ABSTRACT
The protozoan parasite Trypanosoma brucei engages in surface-induced social behavior, termed social motility, characterized by single cells assembling into multicellular groups that coordinate their movements in response to extracellular signals. Social motility requires sensing and responding to extracellular signals, but the underlying mechanisms are unknown. Here we report that T. brucei social motility depends on cyclic AMP (cAMP) signaling systems in the parasite’s flagellum (synonymous with cilium). Pharmacological inhibition of cAMP-specific phosphodiesterase (PDE) completely blocks social motility without impacting the viability or motility of individual cells. Using a fluorescence resonance energy transfer (FRET)-based sensor to monitor cAMP dynamics in live cells, we demonstrate that this block in social motility correlates with an increase in intracellular cAMP levels. RNA interference (RNAi) knockdown of the flagellar PDEB1 phenocopies pharmacological PDE inhibition, demonstrating that PDEB1 is required for social motility. Using parasites expressing distinct fluorescent proteins to monitor individuals in a genetically heterogeneous community, we found that the social motility defect of PDEB1 knockdowns is complemented by wild-type parasites in trans. Therefore, PDEB1 knockdown cells are competent for social motility but appear to lack a necessary factor that can be provided by wild-type cells. The combined data demonstrate that the role of cyclic nucleotides in regulating microbial social behavior extends to African trypanosomes and provide an example of transcomplementation in parasitic protozoa.
IMPORTANCE
In bacteria, studies of cell-cell communication and social behavior have profoundly influenced our understanding of microbial physiology, signaling, and pathogenesis. In contrast, mechanisms underlying social behavior in protozoan parasites are mostly unknown. Here we show that social behavior in the protozoan parasite Trypanosoma brucei is governed by cyclic-AMP signaling systems in the flagellum, with intriguing parallels to signaling systems that control bacterial social behavior. We also generated a T. brucei social behavior mutant and found that the mutant phenotype is complemented by wild-type cells grown in the same culture. Our findings open new avenues for dissecting social behavior and signaling in protozoan parasites and illustrate the capacity of these organisms to influence each other’s behavior in mixed communities.
INTRODUCTION
Recognition of social behavior and cell-cell communication as ubiquitous among bacteria transformed our view of microbiology (1, 2). Examples of microbial social behavior are widespread and include assembly of biofilms and fruiting bodies, quorum sensing, and various forms of group motility across surfaces (1–6). Social behaviors enable bacteria to function as multicellular entities exhibiting emergent properties not evident in individual cells (1). Pathogenic bacteria and fungi exploit social behaviors to resist host immune defenses and antibiotics, to promote tissue colonization, and to exclude competing microbes from infection sites (7–9). As such, microbial cell-cell communication and social behavior are important for development and virulence, while the underlying mechanisms are potential targets for therapeutic intervention (10, 11).
Parasitic protozoa present a significant threat to global public health and agriculture and pose an economic burden in some of the world’s most impoverished regions (12–16). The paradigm of social behavior can inform questions regarding parasite biology, transmission, and pathogenesis (17–20), but little is known about social behaviors and cell-cell interactions in these organisms. The protozoan parasite Trypanosoma brucei is transmitted by bloodsucking tsetse flies, causing sleeping sickness in humans and related diseases in wild and domestic animals throughout sub-Saharan Africa (21). Trypanosomes are typically considered individual cells but are capable of parasite-parasite communication and social behavior. In the mammalian host’s bloodstream, for example, quorum sensing directs T. brucei development into “short stumpy” forms that are uniquely adapted for transmission through the tsetse fly (22, 23). Here, parasite-derived “stumpy induction factor” (SIF) accumulates in a cell density-dependent fashion and triggers cellular differentiation (22, 23). Procyclic (insect midgut stage) trypanosomes are also capable of social behavior. In this case, surface cultivation causes individual parasites to assemble into multicellular communities that engage in collective motility across the surface and modify their movements in response to signals from nearby parasites (24). This group behavior is termed social motility (SoMo) based on features shared with social motility and swarming motility in bacteria (4, 24).
In bacteria, social motility facilitates rapid surface colonization and promotes survival of bacterial populations in harsh environments (4, 5, 7). Specific features of transmission or pathogenesis that are reflected in T. brucei social motility are not yet known. However, recent work has shown that social motility is a property of a specific life cycle stage that occurs early during colonization of the fly midgut, consistent with the idea that social motility reflects parasite features relevant within the fly transmission stage (25). Moreover, the parasites are in constant contact with tissue surfaces in their natural environment, particularly in the tsetse fly and extravascular spaces in the mammalian host, and would benefit from functions provided by social motility in bacteria. More broadly, social motility presents a complex, group-level behavior that highlights the capacity of trypanosomes for cell-cell communication.
Social behaviors in microbes depend upon cell-cell communication and specific signaling systems in individuals within the population (1, 2, 4, 5). Quorum sensing and cyclic nucleotide signaling via the 2nd messenger cyclic-dimeric-GMP (c-di-GMP) have emerged as important regulators of surface-induced swarming motility in bacteria (26, 27). Quorum sensing and c-di-GMP also regulate biofilm formation, which is another surface-associated group behavior (27, 28). In eukaryotes, cyclic nucleotide signaling via cyclic AMP (cAMP) is critical for the surface motility of the social amoeba Dictyostelium discoideum (6).
T. brucei cAMP levels are controlled by receptor-type adenylate cyclases and cAMP-specific phosphodiesterases (PDEs) (29). T. brucei encodes five PDEs, PDEA, PDEB1, PDEB2, PDEC, and PDED (30, 31). PDEB1 and PDEB2 are essential for maintaining physiological levels of cAMP in T. brucei cells (32). T. brucei PDEB1 (TbPDEB1) is localized to the flagellum, while TbPDEB2 is distributed throughout the cell (32). Flagellum localization is of particular significance, because the eukaryotic flagellum is a signaling center that controls communication with the extracellular environment (33–35) and flagellar motility is required for social motility (24). TbPDEB1 and TbPDEB2 are essential in bloodstream trypanosomes in vitro and in an in vivo mouse infection model (32), making them targets for therapeutic intervention, and drug discovery efforts have led to development of potent and specific T. brucei PDE inhibitors (36–38). These inhibitors validate PDEs as drug targets and novel open avenues for dissecting cAMP signaling in trypanosomes with chemical genetics (39).
Given the widespread use of cyclic nucleotides for regulating surface-associated social behaviors in diverse microbes, we hypothesized that cAMP controls social motility in T. brucei. To test this hypothesis, we targeted TbPDEB1. We show that pharmacological or genetic inhibition of TbPDEB1 blocks social motility without affecting the viability or motility of individual cells. By employing a fluorescence resonance energy transfer (FRET) sensor for monitoring cAMP in live T. brucei cells, we showed that the block in social motility is directly correlated with a rise in intracellular cAMP levels. Finally, using fluorescent markers to monitor genetically distinct individuals in a mixed population, we demonstrated that the social motility defect of TbPDEB1 mutants can be complemented by wild-type (WT) cells in trans. Our findings reveal T. brucei social motility mechanisms, demonstrate that cyclic nucleotide regulation of microbial surface motility extends to parasitic protozoa, and reveal a novel form of cell-cell interaction in these parasites.
RESULTS
Intracellular cAMP levels regulate T. brucei social motility.
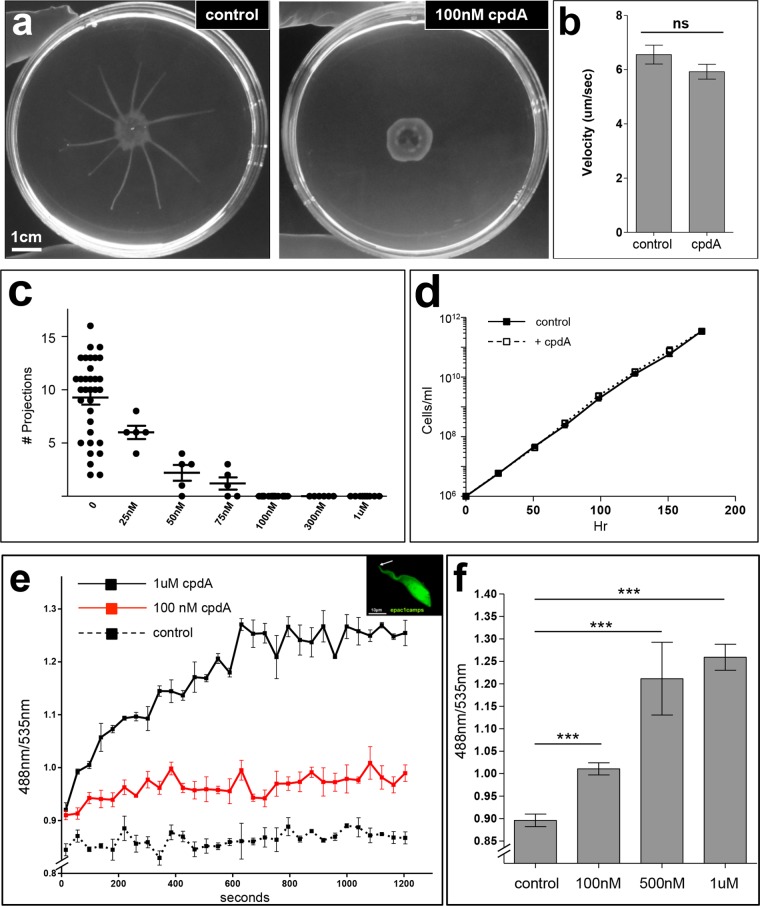
To test the importance of intracellular cAMP dynamics for social motility, we applied a T. brucei PDE inhibitor, cpdA, that inhibits recombinant TbPDEB1 and TbPDEB2 in vitro and causes an increase in cellular cAMP concentrations when added to bloodstream-form T. brucei in culture (37). We tested cpdA at a range of concentrations, from 25nM to 1µM, for its impact on social motility in procyclic T. brucei. When control cells are cultivated on soft agar, parasites assemble at the perimeter of the inoculation site and then move outward as groups, forming projections that radiate away from the center (Fig. 1) (24). cpdA showed dose-dependent inhibition of social motility, and the minimal concentration giving complete inhibition was 100 nM (Fig. 1a and c). Social motility requires active parasite motility (24, 40, 41). We therefore asked if cpdA affects motility of individual cells. We assessed motility using live-video microscopy combined with automated tracking of individual parasites treated with or without 100 nM cpdA in suspension culture. No difference was observed for cpdA-treated versus untreated cells (Fig. 1b). Treatment with 100 nM cpdA did not affect doubling time (Fig. 1d). Therefore, cpdA inhibits social motility in a dose-dependent fashion, with no impact on the proliferation or motility of individual cells.
FIG 1 .
Pharmacological inhibition of T. brucei PDE blocks social motility and elevates cellular cAMP levels. (a) Social motility of cells treated with or without 100 nM cpdA as indicated. (b) Motility of individual cells in suspension cultures treated with or without 100 nM cpdA. (c) Chart showing the number of projections produced upon treatment with the indicated concentrations of cpdA (0 nM [n = 33]; 25 nM [n = 5]; 50 nm [n = 5]; 75 nm [n = 5]; 100 nM [n = 27]; 300 nm [n = 6]; 1 µM [n = 9]). (d) Growth curve for cells treated with or without 100 nM cpdA. (e) Fluorescence ratio (488 nm/535 nm) of epac1camps-expressing cells as a function of time following addition of cpdA at the indicated concentrations. The inset shows that the cAMP FRET sensor epac1camps is expressed throughout the parasite cell, including the flagellum (arrow). (f) Endpoint analysis of 488-nm/535-nm fluorescence ratio of epac1camps cells following 600 s of treatment with cpdA at the indicated concentration. Error bars show ± standard errors. ***, P < 0.0001; ns = not significant.
The inhibitory effect of cpdA on T. brucei social motility is anticipated to be due to a rise in intracellular cAMP concentrations. CpdA has been demonstrated to increase intracellular cAMP concentrations in bloodstream-form T. brucei (37), but its impact on cAMP in procyclic parasites has not been assessed. We therefore tested this by applying a well-characterized fluorescence resonance energy transfer (FRET)-based sensor for cAMP, Epac1-camps (epac1camps), that has been used to monitor intracellular cAMP dynamics in a variety of cell types, including trypanosomes (38, 42, 43). When excited with a wavelength of 436 nm, epac1camps emits 2 distinct wavelengths, one in the cyan (488-nm) spectrum and one in the yellow (535-nm) spectrum, the latter of which is FRET dependent (see Fig. S2a in the supplemental material) (42). Binding of cAMP to the sensor causes a conformational change that reduces FRET, and the 488-nm/535-nm emission ratio therefore increases as a function of increasing intracellular cAMP concentrations (see Fig. S2a) (42). The dynamic range of epac1camps is 0.2 to 10 µM cAMP (42), consistent with the reported intracellular cAMP concentration for T. brucei (30). epac1camps was expressed using a tetracycline (tet)-inducible vector in procyclic T. brucei. Fluorescence microscopy analysis of live, immobilized epac1camps-expressing parasites revealed that epac1camps is evenly distributed throughout the cell, including the cytoplasm and flagellum (Fig. 1e, inset). epac1camps reporter parasites were then used in FRET assays (Fig. 1e and f; see also Fig. S2). When excited at 436 nm, epac1camps parasites showed 488-nm and 535-nm emission signals significantly higher than the background observed for WT controls without the reporter (see Fig. S2b and c). Addition of cpdA epac1camps parasites produced a dose-dependent increase in the 488-nm/535-nm emission ratio, which rose rapidly and plateaued within ~400 to 600 s, indicating a rapid and stable rise in cellular cAMP concentrations (Fig. 1e and f). The 488-nm/535-nm ratio did not change in control epac1camps parasites that did not receive cpdA. Therefore, inhibition of social motility by cpdA is directly correlated with a rise in intracellular cAMP concentrations.
Previous studies suggested that cAMP mediates stumpy formation in BSF T. brucei because addition of cell-permeable cAMP induced stumpy differentiation (23). Subsequent studies showed that differentiation was not induced by hydrolysis-resistant cAMP, even when used at concentrations 20-fold to 200-fold higher than the hydrolyzable cAMP concentrations (44). Moreover, AMP and adenosine metabolic products were 100-fold to 1,000-fold more potent at inducing differentiation than was cAMP, indicating that cAMP downstream products were responsible for stumpy differentiation (44). We tested cAMP analogues for an effect on social motility. In contrast to what was observed for stumpy differentiation, hydrolysis-resistant cAMP inhibited social motility as well as or better than hydrolyzable cAMP, and the effect seen with either compound was equivalent to or more potent than that seen with AMP or adenosine (see Fig. S1 in the supplemental material). Notably, because cAMP added to T. brucei cells is unstable (44), the effective cAMP concentration is even lower than what is added. These results support the view that cAMP mediates inhibition of social motility.
Genetic inhibition of TbPDEB1 phenocopies the effect of the PDE inhibitor cpdA.
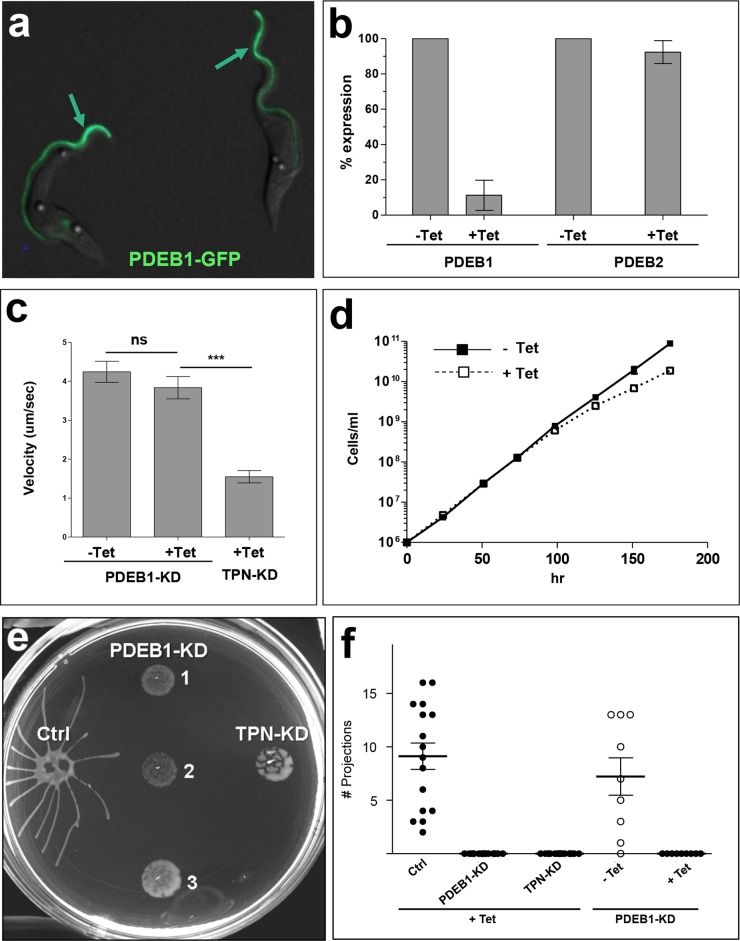
Potent inhibition of TbPDEB1 and TbPDEB2 by cpdA has been demonstrated using recombinant proteins (37). However, there are five PDEs in T. brucei and selectivity of cpdA for any given PDE over the others has not been demonstrated. Likewise, as with any pharmacological treatment, there is potential for off-target effects that may produce a phenotype unrelated to inhibition of the drug’s expected target. Therefore, to determine if the effect of cpdA on social motility is due to inhibition of a specific PDE, we used RNA interference (RNAi) to test the requirement for PDEB1 in social motility. We chose PDEB1 because of its flagellar localization (Fig. 2a) (45) and our goal of interrogating the role of flagellar cAMP signaling in social motility. Upon tet induction, PDEB1 mRNA levels were reduced to 11% of control parasite levels, while the abundance of PDEB2 mRNA was unaffected (Fig. 2b). Therefore, knockdown was potent and specific. PDEB1 knockdown did not affect the motility of individual cells and had only a modest effect on growth (Fig. 2c and d). When assayed for social motility, PDEB1 knockdown completely blocked social motility (Fig. 2e and f). Therefore, genetic ablation of PDEB1 expression phenocopied cpdA treatment, inhibiting social motility while not affecting the motility of individual cells. Social motility is observed in early procyclic but not late procyclic cells (25). Cells treated with cpdA or PDEB1 RNAi expressed the early procyclic marker GPEET procyclin (see Fig. S5 in the supplemental material), demonstrating that inhibition of social motility is not due to differentiation into late procyclics (25).
FIG 2 .
RNAi knockdown of PDEB1 blocks social motility without affecting motility of individual cells. (a) Fluorescence microscopy of procyclic cells expressing a PDEB1-GFP fusion protein, which is localized to the flagellum (arrows). (b) mRNA abundance for PDEB1 and PDEB2, as determined by qRT-PCR, in PDEB1 tetracycline (Tet)-inducible knockdown cells, maintained with or without tetracycline as indicated. Values are normalized to the −Tet expression level. (c) The cell motility of individual cells in suspension culture is shown for Tet-inducible PDEB1 knockdown cells (PDEB-KD), maintained with or without Tet as indicated. Trypanin knockdown (TPN-KD) motility mutants (47) were examined as a control. (d) Growth curve of PDEB1 Tet-inducible knockdown cells grown with or without Tet as indicated. (e) Social motility of 3 independent Tet-inducible PDEB1 knockdown clonal lines (clones 1, 2, and 3), control cells (Ctrl), and trypanin knockdown cells (TPN-KD). PDEB1-KD clone 1 was used for quantitative analysis as shown in panel f. (f) Quantitation of projections formed by control cells (Ctrl), PDEB1 knockdown cells (PDEB1-KD), or trypanin knockdown cells (TPN-KD), grown with or without tet as indicated. Error bars show ± standard errors. ***, P < 0.0001; ns = not significant.
The social motility defect of TbPDEB1 knockdown parasites can be complemented by WT cells provided in trans.
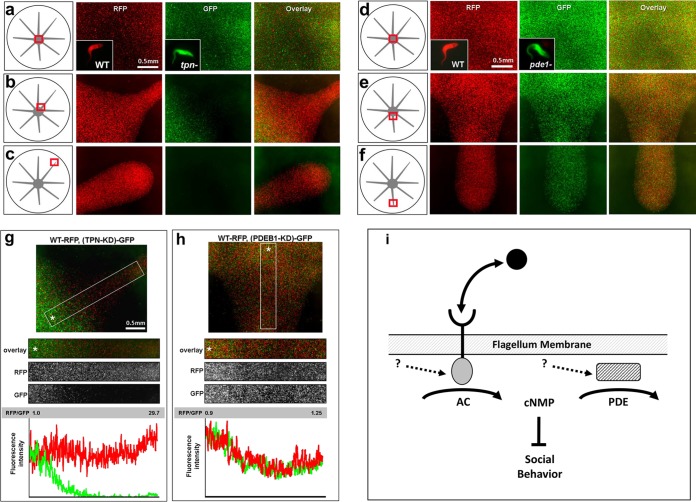
We explored potential mechanisms underlying the role of PDEB1 and cAMP in social motility. We hypothesize at least two alternative models. In model 1, genetic or pharmacological inhibition of PDEB1 leads to misregulation of cAMP signaling, which causes a cell-autonomous inhibition of social motility. In other words, PDEB1-deficient cells are incapable of social motility despite being motile as individuals. In model 2, misregulation of cAMP signaling disrupts an intercellular process that is required for social motility. For example, PDEB1-deficient cells may be competent for social motility but fail to engage in social motility because they lack something that could be provided by other cells. In bacteria, for example, cells with swarming defects can be rescued by wild-type cells in mixed communities, owing to cell-cell transfer of outer membrane proteins important for social behavior (46). To discriminate between these two models, we engineered cell lines with different fluorescent markers to enable monitoring of individual genotypes within a genetically mixed community. We generated WT cells, trypanin knockdowns (47), and PDEB1 knockdowns, with each expressing red fluorescent protein (RFP) or green fluorescent protein (GFP). We then examined these cells in mixed communities. Expression of GFP or RFP did not affect social motility, as WT-RFP and WT-GFP cells are evenly distributed in a mixed social motility community (see Fig. S3d in the supplemental material). Trypanin is a subunit of the flagellar dynein regulatory complex that controls flagellar motility (48). Individual trypanin knockdown cells are incapable of propulsive motility (47) and are consequently defective in social motility (24). We found that the social motility defect of trypanin knockdowns is cell autonomous, as trypanin knockdown-GFP cells were unable to move into radial projections formed by wild-type–RFP cells in a mixed population (Fig. 3a to c and g). The RFP/GFP ratio of the mixed population rose rapidly as a function of the distance from the center, increasing 30-fold within a few millimeters (Fig. 3g). The inability of motility mutants to enter projections formed by wild-type cells demonstrates that parasites are not simply carried into a projection by bulk flow of other cells in the population. Mixing wild-type cells with PDEB1 knockdown cells gave a very different result (Fig. 3d to f and h). In this case, PDEB1 knockdowns, which are incapable of forming projections on their own (Fig. 2), moved into radial projections when cocultured with wild-type cells in a mixed population. Quantitation of relative levels of fluorescence intensity showed that the RFP/GFP ratio remained constant as the population moved outward (Fig. 3h). The result was independent of the status of the cell lines with respect to which harbored GFP versus RFP (see Fig. S4). The ability of the PDEB1 social motility defect to be complemented by wild-type cells in trans favors model 2, indicating that wild-type cells provide a critical factor that the PDEB1 mutants can respond to but cannot generate on their own.
FIG 3 .
The SoMo(−) defect of PDEB1 knockdowns is complemented by wild-type cells provided in trans. (a to c) Social motility assays of mixed communities of wild-type–RFP and trypanin knockdown-GFP cells grown with tetracycline. (d to f) Social motility assays of mixed communities of wild-type–RFP and PDEB1 knockdown-GFP cells grown with tetracycline. Fluorescent images and merged images are shown at the right. The schematic illustrations on the left indicate where the fluorescent images were taken. Insets show representative fluorescent images of individual cells used for the analyses. (g and h) Quantitation of the relative levels of fluorescence of RFP and GFP at the indicated positions of projections formed by communities of wild-type–GFP parasites mixed with either trypanin knockdown-RFP (g) or PDEB1 knockdown-RFP cells (h). Merged and individual fluorescent channels are shown. (i) Generalized model for cyclic nucleotide regulation of social behavior. In T. brucei, receptor-type ACs (AC) in the flagellar membrane catalyze formation of cAMP (cNMP) and are responsive to extracellular ligands. cAMP attenuates social motility and is removed by flagellar phosphodiesterase (PDE). Loss of PDE elevates cAMP levels, blocking social motility, while loss of AC activity reduces cAMP levels, stimulating social motility. Dashed arrows indicate potential regulatory inputs. Similar systems operate in bacteria, except that the cyclic nucleotide produced is cyclic-di-GMP (26).
DISCUSSION
cAMP regulates T. brucei social behavior.
The molecular mechanisms underlying trypanosome social motility are unknown. Here we report that flagellar cAMP signaling systems function in regulation of T. brucei social motility. Our studies here and in recent work (49) provide the first dissection of the mechanisms underlying social motility in trypanosomes and provide new opportunities for investigating cAMP signaling in these pathogens.
Pharmacological inhibition of T. brucei cAMP-specific PDE with cpdA produces a dose-dependent increase in intracellular cAMP concentrations in live trypanosomes that is directly correlated with a dose-dependent block in social motility. These results indicate that PDE activity is required for social motility. Gene-specific knockdown of PDEB1 phenocopies the SoMo(−) defect of cpdA, demonstrating that PDEB1 is specifically required for SoMo. This is in agreement with strong inhibition of recombinant PDEB1 by cpdA at low nanomolar concentrations (50% inhibitory concentration [IC50] = 3.98 nM) (37). T. brucei encodes five PDEs, and it is possible that other PDEs may also participate in regulation of social motility, but our data indicate they are not able to substitute for PDEB1.
By exploiting a FRET-based cAMP sensor, we were able to directly visualize changes in intracellular cAMP concentrations following cpdA inhibition of PDE activity. Exact measurement of absolute intracellular cAMP levels is challenging. However, the cAMP concentration-dependent response of the epac1camps sensor has been titrated in vitro, allowing crude estimates of cAMP levels in vivo (42, 43). Based on titration of the epac1camps sensor in vitro (42, 43), the FRET ratio of ~0.9 in control T. brucei cells corresponds to a cAMP concentration below the 200nM cAMP detection limit of the sensor. Addition of 100 nM cpdA increases cAMP levels to approximately 200 to 400 nM and completely blocks social motility, while addition of 500 nM or 1 µM cpdA increases cAMP levels to above 2 µM. These values reflect total cellular cAMP concentrations, and the changes within the flagellum are likely smaller. The combined pharmacological, gene knockdown, and FRET data indicate that flux through the cAMP signaling pathway controls T. brucei social behavior. In addition, our findings provide the first demonstration, to our knowledge, of a specific function for an individual T. brucei phosphodiesterase, an enzyme that is the focus of current drug development efforts (36–38).
Several independent studies have implicated cAMP signaling as critical for T. brucei biology, development, and pathogenesis (22, 23, 32, 39, 50, 51), but the individual functions of adenylate cyclases (ACs) and PDEs are mostly unknown. Recent work provided an important advance by demonstrating a requirement for T. brucei bloodstream-specific ESAG4 adenylate cyclase function in host-parasite interaction and virulence in mice (51), and elegant genetic studies have identified candidate cAMP downstream effectors (22, 39). Notably, these studies focused exclusively on bloodstream parasites and even less is known regarding cAMP functions in procyclic forms. Recent studies identified procyclic-specific adenylate cyclases, consistent with the suggestion that cAMP functions in parasite differentiation in the fly (52, 53), and at least two of these procyclic-specific ACs have been shown to regulate social motility (49). A primary contributor to the limited understanding of cAMP signaling in trypanosomes is the lack of convenient assays for cAMP function in live cells. Studies of surface-associated group behaviors have provided insights into cyclic nucleotide signaling in other microbes (26, 27, 54, 55). Thus, in addition to demonstrating a role for cAMP in social motility, our studies provide an important advance by establishing a convenient biological assay for dissecting cAMP signaling in T. brucei.
A microdomain model for flagellar cAMP signaling in T. brucei.
cAMP signaling in eukaryotes is restricted to subcellular microdomains 50 to 100 nm in diameter (56–58). Compartmentalization is important for successful signal transduction, as it limits interference between pathways that use the same signal output, increases sensitivity through higher local cAMP concentrations, and enables transient activation and a rapid response to small changes in cAMP levels (56–58). PDE activity is critical for maintaining cAMP microdomains because it limits diffusion of cAMP to the immediate vicinity of synthesis (56–58). As such, only the proteins that sample these microdomains at the right time can transmit the cAMP-dependent signal. A microdomain model for cAMP operating within the trypanosome flagellum, as proposed by Oberholzer et al. (45), provides a potential explanation for the SoMo(−) phenotype observed upon PDEB1 knockdown or chemical inhibition. PDEB1 is localized throughout the flagellum (32). In separate work, we reported that a specific T. brucei adenylate cyclase, AC6, is localized to the tip of the flagellum and that the loss ofAC6 results in hyperactivated social motility, i.e., in an effect opposite that of PDEB1 knockdown (49). The combined data are consistent with a model postulating that social motility is controlled by fluctuations of cAMP concentrations within a specific microdomain at the flagellum tip and that increased cAMP within this microdomain blocks social motility. In this model, PDEB1 is required to insulate the flagellum tip microdomain from cAMP originating elsewhere, for example, from other AC proteins in the flagellum (59). The diffusion coefficient of cAMP in olfactory cilia is estimated to be 2.7 × 10−6 cm2·s−1 (60). Given a length of approximately 20 µm for the T. brucei flagellum, cAMP originating at any location would diffuse throughout the flagellum in less than 10 ms under unrestricted conditions. When PDEB1 is inhibited or knocked down, cAMP would be free to diffuse throughout the flagellum, thereby disrupting cAMP homeostasis and inhibiting social motility.
Conserved architecture of pathways that control social behavior in trypanosomes and bacteria.
Cyclic nucleotide signaling regulates social behaviors in divergent microbes (6, 27, 61). A classic example is the social amoeba Dictyostelium sp., where cAMP functions as an extracellular chemoattractant and an intracellular signaling molecule that promotes development of multicellular fruiting bodies (55). A more directly comparable system is c-di-GMP regulation of bacterial swarming motility (26, 27, 62–64). In bacteria, c-di-GMP levels are controlled through the coordinate activity of diguanylate cyclases (dGCs) that synthesize the molecule and c-di-GMP-specific PDEs that degrade the molecule (54). Perturbation of either the PDE or dGC alters cellular c-di-GMP homeostasis and perturbs swarming motility (26, 27, 62–64). In Pseudomonas aeruginosa, knockout of the bifA gene, encoding a c-di-GMP-specific PDE, blocks swarming motility, owing to elevated intracellular c-di-GMP concentrations (62). Conversely, knockout of sadC or roeA, encoding dGCs, reduces cellular c-di-GMP concentrations and generates hyperswarmers (63, 64). The reciprocal effect of PDE and dGC mutants on bacterial swarming motility is analogous to what we observe for PDE and AC (49) mutants in T. brucei social motility. As such, our findings indicate a conserved architecture for the signaling pathways that control social behavior in trypanosomes and bacteria (Fig. 3i).
In Pseudomonas spp., it is postulated that c-di-GMP derived from specific dGCs, rather than total cellular levels of the molecule, controls swarming (64), and we suspect this may be analogous to the finding that only specific ACs influence T. brucei social motility (49). Trypanosomal ACs and PDEs contain known and suspected regulatory input domains, such as the GAF-A domain of PDEB1 and PDEB2 (65) and the periplasmic binding protein (PBP) domain of ACs (29, 65, 66). Thus, T. brucei PDE- and AC-mediated control of social motility may be regulated by endogenous molecules, as previously observed for PDEs and dGCs that control swarming motility in bacteria (26, 54).
PDE knockdowns are social motility competent but deficient in an intercellular process that promotes social motility.
The genetic tractability of T. brucei, combined with the ease with which individual cells can be visualized, makes trypanosomes an excellent system for monitoring the behavior of individuals within a mixed population. Capitalizing on this, we found that the social motility defect of PDEB1 knockdowns can be complemented by WT cells provided in trans. To our knowledge, this is the first report of transcomplementation in parasitic protozoa, although such processes are well known in bacteria (46). Notably, individual fluorescent parasites in mixed communities retain either red or green fluorescence, indicating that complementation is not due to exchange of cytoplasmic material. We propose that PDEB1 knockdowns are competent for social motility but fail to produce something extracellularly that is necessary for social motility and that WT cells can provide this factor. The parasite-derived components responsible for transcomplementation of social motility remain unknown. These components might be specific proteins or small molecules, as seen, for example, in rescue of bacterial swarming mutants by outer membrane proteins transferred from wild-type cells in the same community (46, 67). Alternatively, they might be something that alters the physical environment, in analogy to the biosurfactants that promote swarming in bacteria (68). Trypanosomes are known to modify their environment by releasing proteins and uncharacterized low-molecular-weight factors as well as metabolic degradation products (69–71). Regardless of the mechanism, our findings reveal a form of cell-cell communication that was not previously recognized in these organisms.
MATERIALS AND METHODS
Cell culture.
2913 procyclic cells (72) were subjected to two rounds of enrichment by growth on semisolid agarose plates, isolating parasites from the tips of social motility projections (24), cloning by limiting dilution, and repeating. These cells, 2913MO2, were used as controls (WT) for all experiments and were used as the parental line for all transfections. Cell culture, transfections, and isolation of clonal lines by limiting dilution were done as previously described (48).
PDEB1-GFP.
The TbPDEB1 (Tb09.160.3590) open reading frame was PCR amplified from plasmid pCR-TbrPDEB1 (gift of T. Seebeck, Bern, Switzerland) using primers B1GAPRONEf and B1GAPRONEr (primer sequences are provided below). The PCR product was cloned upstream of GFP in pG-eGFP-Blast (gift of Isabel Roditi, University of Bern) (73). The construct was verified by sequencing, linearized with SpeI, and transfected into 2913MO2 cells. Expression of the PDEB1-GFP fusion protein was determined by fluorescence microscopy using an Axioskop II microscope (Zeiss, Inc., Germany).
TbPDEB1 RNAi knockdown cells.
A 279-bp fragment corresponding to bp 150 to bp 428 of TbPDEB1 (Tb09.160.3590) was subcloned from an RNAi knockdown construct previously published (32) into the p2T7-Ti-B vector (74) using HindIII and BamHI. The construct was verified by sequencing, linearized with NotI, and transfected into 2913MO2 cells, and clones were obtained by limiting dilution. Clone B1-1 was used for further analysis. Quantitative reverse transcription-PCR (qRT-PCR) was performed as previously described (75) using mRNA from cells at 72 h postinduction with or without 1 µg/ml tetracycline. Each sample was analyzed in duplicate using three independent RNA preparations. The TbPDEB1-specific primers were qRTPDEB1-f and qRTPDEB1-r. The TbPDEB2-specific primers were qRTPDEB2-f and qRTPDEB2-r (primer sequences are provided below). Values were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Tb927.6.4280) and RPS23 (Tb10.70.7030) as described previously (75).
FRET reporter and RFP and GFP reporter cells.
The epac1camps sensor was PCR amplified from plasmid pcDNA3-YFP-epac1-CFP (42) using primers EPACcamp-f and EPACcamp-r1 (primer sequences are provided below). The PCR product was cloned into pCRII Topo (Invitrogen), and the sequences were verified by sequencing. The epac1camps sequence was then subcloned into the HindIII and XbaI sites of pLew100 (72). The construct was linearized with NotI and transfected into 2913MO2 cells. epac1camps expression was detected following tet induction by fluorescence microscopy. RFP-expressing and GFP-expressing cells were obtained by transfection with SpeI-linearized pG-RFP-Blast and pG-eGFP-Blast (gift of Isabel Roditi, University of Bern) (73), and stable transfectants were selected using 10 µg/ml blasticidin and cloned by limiting dilution.
Primer sequences.
The primers for generating PDEB1-GFP were B1GAPRONEf (5′-ATCTCGAGATGTTCATGAACAAGCCCTTTGG-3′) and B1GAPRONEr (5′-ATACCGGTAAACGAGTACTGCTGTTGTTGCC-3′). The XhoI and AgeI restriction sites are underlined.
The primers for qRT-PCR analysis of PDEB1 RNAi knockdown cells were as follows. The TbPDEB1-specific primers were qRTPDEB1-f (5′-TTCATGAACAAGCCCTTTGG-3′) and qRTPDEB1-r (5′-TGATAGCGAGCGAGGATTG-3′). The TbPDEB2-specific primers were qRTPDEB2-f (5′-CGGTGGTCGTCATCTGCTTG-3′) and qRTPDEB2-r (5′-GGAATCATAAGGGGCGACCA-3′).
The primers for constructing FRET reporter cells were as follows: for the epac1camps sensor, EPACcamp-f (5′-TCACTATAGGGAGACCCAAGCTT-3′) and EPACcamp-r1 (5′-TAACTAGTAGCGGGCGCTTACTTGTAC-3′). The HindIII and SpeI sites are underlined. The reverse primer deletes the internal NotI cloning site at the 3′ end of the cyan fluorescent protein (CFP) moiety.
FRET.
epac1camps expression was induced for 24 to 48 h using 1 µg/ml tetracycline. Cells were washed once in culture medium and resuspended to 1.5 × 108 cells/ml in the same medium. A 100-μl volume of cells was added to each well of a 96-well black polystyrene Microplate (Greiner). An additional 100 µl of medium containing cpdA was added to each well to give final concentrations of 100 nM, 500 nM, and 1 µM cpdA. Several controls were performed for normalization of the reading. (i) epac1camps-expressing cells were treated with just dimethyl sulfoxide (DMSO), which is the solvent for cpdA. (ii) 2913MO2 parental cells, i.e., without epac1camps, were treated with cpdA. Immediately following addition of cpdA or DMSO, emission ratios were determined using a photospectrometer (FlexStation) and SoftMax Pro 4.8 software. Filter settings were as follows: excitation, 436 nm; emission, 480 nm (cutoff, 475 nm) or 535 nm (cutoff, 530 nm). The experiments at each time point (41-s intervals) were performed in triplicate, and the results are reported ± standard deviations.
Motility traces and social motility assays.
Motility trace experiments were done as previously described (76). Movies were exported at a resolution of 1 fps and analyzed using automatic tracking in MetaMorph software (Molecular Devices). For social motility assays, cells were diluted for three consecutive days to 3 × 106 cells/ml in suspension culture. A 4% (wt/vol) solution of SeaPlaque GTG agarose (Lonza) in MilliQ water was sterilized for 20 min at 250°C, evaporated water was replaced with sterile MilliQ water, and the stock solution was cooled to 70°C. Stock was diluted to 0.4% with prewarmed culture medium and then maintained at 37°C and supplemented with cpdA, DMSO, 1 µg/ml tetracycline, or methanol as indicated. Medium (13 ml) was poured into 100-mm-by-15-mm petri dishes (Fisherbrand), and plates were dried open for 1.5 h in a laminar flow hood. A total of 5 × 107 cells at 1.2 × 107 to 1.6 × 107 cell/ml were added to the agarose surface, and plates were sealed with Parafilm and incubated at 27°C and 5% CO2.
For cAMP analogue experiments, social motility plates were supplemented with 8-Br-cAMP (Sigma) as cell-permeable cAMP; Rp-8-Br-cAMPS (Sigma) as cell-permeable, hydrolysis-resistant cAMP; 8-pCPT-2′-O-Me-Ado (BioLog, Germany) as cell-permeable adenosine; or 8-pCPT-2′-O-Me-5′-AMP (BioLog, Germany) as cell-permeable AMP; and assays were performed as described above, with plates kept in the dark. Concentrations to be used were determined by starting with 8-Br-cAMP at 150 µM, the concentration that induces stumpy formation in T. brucei (44), and titrating down to find the minimum concentration that inhibited social motility, which was 1 to 10 µM. The other compounds were then also tested at these concentrations. Inhibition of social motility was not due to inhibition of cell proliferation (not shown). AMP and adenosine gave inhibition when the concentration was raised to 20 µM (not shown), though this was likely due to nonspecific effects, as it is ~200-fold higher than the concentration (84 nM to 125 nM) that blocks proliferation and induces stumpy formation in bloodstream-form cells (44). In contrast, the concentration of externally added cAMP required to inhibit social motility is 10-fold to 100-fold lower than what is required to induce stumpy formation (44). Moreover, because cAMP analogues added to T. brucei cells are unstable (44), the effective cAMP concentration is actually much lower than what was added. It is recognized that cAMP analogues have potential off-target effects and may act as agonists or antagonists of cAMP effector proteins (44, 77), and we therefore consider these experiments to be an adjunct to those with the more specific pharmacological T. brucei PDE inhibitor cpdA (37) and gene-specific PDEB1 RNAi.
Colony lifts to assess GPEET expression.
GPEET expression in cells on plates was assessed by colony lifts. Nitrocellulose membrane was incubated on the surface of social motility plates (day 4 postplating) for 5 min. The membrane was removed, air-dried, and stained with Ponceau S prior to imaging to visualize total protein. The membrane was washed in MilliQ water, blocked in phosphate-buffered saline (PBS) containing 5% powdered milk, and stained with anti-GPEET antibody (1:10,000) overnight at 4°C. The membrane was washed 3 times for 10 min in PBS–0.05% Tween 20 and stained with secondary antibodies coupled to horseradish peroxidase (HRP) (1:2,500) and washed as described above prior to development using an enhanced chemiluminescence (ECL) kit and exposure to film.
Social complementation assay.
GFP-expressing cells and RFP-expressing cells were mixed in a 1:1 ratio and plated as described above for social motility assays. Fluorescence imaging of cells on plates was done 96 to 120 h postplating using an Axioskop II microscope (Zeiss, Inc., Jena, Germany) with a 2.56LD Plan NeoFluor objective and a GFP bandpass emission filter (41017; Chroma Technology) or an RFP bandpass emission filter (41007; Chroma Technology) on a Zeiss Axiovert microscope using an AxioCam camera. Pictures were processed using Adobe Photoshop, fluorescence intensities were determined using ImageJ (NIH), and values were plotted using GraphPad PRISM.
SUPPLEMENTAL MATERIAL
Effect of cAMP analogues on social motility. Data represent the social motility of untreated cells (control) or cells treated with cell-permeable cAMP analogues as indicated. 1, hydrolyzable cAMP:8-Br-cAMP; 2, hydrolysis-resistant cAMP (Rp-8-Br-cAMPS); 3, AMP (8-pCPT-2′-O-Me-5′-AMP); 4, adenosine (8-pCPT-2′-O-Me-Ado).The numbers of projections per plate were counted at 72 h postplating. Samples showing statistically significant differences relative to the control are indicated with asterisks. *, P < 0.0001; **, P < 0.01; ***, P < 0.03 (two-tailed, unpaired t test). Download
FRET assay using epac1camps sensor. (a) Schematic showing the FRET-based cAMP sensor, epac1camps (42), and the effect of adding cAMP. (b) Fluorescence emission at 488 and 535 nm for wild-type (WT) cells and cells expressing the epac1camps sensor (epac). (c) Fluorescence ratio (488 nm/535 nm) for epac1camps-expressing cells normalized to the background fluorescence of wild-type cells without the sensor. Download
Expression of GFP or RFP does not affect the social motility phenotype. (a to d) Social motility assays with the indicated cell lines expressing RFP or GFP. (a to c) Images of the social motility phenotype. (d) Individual and merged fluorescent channels for mixed communities of wild-type cells expressing RFP or GFP as indicated. Schematic at left shows the region of the projection that was imaged for fluorescence. Download
The PDEB1 knockdown social motility defect is rescued by wild-type cells provided in trans. Fluorescent and merged images of social motility assays using mixed populations of WT, trypanin knockdown, and PDEB1 knockdown cells as indicated are shown. See the Fig. 3 legend for details. Here, WT cells express GFP and the knockdown cells express RFP, while in Fig. 3, WT cells express RFP and the knockdown cells express GFP. Download
GPEET is expressed in PDEB1-RNAi and cpdA-treated cells. Colony lifts were performed on social motility colonies of PDEB1 RNAi cells grown in the absence or presence of tetracycline (tet) as indicated or on control cells grown with or without 0.1 µM cpdA as indicated. Nitrocellulose filters were stained with Ponceau S (top) to visualize total protein and then probed with anti-GPEET antibody to visualize GPEET procyclin (bottom). Download
ACKNOWLEDGMENTS
We thank Geert Jan Sterk (Mercachem) for providing cpdA, Isabel Roditi (University of Bern) for GPEET antibody and for providing the pG-eGFP-Blast and pRFP-Blast plasmids prior to publication, Viacheslav O. Nikolaev and Martin J. Lohse (University of Würzburg) for providing the epac1camps construct and advice on its use, and Robert Damoiseaux (Molecular Shared Screening Resource, UCLA) for assistance with the FRET analysis. We thank Thomas Seebeck, Isabel Roditi, and Simon Imhof (University of Bern) for many helpful discussions. We also thank members of the Hill Laboratory, especially Miguel Lopez and Michelle Shimogawa, for helpful discussions and technical assistance.
This work was supported by a Swiss National Science Foundation fellowship to M.O., a Burroughs Wellcome Fund Investigator in Global Infectious Disease award to K.L.H., and an NIH grant (R01AI052348) to K.L.H.
Footnotes
Citation Oberholzer M, Saada EA, Hill KL. 2015. Cyclic AMP regulates social behavior in African trypanosomes. mBio 6(3):e01954-14. doi:10.1128/mBio.01954-14.
Contributor Information
Stephen L. Hajduk, University of Georgia.
Thomas E. Wellems, National Institutes of Health.
REFERENCES
- 1.Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 3.Blankenship JR, Mitchell AP. 2006. How to build a biofilm: a fungal perspective. Curr Opin Microbiol 9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 5.Velicer GJ, Vos M. 2009. Sociobiology of the myxobacteria. Annu Rev Microbiol 63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 6.Firtel RA, Meili R. 2000. Dictyostelium: a model for regulated cell movement during morphogenesis. Curr Opin Genet Dev 10:421–427. doi: 10.1016/S0959-437X(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 7.Butler MT, Wang Q, Harshey RM. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A 107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol 11:30–36. doi: 10.1016/S0966-842X(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 10.Antunes LC, Ferreira RB, Buckner MM, Finlay BB. 2010. Quorum sensing in bacterial virulence. Microbiology 156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 11.Landini P, Antoniani D, Burgess JG, Nijland R. 2010. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl Microbiol Biotechnol 86:813–823. doi: 10.1007/s00253-010-2468-8. [DOI] [PubMed] [Google Scholar]
- 12.Stuart K, Brun R, Croft S, Fairlamb A, Gürtler RE, McKerrow J, Reed S, Tarleton R. 2008. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest 118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. 2014. Malaria. Lancet 383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 14.Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AM, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR. 2009. Genome sequence and analysis of the Irish potato famine pathogen phytophthora infestans. Nature 461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 15.Laishram S, Kang G, Ajjampur SS. 2012. Giardiasis: a review on assemblage distribution and epidemiology in India. Indian J Gastroenterol 31:3–12. doi: 10.1007/s12664-012-0161-9. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher SM, Stark D, Harkness J, Ellis J. 2012. Enteric protozoa in the developed world: a public health perspective. Clin Microbiol Rev 25:420–449. doi: 10.1128/CMR.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacGregor P, Savill NJ, Hall D, Matthews KR. 2011. Transmission stages dominate trypanosome within-host dynamics during chronic infections. Cell Host Microbe 9:310–318. doi: 10.1016/j.chom.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez MA, Nguyen HT, Oberholzer M, Hill KL. 2011. Social parasites. Curr Opin Microbiol 14:642–648. doi: 10.1016/j.mib.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reece SE, Drew DR, Gardner A. 2008. Sex ratio adjustment and kin discrimination in malaria parasites. Nature 453:609–614. doi: 10.1038/nature06954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollitt LC, MacGregor P, Matthews K, Reece SE. 2011. Malaria and trypanosome transmission: different parasites, same rules? Trends Parasitol 27:197–203. doi: 10.1016/j.pt.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brun R, Blum J, Chappuis F, Burri C. 2010. Human African trypanosomiasis. Lancet 375:148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 22.Mony BM, MacGregor P, Ivens A, Rojas F, Cowton A, Young J, Horn D, Matthews K. 2014. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature 505:681–685. doi: 10.1038/nature12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassella E, Reuner B, Yutzy B, Boshart M. 1997. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci 110:2661–2671. [DOI] [PubMed] [Google Scholar]
- 24.Oberholzer M, Lopez MA, McLelland BT, Hill KL. 2010. Social motility in African trypanosomes. PLoS Pathog 6:e1000739. doi: 10.1371/journal.ppat.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imhof S, Knüsel S, Gunasekera K, Vu XL, Roditi I. 2014. Social motility of African trypanosomes is a property of a distinct life-cycle stage that occurs early in tsetse fly transmission. PLoS Pathog 10:e1004493. doi: 10.1371/journal.ppat.1004493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trimble MJ, McCarter LL. 2011. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc Natl Acad Sci U S A 108:18079–18084. doi: 10.1073/pnas.1113790108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 28.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seebeck T, Schaub R, Johner A. 2004. cAMP signalling in the kinetoplastid protozoa. Curr Mol Med 4:585–599. doi: 10.2174/1566524043360113. [DOI] [PubMed] [Google Scholar]
- 30.Zoraghi R, Seebeck T. 2002. The cAMP-specific phosphodiesterase TbPDE2C is an essential enzyme in bloodstream form Trypanosoma brucei. Proc Natl Acad Sci U S A 99:4343–4348. doi: 10.1073/pnas.062716599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rascón A, Soderling SH, Schaefer JB, Beavo JA. 2002. Cloning and characterization of a cAMP-specific phosphodiesterase (TbPDE2B) from Trypanosoma brucei. Proc Natl Acad Sci U S A 99:4714–4719. doi: 10.1073/pnas.002031599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberholzer M, Marti G, Baresic M, Kunz S, Hemphill A, Seebeck T. 2007. The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. FASEB J 21:720–731. doi: 10.1096/fj.06-6818com. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes JM, Davis EE, Katsanis N. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell 137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotureau B, Morales MA, Bastin P, Späth GF. 2009. The flagellum-mitogen-activated protein kinase connection in trypanosomatids: a key sensory role in parasite signalling and development? Cell Microbiol 11:710–718. doi: 10.1111/j.1462-5822.2009.01295.x. [DOI] [PubMed] [Google Scholar]
- 35.Maric D, Epting CL, Engman DM. 2010. Composition and sensory function of the trypanosome flagellar membrane. Curr Opin Microbiol 13:466–472. doi: 10.1016/j.mib.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seebeck T, Sterk GJ, Ke H. 2011. Phosphodiesterase inhibitors as a new generation of antiprotozoan drugs: exploiting the benefit of enzymes that are highly conserved between host and parasite. Future Med Chem 3:1289–1306. doi: 10.4155/fmc.11.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Koning HP, Gould MK, Sterk GJ, Tenor H, Kunz S, Luginbuehl E, Seebeck T. 2012. Pharmacological validation of Trypanosoma brucei phosphodiesterases as novel drug targets. J Infect Dis 206:229–237. doi: 10.1093/infdis/jir857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orrling KM, Jansen C, Vu XL, Balmer V, Bregy P, Shanmugham A, England P, Bailey D, Cos P, Maes L, Adams E, van den Bogaart E, Chatelain E, Ioset JR, van de Stolpe A, Zorg S, Veerman J, Seebeck T, Sterk GJ, de Esch IJ. 2012. Catechol pyrazolinones as trypanocidals: fragment-based design, synthesis, and pharmacological evaluation of nanomolar inhibitors of trypanosomal phosphodiesterase B1. J Med Chem 55:8745–8756. doi: 10.1021/jm301059b. [DOI] [PubMed] [Google Scholar]
- 39.Gould MK, Bachmaier S, Ali JA, Alsford S, Tagoe DN, Munday JC, Schnaufer AC, Horn D, Boshart M, de Koning HP. 2013. Cyclic AMP effectors in African trypanosomes revealed by genome-scale RNA interference library screening for resistance to the phosphodiesterase inhibitor CpdA. Antimicrob Agents Chemother 57:4882–4893. doi: 10.1128/AAC.00508-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freire ER, Vashisht AA, Malvezzi AM, Zuberek J, Langousis G, Saada EA, Nascimento Jde F, Stepinski J, Darzynkiewicz E, Hill K, De Melo Neto OP, Wohlschlegel JA, Sturm NR, Campbell DA. 2014. eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA 20:1272–1286. doi: 10.1261/rna.045534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freire ER, Malvezzi AM, Vashisht AA, Zuberek J, Saada EA, Langousis G, Nascimento JD, Moura D, Darzynkiewicz E, Hill K, de Melo Neto OP, Wohlschlegel JA, Sturm NR, Campbell DA. 2014. Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot Cell 13:896–908. doi: 10.1128/EC.00071-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolaev VO, Bünemann M, Hein L, Hannawacker A, Lohse MJ. 2004. Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem 279:37215–37218. doi: 10.1074/jbc.C400302200. [DOI] [PubMed] [Google Scholar]
- 43.Börner S, Schwede F, Schlipp A, Berisha F, Calebiro D, Lohse MJ, Nikolaev VO. 2011. FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells. Nat Protoc 6:427–438. doi: 10.1038/nprot.2010.198. [DOI] [PubMed] [Google Scholar]
- 44.Laxman S, Riechers A, Sadilek M, Schwede F, Beavo JA. 2006. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc Natl Acad Sci U S A 103:19194–19199. doi: 10.1073/pnas.0608971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oberholzer M, Bregy P, Marti G, Minca M, Peier M, Seebeck T. 2007. Trypanosomes and mammalian sperm: one of a kind? Trends Parasitol 23:71–77. doi: 10.1016/j.pt.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Nudleman E, Wall D, Kaiser D. 2005. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 47.Hutchings NR, Donelson JE, Hill KL. 2002. Trypanin is a cytoskeletal linker protein and is required for cell motility in African trypanosomes. J Cell Biol 156:867–877. doi: 10.1083/jcb.200201036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ralston KS, Lerner AG, Diener DR, Hill KL. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell 5:696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez MA, Saada EA, Hill KL. 2015. Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes. Eukaryot Cell 14:104–112. doi: 10.1128/EC.00217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmon D, Bachmaier S, Krumbholz C, Kador M, Gossmann JA, Uzureau P, Pays E, Boshart M. 2012. Cytokinesis of Trypanosoma brucei bloodstream forms depends on expression of adenylyl cyclases of the ESAG4 or ESAG4-like subfamily. Mol Microbiol 84:225–242. doi: 10.1111/j.1365-2958.2012.08013.x. [DOI] [PubMed] [Google Scholar]
- 51.Salmon D, Vanwalleghem G, Morias Y, Denoeud J, Krumbholz C, Lhommé F, Bachmaier S, Kador M, Gossmann J, Dias FB, De Muylder G, Uzureau P, Magez S, Moser M, De Baetselier P, Van Den Abbeele J, Beschin A, Boshart M, Pays E. 2012. Adenylate cyclases of Trypanosoma brucei inhibit the innate immune response of the host. Science 337:463–466. doi: 10.1126/science.1222753. [DOI] [PubMed] [Google Scholar]
- 52.Saada EA, Kabututu ZP, Lopez M, Shimogawa MM, Langousis G, Oberholzer M, Riestra A, Jonsson ZO, Wohlschlegel JA, Hill KL. 2014. Insect stage-specific receptor adenylate cyclases are localized to distinct subdomains of the Trypanosoma brucei flagellar membrane. Eukaryot Cell 13:1064–1076. doi: 10.1128/EC.00019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rolin S, Paindavoine P, Hanocq-Quertier J, Hanocq F, Claes Y, Le Ray D, Overath P, Pays E. 1993. Transient adenylate cyclase activation accompanies differentiation of Trypanosoma brucei from bloodstream to procyclic forms. Mol Biochem Parasitol 61:115–125. doi: 10.1016/0166-6851(93)90164-S. [DOI] [PubMed] [Google Scholar]
- 54.Boyd CD, O’Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manahan CL, Iglesias PA, Long Y, Devreotes PN. 2004. Chemoattractant signaling in Dictyostelium discoideum. Annu Rev Cell Dev Biol 20:223–253. doi: 10.1146/annurev.cellbio.20.011303.132633. [DOI] [PubMed] [Google Scholar]
- 56.Willoughby D, Wong W, Schaack J, Scott JD, Cooper DM. 2006. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J 25:2051–2061. doi: 10.1038/sj.emboj.7601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baillie GS, Houslay MD. 2005. Arrestin times for compartmentalised cAMP signalling and phosphodiesterase-4 enzymes. Curr Opin Cell Biol 17:129–134. doi: 10.1016/j.ceb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Karpen JW, Rich TC. 2005. High-resolution measurements of cyclic adenosine monophosphate signals in 3D microdomains. Methods Mol Biol 307:15–26. doi: 10.1385/1-59259-839-0:015. [DOI] [PubMed] [Google Scholar]
- 59.Paindavoine P, Rolin S, Van Assel S, Geuskens M, Jauniaux JC, Dinsart C, Huet G, Pays E. 1992. A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol Cell Biol 12:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C, Nakamura T, Koutalos Y. 1999. Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophys J 76:2861–2867. doi: 10.1016/S0006-3495(99)77440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Mühlschlegel FA. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merritt JH, Brothers KM, Kuchma SL, O’Toole GA. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 189:8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O’Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:00183-10. doi: 10.1128/mBio.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laxman S, Rascón A, Beavo JA. 2005. Trypanosome cyclic nucleotide phosphodiesterase 2B binds cAMP through its GAF-A domain. J Biol Chem 280:3771–3779. doi: 10.1074/jbc.M408111200. [DOI] [PubMed] [Google Scholar]
- 66.Kunz S, Luginbuehl E, Seebeck T. 2009. Gene conversion transfers the GAF-A domain of phosphodiesterase TbrPDEB1 to one allele of TbrPDEB2 of Trypanosoma brucei. PLoS Negl Trop Dis 3:e455. doi: 10.1371/journal.pntd.0000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. 2012. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet 8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daniels R, Reynaert S, Hoekstra H, Verreth C, Janssens J, Braeken K, Fauvart M, Beullens S, Heusdens C, Lambrichts I, De Vos DE, Vanderleyden J, Vermant J, Michiels J. 2006. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci U S A 103:14965–14970. doi: 10.1073/pnas.0511037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Proto WR, Castanys-Munoz E, Black A, Tetley L, Moss CX, Juliano L, Coombs GH, Mottram JC. 2011. Trypanosoma brucei metacaspase 4 is a pseudopeptidase and a virulence factor. J Biol Chem 286:39914–39925. doi: 10.1074/jbc.M111.292334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geiger A, Hirtz C, Bécue T, Bellard E, Centeno D, Gargani D, Rossignol M, Cuny G, Peltier JB. 2010. Exocytosis and protein secretion in Trypanosoma. BMC Microbiol 10:20. doi: 10.1186/1471-2180-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnwell EM, van Deursen FJ, Jeacock L, Smith KA, Maizels RM, Acosta-Serrano A, Matthews K. 2010. Developmental regulation and extracellular release of a VSG expression-site-associated gene product from Trypanosoma brucei bloodstream forms. J Cell Sci 123:3401–3411. doi: 10.1242/jcs.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knockouts and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99:89–101. doi: 10.1016/S0166-6851(99)00002-X. [DOI] [PubMed] [Google Scholar]
- 73.Urwyler S, Studer E, Renggli CK, Roditi I. 2007. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol 63:218–228. doi: 10.1111/j.1365-2958.2006.05492.x. [DOI] [PubMed] [Google Scholar]
- 74.LaCount DJ, Barrett B, Donelson JE. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J Biol Chem 277:17580–17588. doi: 10.1074/jbc.M200873200. [DOI] [PubMed] [Google Scholar]
- 75.Kabututu ZP, Thayer M, Melehani JH, Hill KL. 2010. CMF70 is a subunit of the dynein regulatory complex. J Cell Sci 123:3587–3595. doi: 10.1242/jcs.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baron DM, Ralston KS, Kabututu ZP, Hill KL. 2007. Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J Cell Sci 120:478–491. doi: 10.1242/jcs.03352. [DOI] [PubMed] [Google Scholar]
- 77.Poppe H, Rybalkin SD, Rehmann H, Hinds TR, Tang XB, Christensen AE, Schwede F, Genieser HG, Bos JL, Doskeland SO, Beavo JA, Butt E. 2008. Cyclic nucleotide analogs as probes of signaling pathways. Nat Methods 5:277–278. doi: 10.1038/nmeth0408-277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of cAMP analogues on social motility. Data represent the social motility of untreated cells (control) or cells treated with cell-permeable cAMP analogues as indicated. 1, hydrolyzable cAMP:8-Br-cAMP; 2, hydrolysis-resistant cAMP (Rp-8-Br-cAMPS); 3, AMP (8-pCPT-2′-O-Me-5′-AMP); 4, adenosine (8-pCPT-2′-O-Me-Ado).The numbers of projections per plate were counted at 72 h postplating. Samples showing statistically significant differences relative to the control are indicated with asterisks. *, P < 0.0001; **, P < 0.01; ***, P < 0.03 (two-tailed, unpaired t test). Download
FRET assay using epac1camps sensor. (a) Schematic showing the FRET-based cAMP sensor, epac1camps (42), and the effect of adding cAMP. (b) Fluorescence emission at 488 and 535 nm for wild-type (WT) cells and cells expressing the epac1camps sensor (epac). (c) Fluorescence ratio (488 nm/535 nm) for epac1camps-expressing cells normalized to the background fluorescence of wild-type cells without the sensor. Download
Expression of GFP or RFP does not affect the social motility phenotype. (a to d) Social motility assays with the indicated cell lines expressing RFP or GFP. (a to c) Images of the social motility phenotype. (d) Individual and merged fluorescent channels for mixed communities of wild-type cells expressing RFP or GFP as indicated. Schematic at left shows the region of the projection that was imaged for fluorescence. Download
The PDEB1 knockdown social motility defect is rescued by wild-type cells provided in trans. Fluorescent and merged images of social motility assays using mixed populations of WT, trypanin knockdown, and PDEB1 knockdown cells as indicated are shown. See the Fig. 3 legend for details. Here, WT cells express GFP and the knockdown cells express RFP, while in Fig. 3, WT cells express RFP and the knockdown cells express GFP. Download
GPEET is expressed in PDEB1-RNAi and cpdA-treated cells. Colony lifts were performed on social motility colonies of PDEB1 RNAi cells grown in the absence or presence of tetracycline (tet) as indicated or on control cells grown with or without 0.1 µM cpdA as indicated. Nitrocellulose filters were stained with Ponceau S (top) to visualize total protein and then probed with anti-GPEET antibody to visualize GPEET procyclin (bottom). Download