*) representing a P value of <0.05 compared to the WT as determined with a Student’s t test. (B) The indicated strains were grown on a glass coverslip under biofilm-inducing conditions, stained with the BacLight cell viability kit, and visualized at the air-liquid interface (ALI) at 6 h. (C) Quantification of the ALI images displayed as the mean intensity of viable cells (Syto-9; green cells in panel B) over the mean intensity of the membrane-compromised cells (propidium iodide; red cells in panel B) for the indicated strains. Error bars represent standard deviations of the results from three biological replicates. (D) The indicated strains were grown in 2 ml M63 supplemented with 0.4% arginine and 1 mM MgSO4 (biofilm-inducing media) in a 12-well plate, and the planktonic cells were removed directly from the well at 6 h, stained using a BacLight cell viability kit, washed, and transferred to a glass coverslip for visualization. (E) Quantification of the images displayed as the direct counts of viable cells (Syto-9 stained) over direct counts of the membrane-compromised cells (propidium iodide stained) of the indicated strains. Error bars represent standard deviations of the results from three biological replicates with 10 randomly acquired images each. There is no significant difference between the strains as concluded from a Student’s t test. (F) The indicated strains were grown in a 96-well plate in 100 µl of biofilm-inducing media with 50 µl of the planktonic cells removed directly from each well and plated for determinations of levels of colony-forming units at the indicated time points. Error bars represent standard deviations of the results from three biological replicates, with an asterisk (*) indicating a P value of <0.05 compared to the WT as determined with a Student’s t test. Download
Quantitative RT-PCR of pyocin and PA14_52530-repressed genes using alternative normalization controls. RNA was harvested from both planktonic and surface (swarm) populations of the indicated strains and used for quantitative reverse transcriptase PCR to measure the transcript of PA14_08300 (a predicted F pyocin tail protein) normalized to rplU (A), PA14_07990 (a predicted pyocin-encoded holin) normalized to proC (B), PA14_52490 normalized to rplU (C), PA14_52480 normalized to proC (D), rplU normalized to fabD (E), and proC normalized to fabD (F). All values are displayed relative to the surface population wild-type expression. Black horizontal bars display the fold increase in relative expression of the indicated transcript from the wild type compared to the PS-CKOn strains under both planktonic and surface conditions. Error bars represent standard deviations of the results from at least three biological replicates. For all panels, “a” indicates a P value of <0.05 compared to the WT grown planktonically and “b” indicates a P value of <0.05 compared to the WT grown on a surface. Download
RecA is required for normal growth in the PS-CKOn construct. The indicated strains were grown in LB with shaking at 37°C for 8 h, with OD600 readings measured every hour. Error bars represent standard deviations of the results from three biological replicates. Download
The CRISPR system is functional in the PS-CKOn ΔR/F mutant, the PS-CKOn ΔPA14_52530 operon mutant, and the PS-CKOn ΔR/F ΔPA14_52530 operon double mutant. ΔPS-CKOn operon mutants and deletion of the S-pyocins do not increase biofilm formation in the PS-CKOn ΔR/F background. (A) Lawns of the indicated bacterial strains were prepared in 0.5% LB top agar as described in Materials and Methods. Either WT (CRISPR-insensitive) or T255C (CRISPR-sensitive) DMS3 was 10-fold serially diluted with 5 µl of each dilution prepared as plaques on the indicated bacterial lawn and incubated at 37°C for 16 h. (B) The indicated strains were grown in M63 supplemented with 0.4% arginine and 1 mM MgSO4 and stained with CV. The quantification of the CV stain as measured by absorbance at 550 nm is shown with a representative image displayed above. Error bars represent standard deviations of the results of three biological replicates. Download
Differentially expressed genes as assessed by RNA-Seq. The PAO1 ortholog is the homolog of the gene in P. aeruginosa PAO1. LogFC is the log2 fold change of the level of PS-CKOn compared to the WT level. The P value is the P value calculated with an exact test for the negative binomial distribution (see Materials and Methods). CPM WT and CPM PS-CKOn are the specific reads of the transcript (in counts per million) after edgeR normalization in the WT strain sequence and the PS-CKOn sequence, respectively. Functions and predicted functions were assigned based on information from http://www.pseudomonas.com
Strains, plasmids, and primers used in this study. All strains, plasmids, and primers used in this study are presented with name, genotype or relevant characteristic(s), and source.